Abstract
Members of the serpin (serine protease inhibitor) superfamily have been identified in higher multicellular eukaryotes, as well as in bacteria, although examination of available genome sequences has indicated that homologs of the bacterial serpin-encoding gene (ser) are not widely distributed. In members of the genus Bifidobacterium this gene appears to be present in at least 5, and perhaps up to 9, of the 30 species tested. Moreover, phylogenetic analysis using available bacterial and eukaryotic serpin sequences revealed that bifidobacteria produce serpins that form a separate clade. We characterized the ser210B locus of Bifidobacterium breve 210B, which encompasses a number of genes whose deduced protein products display significant similarity to proteins encoded by corresponding loci found in several other bifidobacteria. Northern hybridization, primer extension, microarray, reverse transcription-PCR (RT-PCR), and quantitative real-time PCR (qRT-PCR) analyses revealed that a 3.5-kb polycistronic mRNA encompassing the ser210B operon with a single transcriptional start site is strongly induced following treatment of B. breve 210B cultures with some proteases. Interestingly, transcription of other bifidobacterial ser homologs appears to be triggered by different proteases.
Bifidobacteria are Gram-positive microorganisms that naturally reside in the gastrointestinal tract (GIT) of mammals (44). Recently, there has been growing interest in bifidobacteria due to their perceived role in the maintenance of gastrointestinal health (23) and other beneficial or probiotic properties (16). For this reason various bifidobacterial strains are incorporated as viable bacteria into dairy products (e.g., yoghurt) or infant foods. Such bifidobacteria should be selected on the basis of a range of beneficial activities that, for example, promote restoration or stability of a normal intestinal microbiota (i.e., a microbiota present in a healthy individual), stimulate the immune response, protect against infection by pathogens, and/or reduce the activities of bacterial enzymes associated with development of colonic cancer (for reviews, see references 18 and 21).
The advent of genomic technologies has allowed determination of the complete sequences of a number of bifidobacterial genomes (1, 17, 30, 31; for a review, see reference 39). Analysis of the genomic data generated has indicated that the chromosomal content of bifidobacteria is tuned so that it provides optimal genetic adaptation for successful competition in the GIT environment (38, 41). However, little is known about the mechanisms for interaction between bifidobacteria and their hosts, and only a few genetic loci have been putatively implicated in sustaining interactions between bifidobacteria and human host cells (38, 41). The Bifidobacterium longum subsp. longum NCC2705 genome was shown to encode a predicted secreted serine protease inhibitor, or serpin-like protein (30), which acts as an efficient inhibitor of pancreatic elastase and neutrophil elastase, which are enzymes that enteric bifidobacteria would be expected to encounter in their natural habitat or, in the case of neutrophil elastase, during inflammatory conditions with a breach of the intestinal barrier (14). Members of this protein superfamily are widely distributed throughout all kingdoms of life, including the Eukarya, Prokarya, Archaea, and certain viruses (13, 33). The widespread distribution of serpins, together with their common structural features and inhibitory mechanism, suggests that they originated at an early stage of evolution (13). Very little is known about the function of serpins in bacteria, even though some studies identified their role as broad-spectrum protease inhibitors (14, 25). It has been hypothesized that intestinal bacteria, such as bifidobacteria, produce serpins to protect themselves against host-derived proteases and to provide an advantage for survival in a highly complex and competitive environment (14).
In this report we describe the findings of transcriptional profiling studies aimed at identifying Bifidobacterium breve 210B genes that are induced following exposure to proteases. The results obtained revealed that the serpin-encoding gene of B. breve 210B, ser210B, is subject to specific protease induction, which was investigated further by performing Northern blot hybridizations, primer extension experiments, and quantitative real-time PCR (qRT-PCR).
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The strain used in this study, B. breve 210B, is a commensal bacterium that was isolated from a human mucosal sample, as described previously (35). Cultures of B. breve 210B, Bifidobacterium longum subsp. infantis ATCC 15697, B. longum subsp. longum ATCC 15707, and Bifidobacterium dentium Bd1 (43) were grown anaerobically in the DeMan-Rogosa-Sharpe (MRS) medium (Scharlau Chemie, Barcelona, Spain) supplemented with 0.05% (wt/vol) l-cysteine hydrochloride and incubated at 37°C for 16 h.
Serpin 3D structure.
Analysis of the folds of two bifidobacterial serpin proteins was performed using the protein structure prediction Meta server (http://bioinfo.pl/Meta/). Using the crystal structure of tengpinD31 (PDB entry 2PEE) (47) as the structure template, individual three-dimensional (3D) models of the serpin proteins of B. breve 210B and B. dentium Bd1 were generated using the modeler module (26) of the Accelrys Discovery Studio software.
Phylogenetic analyses.
Phylogeny analyses, including distance calculations and generation of phylogenetic trees, were performed using PHYLIP (Phylogeny Inference Package) (7), version 3.5c (kindly provided by J. Felsenstein, University of Washington, Seattle, WA). Trees were calculated using the neighbor-joining method with Kimura's two-parameter substitution model (15). Bootstrap values were computed by performing 1,000 resamplings. Dendrograms of gene sequences were drawn using the ClustalW program (National Center for Biotechnology Information; http://www.ebi.ac.uk/Tools/clustalw2/index.html) and were visualized with the TreeView program (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html).
DNA amplification of ser genes from bifidobacteria.
Genomic DNA that was used as a template for PCRs was extracted by using a previously described protocol (42). PCR amplification to obtain ser homologs from various Bifidobacterium strains was performed using oligonucleotides Ser-fwd-univ (5′-CTTCGNTGNNGATGGCGTTG-3′) and Ser-rev-univ-(5′-TCACNNCGATGCGCGTGC-3′). Each PCR consisted of an initial denaturation step of 3 min at 95°C, followed by 35 cycles of denaturation for 30 s at 95°C, annealing for 30 s at 55°C, and extension for 1 min at 72°C and then elongation for 5 min at 72°C. Electrophoresis on a 1.5% agarose gel and visualization by ethidium bromide staining were used to determine the presence of amplicons.
Slot blot hybridization.
A 15-μg sample of bacterial DNA was spotted onto a Biodyne nylon membrane (Pall, NY) using a Bio-Dot SF microfiltration apparatus (Bio-Rad, CA) as specified by the manufacturer and was treated with one UV autocross-linking cycle using a UV Stratalinker 1800 (Stratagene). Filters were hybridized with a probe mixture that consisted of ser-containing amplicons (see above) obtained from B. breve 210B, B. longum subsp. suis LMG 21814, B. longum subsp. longum 15707, B. longum subsp. infantis ATCC 15697, and B. dentium Bd1. This probe mixture was labeled by incorporation of biotin 16-dUTP (Roche, United Kingdom) using a PCR strategy, as described by the manufacturer. Subsequent prehybridization, hybridization, and detection steps were carried out using the Southern blot hybridization protocol (Li-Cor, United Kingdom). The hybridized filters were finally scanned using the Odyssey infrared imaging system at an intensity level of 5 (Li-Cor, United Kingdom).
RNA isolation and Northern blot analyses.
B. breve 210B, B. dentium Bd1, B. longum subsp. infantis ATCC 15697, and B. longum subsp. longum ATCC 15707 cultures were grown at 37°C in MRS medium to an optical density at 600 nm (OD600) of 0.6, after which enzyme treatment was initiated by addition of pancreatic elastase (final concentration, 1 mg/ml), human neutrophil elastase (4 mg/ml), thrombin (1 mg/ml), papain (0.5 mg/ml), kallikrein (0.55 mg/ml), trypsin (0.1 mg/ml), α-antitrypsin (0.25 mg/ml), chymotrypsin (0.16 mg/ml), or plasmin (3 mg/ml) (all enzymes were obtained from Sigma, Italy). Cultures were incubated at 37°C for various times (90 and 180 min), after which samples (30 ml) were collected and briefly centrifuged to harvest cells and total RNA was extracted.
Total RNA was isolated using the macaloid acid method (48) and then treated with DNase (Roche, United Kingdom). Briefly, each cell pellet was resuspended in 1 ml of QUIAZOL (Qiagen, United Kingdom) and placed in a tube containing 0.8 g of glass beads (diameter, 106 μm; Sigma). The cells were lysed by shaking the mixture on a BioSpec homogenizer at 4°C for 2 min at the maximum setting. The mixture was then centrifuged at 12,000 rpm for 15 min, and the upper phase containing the RNA was recovered. The RNA sample was purified further by phenol extraction and ethanol precipitation using a previously described method (27). Slot blot hybridization was carried out as previously described (40). RNA electrophoresis and Northern blot hybridization were carried out using the Northern blot hybridization protocol for the Odyssey infrared imaging system (Li-CoR, United Kingdom). The hybridized filters were scanned using the Odyssey infrared imaging system at an intensity level of 5 (Li-Cor, United Kingdom). All slot blot and Northern hybridization experiments were performed at least twice.
The probes corresponding to the ser210B and ORF1502 genes were generated by PCR using primers Ser-fwd-univ and Ser-rev-univ and primers 1502-fwd (5′-CACAAGCACTCAGCAAGCTC-3′) and 1502-rwd (5′-CACGATGACGAAATGCAAGAC-3′) respectively.
Primer extension analysis.
The 5′ end of the ser210B RNA transcript was determined using a protocol described in a previous study (45) The synthetic oligonucleotide used was Ser-prom (5′-CACGCGAATAATCGTTGCGTC-3′).
RT-PCR analysis.
Five micrograms of mRNA was treated with DNase (Roche, United Kingdom) and used as the template in a 100-μl reaction mixture containing 20 ng of random primers, each deoxyribonucleoside triphosphate at a concentration of 0.125 mM, and the Superscript enzyme (Invitrogen, Paisley, United Kingdom) used according the manufacturer's instructions to produce cDNA. The cDNA generated was used as a template for reverse transcription-PCRs (RT-PCRs) to determine the arrangement of the transcript encompassing the ser210B locus with primers lacI rev (5′-CACTGATGAAACCGAGGATG-3′) and lacI-uni (5′-GACGGCATACCATTATTCATC-3′), primers lacI-rev (5′-CTCGATGGATCAGGTCTTG-3′) and ser1-rev (5′-GAGCCCAGCAGCTCATTC-3′), and primers lac2-uni (5′-GAAGGACGCTCGATGGAC-3′) and lac2-rev (5′-CATCTTCAGAACGGAGATC-3′).
qRT-PCR.
Quantitative real-time reverse transcription-PCR (qRT-PCR) primers (see Table S1 in the supplemental material) were used to amplify ser210B, the ser genes of B. dentium Bd1 (serBd1), B. longum subsp. infantis ATCC 15697 (ser15796), and B. longum subsp. longum ATCC 15707 (ser15707), and the reference genes atpD, tufA, rpoB, and ldh. Primer design criteria were based on desired melting temperatures between 58 and 60°C and an amplicon size of approximately 100 bp. qRT-PCR was performed using the CFX96 system (Bio-Rad, CA). PCR products were detected with SYBR green fluorescent dye and were amplified using the following protocol: one cycle of 95°C for 3 min, followed by 39 cycles of 95°C for 5 s and 66°C for 20 s. A melting curve was obtained by using temperatures of 65°C to 95°C that increased at a rate of 0.5°C/s.
Each PCR mixture contained 12.5 μl 2× SYBR green SuperMix (Bio-Rad, CA), 1 μl of diluted cDNA, the forward and reverse primers each at a concentration of 0.5 μM, and enough nuclease-free water so that the final volume was 20 μl. In each run, negative controls (no cDNA) for each primer set were included.
DNA microarray preparation, hybridization, and data analysis.
Microarray analysis was performed with a B. breve 210B array. A total of 2,110 probes, which represented all open reading frame (ORFs) of the B. breve 210B genome that have been identified (M. Ventura, F. Turroni, E. Foroni, F. Bottacini, V. Giubellini, and D. van Sinderen, unpublished data) and were 35 to 40 bp long, were designed using the OligoArray 2.1 software (J. M. Rouillard et al., unpublished data). Eighteen replicates of oligonucleotides were synthesized on a 2x40k CombiMatrix array (CombiMatrix, Mulkiteo, WA). Replicates were distributed on the chip at random, nonadjacent positions. Sixty replicates of a set of 29 negative-control probes designed by using phage and plant sequences were also included on the chip at randomly distributed positions.
Reverse transcription and amplification of 500 ng of total RNA were performed with a MessageAmp II-Bacteria kit (Ambion, Austin, TX) used according to the manufacturer's instructions. Five micrograms of mRNA was then labeled with a ULS labeling kit for Combimatrix arrays with Cy5 (Kreatech, Netherlands). Hybridization of labeled DNA to B. breve 210B arrays was performed using CombiMatrix protocols (CombiMatrix, Mulkiteo, WA).
Microarray data acquisition and treatment.
Fluorescence scanning was performed with an Axon GenePix 4400A microarray scanner (Molecular Devices, United States). The signal intensity for each spot was determined using GenePix Pro 7 software (Molecular Devices, United States). The signal background was calculated by using the mean of negative controls plus 2 standard deviations (3). Global quantile normalization was performed (4), and log2 ratios for a reference sample (a B. breve 210B culture not subjected to protease treatment) and the test samples were calculated. The distribution of the log2-transformed ratios was separately calculated for each hybridization reaction (34).
Microarray accession number.
The microarray data have been deposited in the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE20626.
Nucleotide sequence accession numbers.
All nucleotide sequences reported here have been deposited in the GenBank database under the following accession numbers: B. breve 210B ser210B locus, GU183262; B. dentium Bd1 serBd1 locus, CP001750; B. longum subsp. suis LMG21814 ser21814 gene, GU183261; B. breve 210B BR_0049, GU942824; BR_0081, GU942825; BR_0962, GU942826; BR_0521, GU942827; BR_1432, GU942828; BR_2052, GU942829; BR_0140, GU942830; BR_142, GU942831; BR_0143, GU942832; and BR_0144, GU942833.
RESULTS AND DISCUSSION
Genome-wide transcriptional response to protease treatments.
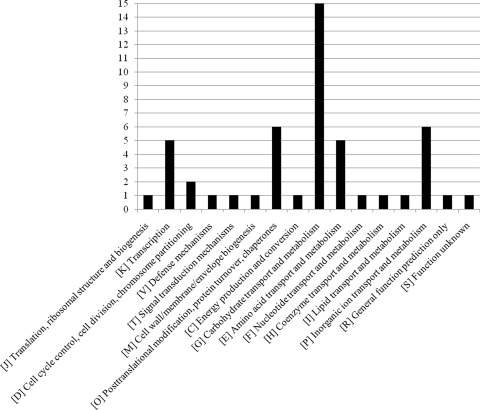
In order to investigate whether and how B. breve 210B cells respond to protease treatments, transcriptional profiling experiments were carried out. B. breve 210B cells were grown to an optical density of 0.6, and this was followed by addition of a protease, papain, chymotrypsin, or kallikrein, and then incubation for 180 min, after which RNA was harvested and labeled for microarray analysis. These proteases were selected because preliminary observations indicated that papain, chymotrypsin, and kallikrein treatments were the most effective treatments for eliciting changes in expression of a number of B. breve 210B genes (e.g., the serpin gene). Table 1 shows the results for B. breve 210B genes that exhibited at least a 2.0-fold increase in expression following various protease treatments. The transcriptional profiling analysis indicated that 42 genes were upregulated after treatment with at least one protease (Table 1), and 12 of these genes were shown to be upregulated by all three protease treatments (Table 1) and represented a core set of genes that respond to the presence of proteases. Among these 12 genes we identified the serpin-encoding locus (see below), as well as genes encoding a membrane protein displaying a DUF2194 motif, a transpeptidase, a transporter, a NagC-type transcriptional regulator, and an extracellular solute binding protein (Table 1). Other interesting genes that were upregulated following protease treatment were annotated as genes encoding (i) transport-related proteins, (ii) signal transduction proteins, (iii) molecular chaperones (e.g., DnaK and ClpB), and (iv) enzymes involved in carbohydrate degradation. The predicted products of the genes that were significantly upregulated by protease treatments were distributed in 16 cluster of orthologous group (COG) functional categories (Fig. 1). These results suggest that proteases, to which bifidobacteria are often exposed in their natural environment (28), induce a complex physiological response rather than a single event in which proteins belonging to many different functional categories take part. Aside from genes involved in carbohydrate transport and metabolism, the most abundant genes that were shown to be upregulated by protease treatment were associated with amino acid metabolism and transport, inorganic ion transport and metabolism, and posttranslational modification and transcription. On the basis of these data, we propose that B. breve 210B employs an adaptive mechanism to counteract the negative effects of GIT-derived protease activities. The scenario that we envisage assumes that B. breve 210B detects the presence of and/or an increase in protease activity by using a sensor system, which triggers expression of the protease inhibitor (i.e., the serpin product), which in turn antagonizes the action of extracellular proteases. Furthermore, we speculate that the proteases, before their activity is counteracted by serpin, degrade bacterial proteins into amino acids or peptides that are imported into the cell by several transporters (e.g., extracellular binding protein, His permease, and ABC-type dipeptide transport system). The induction of general stress response genes (e.g., genes of the dnaK operon and clpB), as well as genes involved in carbon metabolism, indicates that these genes assist in protein folding and energy foraging, respectively, as a direct or indirect response to protease treatment. Finally, induction of members of these functional categories has previously been shown to occur in intestinal bifidobacteria as a general response to natural stress agents, such as bile salts (28).
TABLE 1.
Selected transcripts upregulated in cells exposed to protease treatment (treatment with papain, chymotrypsin, or kallikrein) compared to B. breve 210B cells grown in MRS mediuma
| Gene | Change (fold) after treatment with: |
Predicted function | ||
|---|---|---|---|---|
| Papain | Chymotrypsin | Kallikrein | ||
| BR_1502 | 9.8 (1.1E-10) | 6.5 (2.0E-10) | 5.0 (8.33E-01) | Hypothetical protein |
| BR_1503 | 5.3 (1.4E-04) | 3.0 (2.32E-04) | 2.4 (2.0E-06) | Serpin protein |
| BR_0049 | 3.1 (3.2E-16) | 2.2 (3.0E-05) | 2.4 (4.1E-12) | Membrane protein |
| BR_0081 | 5.3 (2.1E-10) | 2.1 (2.0E-15) | 3.7 (2.0E-08) | Transpeptidase |
| BR_0962 | 2.8 (2.3E-16) | 4.7 (2.6E-15) | 2.1 (1.36E-01) | RelB antitoxin |
| BR_0521 | 2.2 (2.4E-08) | 4.6 (1.0E-05) | 2.1 (0.0E + 00) | Transporter system for sugar |
| BR_1432 | 17.3 (1.1E-09) | 18.6 (1.5E-08) | 2.3 (5.00E-06) | NagC-type transcriptional regulator |
| BR_2052 | 2.4 (1.7E-09) | 3.2 (2.1E-11) | 2.6 (1.4E-05) | Extracellular solute-binding protein |
| BR_0140 | 14.4 (1.8E-10) | 8.8 (2.2E-12) | 1.8 (3.19E-03) | DnaK chaperone |
| BR_0142 | 8.6 (1.5E-03) | 11.2 (2.1E-04) | 1.7 (2.00E-06) | DnaJ chaperone |
| BR_0143 | 7.6 (1.0E-04) | 7.7 (2.2E-07) | 1.5 (8.70E-05) | GrpE chaperone |
| BR_0144 | 2.3 (2.0E-04) | 2.4 (1.2E-05) | 1.5 (9.60E-05) | HspR repressor |
| BR_0010 | 2.0 (1.0E-03) | 2.5 (1.3E-05) | 1.0 (9.05E-02) | β-Galactosidase |
| BR_0017 | 4.5 (1.0E-06) | 2.7 (1.2E-07) | 1.1 (1.16E-01) | DNA-binding ferritin-like protein for protection from oxidation |
| BR_0037 | 2.6 (2.0E-07) | 7.4 (9.5E-08) | 1.2 (4.75E-04) | ABC transporter solute-binding protein |
| BR_0039 | 6.4 (3.0E-07) | 7.1 (2.3E-07) | 1.3 (1.00E-06) | Extracellular binding protein, LysR superfamily |
| BR_0040 | 2.9 (3.00E-06) | 3.3 (1.00E-06) | 0.3 (8.77E-04) | Ferritin, Dps family protein |
| BR_0045 | 3.9 (4.1E-06) | 1.7 (1.0E-01) | 3.4 (2.0E-08) | Thioredoxin disulfide reductase |
| BR_0067 | 3.4 (2.3E-08) | 1.7 (2.8E-01) | 3.1 (3.0E-09) | Major facilitator superfamily MFS_1 transporter |
| BR_0117 | 2.3 (3.5E-05) | 2.4 (4.1E-10) | 1.5 (3.6E-06) | Sucrose phosphorylase |
| BR_0175 | 2.1 (2.1E-03) | 2.2 (3.3E-09) | 1.3 (9.15E-02) | ABC-type dipeptide transport system |
| BR_0235 | 2.4 (3.3E-05) | 4.7 (0.0E+00) | 1.3 (4.28E-03) | Ammonium transporter |
| BR_0236 | 2.1 (1.0E-10) | 4.1 (2.0E-10) | 1.0 (2.48E-01) | Ammonium transporter |
| BR_0263 | 4.2 (1.1E-04) | 27.2 (3.2E-05) | 1.0 (7.52E-02) | Hypothetical protein |
| BR_0264 | 2.8 (2.2E-04) | 18.9 (5.1E-07) | 1.4 (1.80E-05) | CopG/Arc/MetJ family transcriptional regulator |
| BR_0267 | 2.7 (2.8E-08) | 1.3 (1.48E-02) | 3.2 (2.8E-11) | Glycerol uptake facilitator-related permease |
| BR_0385 | 2.6 (3.3E-05) | 1.3 (6.00E-06) | 2.9 (3.2E-04) | Hypothetical protein |
| BR_0393 | 2.2 (5.1E-06) | 2.2 (4.1E-03) | 1.2 (5.91E-01) | Binding protein-dependent transport system, inner |
| BR_0564 | 3.8 (3.3E-09) | 1.9 (1.0E-03) | 3.4 (1.1E-04) | 1-Deoxy-d-xylulose-5-phosphate synthase |
| BR_0579 | 2.4 (0.0E+00) | 4.3 (0.0E+00) | 1.5 (2.00E-06) | Putative 50S ribosomal L32 |
| BR_0582 | 7.2 (0.0E+00) | 9.5 (2.0E-04) | 1.5 (7.1E-06) | Putative 50S ribosomal L33 |
| BR_0586 | 6.3 (8.1E-05) | 6.8 (7.7E-10) | 1.3 (1.73E-03) | ABC transporter solute-binding protein |
| BR_0624 | 3.7 (6.1E-06) | 1.7 (5.2E-05) | 5.0 (3.1E-05) | Integral membrane protein involved in cell division |
| BR_0897 | 2.3 (2.0E-01) | 3.2 (2.1E-03) | 1.0 (3.06E-01) | Transcriptional regulator, PemK growth transduction signal |
| BR_0898 | 2.5 (3.0E-03) | 2.8 (6.9E-05) | 1.3 (5.00E-06) | RelB antitoxin |
| BR_0914 | 2.1 (1.0E-10) | 2.4 (2.0E-06) | 1.3 (3.29E-01) | Aldose 1-epimerase-like protein |
| BR_0933 | 3.7 (3.0E-05) | 1.5 (3.0E-06) | 3.8 (5.2E-05) | Cation-transporting ATPase |
| BR_0980 | 2.4 (4.4E-05) | 1.4 (8.50E-05) | 2.1 (2.2E-06) | Aminotransferase |
| BR_1067 | 2.1 (1.1E-10) | 1.1 (8.04E-01) | 2.5 (3.0E-05) | Dihydroorotase dehydrogenase, nucleotide metabolism and transport |
| BR_1091 | 2.8 (0.0E+00) | 1.1 (5.76E-01) | 3.1 (0.0E+00) | Asp aminotransferase |
| BR_1252 | 5.0 (0.0E+00) | 5.0 (0.0E+00) | 1.3 (7.50E-05) | His permease |
| BR_1359 | 10.0 (0.0E+00) | 4.1 (0.0E+00) | 0.8 (4.94E-01) | Transcriptional regulator |
| BR_1408 | 3.1 (0.0E+00) | 1.3 (1.61E-03) | 3.3 (0.0E+00) | Transcriptional regulator |
| BR_1430 | 3.2 (0.0E+00) | 2.5 (0.0E+00) | 1.5 (1.10E-05) | Glucosamine-6-phosphate deaminase |
| BR_1459 | 2.5 (0.0E+00) | 1.1 (6.03E-01) | 3.2 (0.0E+00) | Major facilitator superfamily MFS_1 |
| BR_1514 | 2.1 (0.0E+00) | 0.8 (3.50E-04) | 3.1 (0.0E+00) | Multisensor signal transduction histidine kinase |
| BR_1547 | 3.5 (2.2E-10) | 1.5 (1.1E-08) | 3.3 (2.0E-07) | Two-component system |
| BR_1585 | 2.2 (0.0E+00) | 1.3 (2.75E-04) | 2.9 (0.0E+00) | S-Adenosylmethionine synthetase |
| BR_1587 | 2.1 (0.0E+00) | 0.9 (1.48E-01) | 4.0 (0.0E+00) | Hypothetical protein |
| BR_1614 | 3.1 (0.0E+00) | 2.3 (0.0E+00) | 1.6 (0.0E+00) | Acetyltransferase, GNAT family |
| BR_1731 | 2.2 (0.0E+00) | 4.3 (0.0E+00) | 1.4 (0.0E+00) | Major permease |
| BR_1767 | 2.1 (2.0E-06) | 2.8 (1.2E-06) | 1.1 (7.24E-02) | ABC superfamily ATP binding cassette transporter, membrane protein |
| BR_1798 | 3.5 (1.00E-06) | 2.2 (2.00E-04) | 0.5 (5.42E-01) | UDP-4-glucose-epimerase |
| BR_1809 | 2.3 (11E-10) | 5.8 (2.2E-08) | 0.6 (1.1E-09) | Phosphotransferase system, glucose subfamily, IIA subunit |
| BR_1814 | 3.0 (1.2E-08) | 3.0 (1.0E-10) | 1.0 (8.65E-01) | Transcriptional regulator |
| BR_1885 | 2.2 (1.0E+06) | 1.2 (8.01E-02) | 2.6 (1.1E+06) | Threonine/homoserine efflux transporter |
| BR_1890 | 2.3 (1.8E-08) | 1.5 (7.0E-11) | 2.4 (1.1E-06) | Cationic amino acid transporter |
| BR_1926 | 3.0 (1.0E-03) | 3.1 (1.2E-10) | 1.1 (4.95E-01) | Ribokinase family sugar kinase |
| BR_1965 | 2.2 (0.0E+00) | 0.8 (6.00E-06) | 3.1 (0.0E+00) | Sugar transporter |
| BR_2013 | 4.5 (0.0E+00) | 4.5 (0.0E+00) | 1.7 (0.0E+00) | ClpB chaperon |
| BR_2062 | 2.1 (0.0E+00) | 0.8 (1.80E-05) | 4.4 (0.0E+00) | Oligopeptide ABC superfamily ATP binding cassette transporter |
| BR_2132 | 2.3 (0.0E+00) | 5.4 (0.0E+00) | 0.6 (1.91E-02) | Phosphotransferase system |
Bold type indicates genes that were upregulated at least 2-fold following treatment with all three proteases. All other genes are indicated by their ORF numbers and were upregulated at least 2-fold in at least one sample analyzed. The results are expressed as fold changes compared with the reference sample. The values in parentheses are adjusted P values, which were calculated as described by Benjamini and Hochberg (2) using replicate measurements from two different hybridization experiments.
FIG. 1.
Distribution of genes that are upregulated upon protease treatment based on COG functional categories. The y axis indicates the numbers of genes identified for the COG functional categories.
When the COG distribution of the genes that were shown to be differentially regulated in response to protease treatment of B. breve 210B was compared to that of the entire genome, no clear differences were found, except for slight overrepresentation of the COG categories corresponding to carbohydrate metabolism and transport, as well as chaperone production, and underrepresentation of the COG family corresponding to amino acid metabolism.
Sequence analysis of B. breve 210B ser locus.
The B. breve 210B genome (M. Ventura et al., unpublished data) contains a single serpin-like gene, designated ser210B, whose predicted secreted protein product displays a high level of similarity (>30%) with serine protease inhibitors (serpins) of a broad range of bacteria, including Actinobacteria (e.g., B. longum subsp. longum NCC2705, B. longum subsp. longum DJO10A, B. longum subsp. infantis ATCC 15697, B. longum subsp. infantis CCUG52486, B. dentium Bd1, and Eggerthella lenta DSM2243), Firmicutes (e.g., Desulfitobacterium hofniense DCB-2), cyanobacteria (e.g., Nostoc spp.), and Chloroflexi (e.g., Dehalococcoides spp.), and even with serpins produced by various organisms belonging to other kingdoms, such as Eukarya (e.g., Sus scrofa). The deduced Ser210B sequence contains all functional domains typically found in other members of the serpin superfamily (accession no. PFAM00079). Generally, serpin proteins react with tissue-type plasminogen activator (tPA) and, in the case of neuroserpins (12), with neurons in regions of the brain where tPA is detected, which suggests that neuroserpin is the selective inhibitor of tPA in the central nervous system (12). Moreover, Ser210B contains domains identified in alpha-1 antitrypsin, as well as antithrombin, which controls blood coagulation (6, 12, 20).
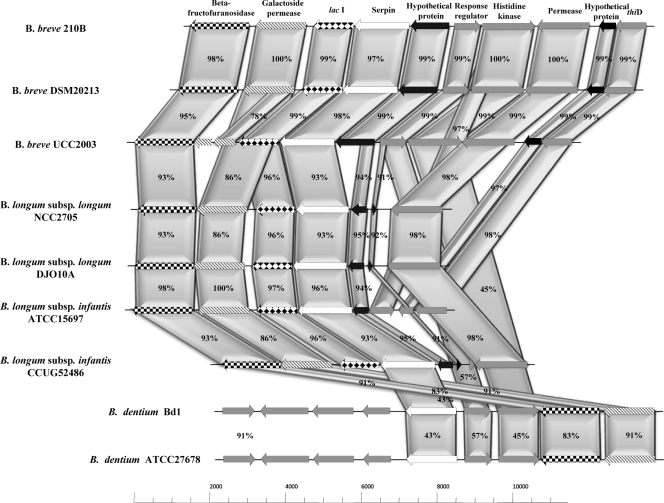
The genetic organization and location of the ser210B gene on the chromosome of B. breve 210B and genetic organization and location of its homologs in other bacteria are shown schematically in Fig. 2, in which the deduced amino acid sequence encoded by the ser210B locus is aligned with those of other bifidobacteria. This comparative analysis revealed a high level of identity (>70%) between the serine protease inhibitor proteins of bifidobacteria belonging to the B. longum phylogenetic group (37). In contrast, the flanking DNA regions of the bifidobacterial ser homologs were shown to be variable with respect to gene synteny, except at the intraspecies level. The ser genes in the B. breve and B. longum genomes are preceded by a gene encoding a membrane-associated protein and are followed by a lacI-type gene that is predicted to encode a transcriptional regulator. Furthermore, the ser locus in the B. breve strain analyzed, as well as in B. dentium genomes, is flanked by a putative response regulator-encoding gene, followed by a gene that encodes a predicted histidine protein kinase, whereas in the chromosomes of the B. longum strains analyzed this two-component system is completely (e.g., B. longum subsp. infantis ATCC 15697) or partially absent (Fig. 2).
FIG. 2.
Schematic diagram of ser loci of B. breve 210B and other bifidobacterial strains. Each arrow indicates an ORF, and the length of the arrow is proportional to the size of the ORF. The predicted protein functions are indicated above the arrows. The levels of amino acid identity (expressed as percentages) are also indicated.
Structural investigation of serpin proteins produced by bifidobacteria.
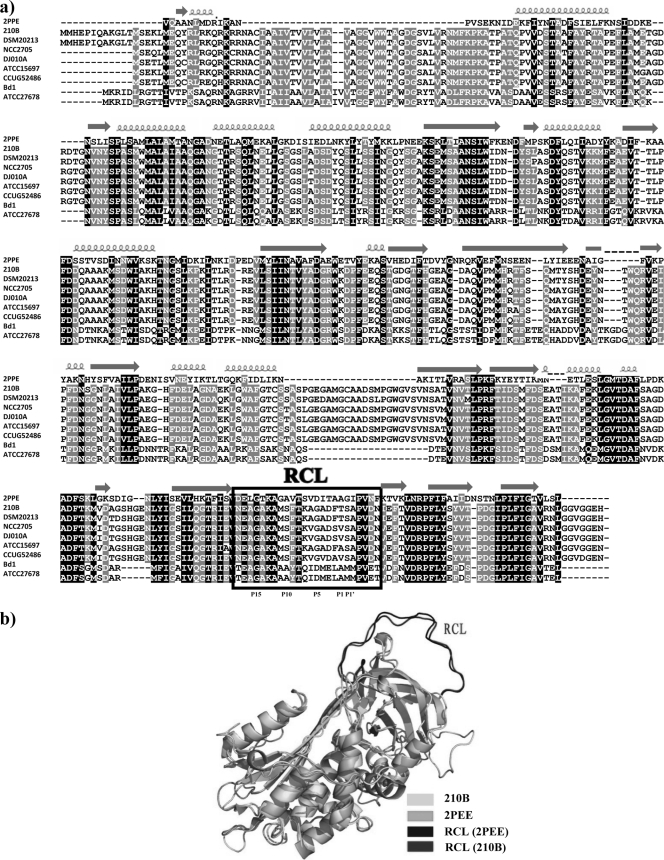
In our analysis of the 3D structure of bifidobacterial serpin proteins we included the serpin proteins produced by B. breve 210B and B. dentium Bd1. A BLASTP analysis of the serpin protein sequences produced by B. breve 210B (Ser210B) and B. dentium Bd1 (SerBd1) with the PDB database resulted in identification of a reliable structural template (tengpinD31) (47), which is an active protease inhibitor. Although the levels of sequence identity between the serpin protein sequences produced by these two bifidobacterial species and the template are low (between 20.3% and 19.6%) (Fig. 3a), the corresponding E values are significant (2e−32 and 1e−36 for Ser210B and SerBd1, respectively). Similar to most other serpin proteins, these two model proteins are composed of at least three beta-sheets and at least seven alpha-helices (33). The 3D structures of Ser210B and SerBd1 are predicted to contain a reactive center loop (RCL) as an important structural component (33), which is responsible for the interaction with the target proteases and which forms an extended, exposed area above the core domain of the serpin structure (Fig. 3b). From other work it is known that when the serpin protein interacts with a target protease, a scissile bond between residues P1 and P1′ in the RCL region is cleaved by the target protease (11). Once cleaved, the RCL moves to the opposite pole of the serpin, trapping the protease. This whole process is irreversible, and once the serpin interacts with the target protease, its conformation cannot change back to the original form. Based on this single-use substrate mechanism, serpins inhibit different serine proteases. In general, the RCL region contains approximately 20 residues near the C terminus (11). In the two predicted models, both RCLs are composed of 24 residues (Fig. 3a). It has previously been established that the residues at the P1 position are involved in the protease specificity of serpins (9). The serpin proteins encoded by B. breve 210B and B. dentium Bd1 contain a charged residue (Thr) and a hydrophobic residue (Ala) at the P1 position, respectively, which suggests that they have different substrates. From P9 to P14, the B. dentium Bd1 Ser protein contains more alanine residues, which may assist insertion of its RCL into the serpin body (9). This suggests that the serpin proteins produced by different bifidobacterial species and subspecies (e.g., B. breve, B. dentium, B. longum subsp. longum, and B. longum subsp. infantis) exhibit different substrate behaviors and thus have differential inhibitory spectra. This may be a consequence of the specific ecological niches where these bifidobacteria reside (e.g., the intestine in the case of B. breve and the oral cavity in the case of B. dentium), where they are faced with different repertoires of proteases.
FIG. 3.
(a) Alignment of the Ser proteins of B. breve 210B, B. breve DSM20213, B. longum subsp. longum DJO10A, B. longum subsp. longum NCC2705, B. longum subsp. infantis ATCC 15697, B. longum subsp. infantis CCUG52486, B. dentium Bd1, and B. dentium ATCC 27678 with the structural template tengpinD31 (2PPE). Shading indicates conservation at a position in at least 50% of the amino acid residues in the alignment (black, identical residues; gray, similar residues). The α-helices and turns are indicated. The RCL region is enclosed in a box. (b) 3D structure of B. breve 210B serpin. The 3D structure of the B. breve 210B serpin (light gray) is superimposed on the predicted serpin model (PDB entry 2PEE) (gray). The potential residues in the various serpins that are predicted to form the RCL regions are indicated.
Prevalence of ser homologs across Bifidobacterium genomes.
In order to determine whether and to what extent ser homologs are present in bifidobacterial genomes, a PCR-based screening strategy with primers Ser-uni-univ and Ser-rev-univ based on two conserved regions (positions 290 to 309 and positions 1232 to 1249 in ser210B) of the ser genes identified in the genomes of B. breve, B. longum subsp. longum, and B. longum subsp. infantis was used. In this way we were able to obtain amplification products that indicated the presence of a ser homolog in strains representing five bifidobacterial species and subspecies (B. longum subsp. longum, B. longum subsp. infantis, B. longum subsp. suis, B. breve, and B. dentium). Moreover, DNA sequencing of the PCR products obtained confirmed the identity of the ser gene (data not shown). To expand and verify these results, amplified ser-containing DNA was used as a probe in a slot blot hybridization experiment to detect ser homologs in the genomic DNA of 34 bifidobacterial strains representing 30 different species (Fig. 4). Interestingly, using different stringency conditions, only B. breve, B. longum subsp. longum, B. longum subsp. suis, B. longum subsp. infantis, B. dentium, Bifidobacterium cuniculi, and Bifidobacterium scardovi yielded a strong positive hybridization signal (Fig. 4a). Weaker hybridization signals were observed for Bifidobacterium mericycum and for bifidobacterial species belonging to the Bifidobacterium pullorum phylogenetic group (Bifidobacterium gallinarum, Bifidobacterium pullorum, and Bifidobacterium saeculare) (Fig. 4 and data not shown). This clearly indicates that ser homologs are not widely distributed in bifidobacterial genomes, which is similar to findings described for other bacterial genera (e.g., Nostoc), where the serpin-encoding gene is not uniformly present in all species of a bacterial genus but is restricted to a small number of species in the genus (13).
FIG. 4.
Slot blot hybridization using DNA extracted from different bifidobacterial species and hybridized using a mixed ser probe. The spots contained DNA from the following strains: 1, B. longum subsp. longum ATCC 15707T; 2, B. bifidum LMG 11041T; 3, B. pseudolongum subsp. globosum LMG11569T; 4, B. asteroides LMG 10735T; 5, B. saeculare LMG 14934T; 6, B. gallinarum 11586T; 7, B. angulatum ATCC 27535T; 8, B. choerinum LMG 10510T; 9, B. gallicum LMG 11596T; 10, B. subtile LMG 11597T; 11, B. dentium LMG 11045T; 12, B. longum subsp. infantis ATCC 15697T; 13, B. animalis subsp. lactis DSM 10140T; 14, B. animalis subsp. animalis ATCC 25527T; 15, B. ruminatium LMG21811T; 16, B. thermacidophilum subsp. thermacidophilum LMG 21395T; 17, B. thermacidophilum subsp. porcinum LMG21689T; 18, B. longum subsp. suis LMG 21814T; 19, B. pullorum LMG 21816T; 20, B. boum LMG 10736T; 21, B. pseudolongum subsp. pseudolongum LMG 11571T; 22, B. minimum LMG 11592T; 23, B. catenulatum LMG 11591T; 24, B. merycicum LMG 11341T; 25, B. pseudocatenulatum LMG 10505T; 26, B. psychraerophilum LMG 21775T; 27, B. indicum LMG 11587T; 28, B. coryneforme LMG 18911T; 29, B. adolescentis ATCC 15703T; 30, B. magnum LMG 11591T; 31, B. thermophilum JCM 7027T; 32, B. scardovii LMG 21589T; 33, B. animalis subsp. animalis ATCC 27536; 34, B. bifidum 317B; 35, B. pseudocatenulatum 318B; 36, B. cuniculi LMG 10738T; and 37, B. breve 210B.
To further assess the conservation of the ser gene in different strains of B. breve, B. longum, and B. dentium, we performed PCRs using ser-specific, conserved PCR primers Ser-uni-univ and Ser-rev-univ and, as the template, chromosomal DNA derived from 734 bifidobacterial isolates which had previously been shown to belong to the species B. longum, B. breve, Bifidobacterium adolescentis, Bifidobacterium pseudocatenulatum, Bifidobacterium bifidum, Bifidobacterium pseudolongum, Bifidobacterium animalis, and B. dentium (35). The results of this large-scale survey showed that all PCRs involving DNA from isolates belonging to B. breve, B. longum, and B. dentium generated an amplicon of the expected size, whose identity was also verified by DNA sequencing, suggesting that the ser gene is universally conserved in strains belonging to these species (data not shown). In contrast, strains belonging to other bifidobacterial species did not yield any specific amplicons.
Phylogenetic analysis based on serpin protein sequences.
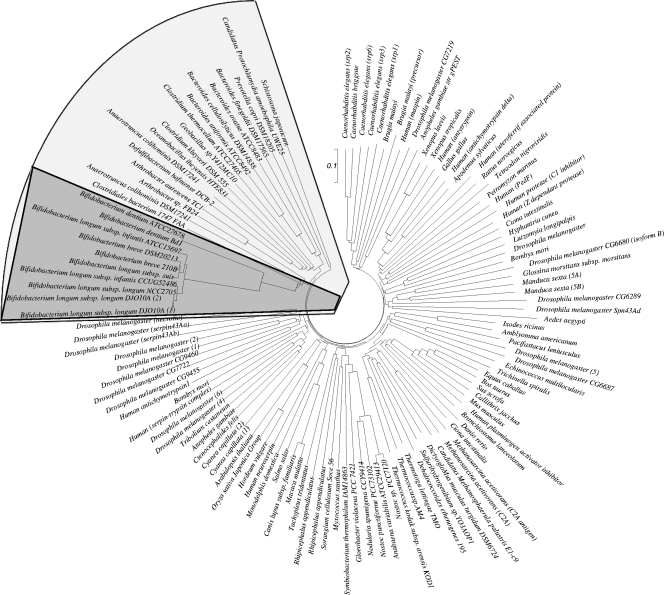
In order to assess the distribution of ser homologs across bacteria, we surveyed available genomic data for both prokaryotes and eukaryotes to determine the presence of serpin-encoding genes. Alignment of the serpin proteins was performed using ClustalW, which resulted in an unrooted neighbor-joining phylogenetic tree (Fig. 5). The serpin orthologs identified in bacteria clustered separately from those identified in eukaryotes, suggesting that the bacterial serpin protein subfamilies either diverged after the serpin genes evolved from one ancestral gene or resulted from convergent evolution from more than one ancestral gene (Fig. 5). Notably, bifidobacterial serpin proteins form a monophyletic group in the clustered bacterial serpins (Fig. 5). This phylogenetic distribution in bifidobacteria suggests that the ser gene in this group of bacteria was acquired through horizontal gene transfer (HGT). However, this suspected HGT origin is not supported by data for sequence features used to detect HGT events, such as analyses of G+C content and codon usage, which did not reveal any bias different from that of the genome.
FIG. 5.
Phylogenetic tree obtained using serpin homologs from various bacteria. The scale bar indicates phylogenetic distance. The serpin protein sequences of bacteria are enclosed in a box, while the serpin proteins of bifidobacteria are indicated by dark shading.
Transcriptional analysis of bifidobacterial ser loci.
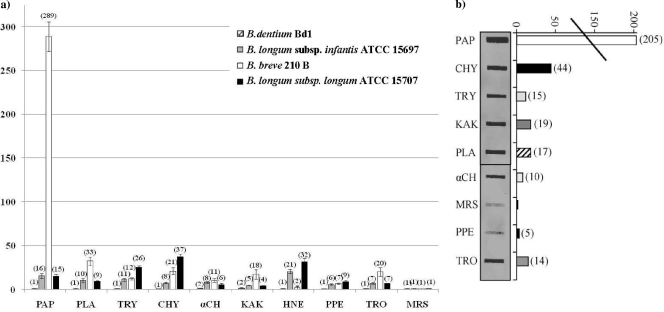
In order to determine if, besides the ser210B gene, ser genes of other intestinal bifidobacterial strains, such as B. longum subsp. infantis ATCC 15697, B. longum subsp. longum ATCC 15707, and the oral cavity inhabitant B. dentium Bd1, are differentially induced when they are exposed to different protease activities, the amounts of ser gene-specific mRNAs were determined by quantitative real-time PCR (qRT-PCR) assays. These experiments were performed using samples of mRNA extracted from exponential cultures of B. breve 210B, B. longum subsp. infantis ATCC 15697, B. longum subsp. longum ATCC 15707, and B. dentium Bd1 which had been resuspended in prewarmed MRS medium containing one protease (pancreatic elastase, human neutrophil elastase, thrombin, papain, kallikrein, trypsin, α-antitrypsin, chymotrypsin, or plasmin) at a concentration simulating the concentration encountered by bacteria in their natural environment (e.g., human gut) (19). The observed levels of induction of ser210B in response to these serine proteases ranged from 7-fold with pancreatic elastase to more than 250-fold when cultures were exposed to papain. A lower level of induction of ser210B transcription was observed when the cultures were exposed to other proteases, including chymotrypsin, plasmin, kallikrein, thrombin, and trypsin (Fig. 6a). Conversely, the level of ser210B-containing mRNA did not change considerably after treatment with human neutrophil elastase (Fig. 6a). These findings are different from those previously obtained for the serpin-encoding gene of B. longum subsp. longum NCC2705 (14) (designated ser2705), for which the human serine proteases that were most effective for increasing the amount of the Ser2705 protein were human elastase and pancreatic elastase (14). These findings were partially corroborated by analysis of the induction of the serpin-encoding gene of another B. longum strain, B. longum subsp. longum ATCC 15707, in which the highest level of induction occurred in the presence of human elastase (Fig. 6a). In accordance with this previously described observation, we showed that the most effective induction of ser gene expression in B. longum subsp. infantis ATCC 15697 (designated ser15697) also occurred upon treatment with human elastase or papain (Fig. 6a). This suggests that the regulatory signals and/or mechanisms of responses of bifidobacteria to the host's serine proteases are different for the phylogenetically closely related species B. breve and B. longum (38). It is possible that the different patterns of ser gene induction in B. breve and B. longum are linked to the different ecological origins of these bacteria (e.g., different niches in the colon) (35, 44). Consequently, the different human proteases present in infants and adults or in different locations of the gut may have induced different responses to provide protection against the negative effects of human gut serine proteases. Furthermore, in order to validate this age- or niche-specific hypothesis, we determined the levels of induction of the ser gene of B. dentium Bd1 (designated serBd1) in response to the same set of serine proteases that were examined for ser210B, ser15707, and ser15697 expression. Surprisingly, no significant induction of serBd1 was found (Fig. 6a) for any of the proteases tested. This suggests that this bacterium requires different proteases for induction, that transcription of serBd1 depends on other environmental factors, or that the expression of serBd1 is not subject to transcriptional control.
FIG. 6.
Plots of relative levels of transcription of B. breve 210B, B. longum subsp. infantis ATCC 15697, B. longum subsp. longum ATCC 15707, and B. dentium Bd1 serpin-encoding genes after serine protease treatment versus growth in MRS-based media as analyzed by quantitative real-time PCR assays (a) and by slot blot hybridization (b). The bars indicate the relative amounts of ser mRNAs for the specific samples. The cDNAs used were synthesized using RNA collected from bifidobacterial cultures exposed for 150 min to α-chymotrypsin (αCH), chymotrypsin (CHY), neutrophil elastase (HNE), kallikrein (KAK), pancreatic elastase (PPE), papain (PAP), plasmin (PLA), trypsin (TRY), or thrombin (TRO). For panel b, induction of the ser210B gene was evaluated by slot blot hybridization. Total RNA (25 μg per slot) was isolated from B. breve 210B cells exposed for up to 150 min to different serine proteases (see above) and probed with biotin-labeled ser. The resulting hybridization signals obtained in the autoradiograms were quantified. The amount of mRNA synthesized under the conditions described above was normalized to the amount present in cultures grown in MRS medium.
In order to verify the results obtained with qRT-PCR, we performed slot blot hybridization involving mRNA isolated from B. breve 210B cultures exposed to the same set of proteases (Fig. 6b). Based on the intensity of the hybridization signal, the highest level of transcription of the B. breve 210B ser210B gene was found to occur following exposure to chymotrypsin and papain for 150 min, conditions which increased the ser210B mRNA levels approximately 44- and 200-fold, respectively (Fig. 6b). These and other protease-induced levels of transcription were consistent with those determined by qRT-PCR (Fig. 6a).
Transcriptional mapping of the ser210B locus.
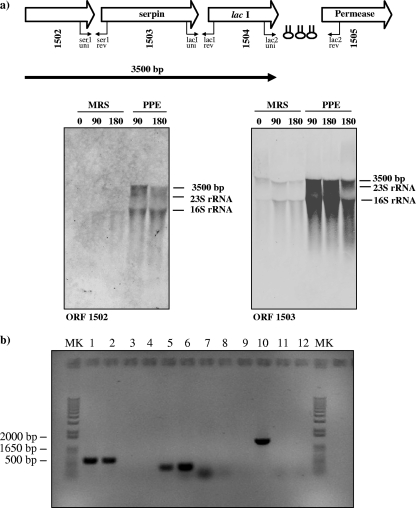
In order to analyze transcription of the ser210B locus further, total RNA that was extracted from B. breve 210B cells grown in complex media, such as MRS medium, in the presence or absence of a human serine protease (i.e., pancreatic elastase) that induces expression of the ser210B gene, as demonstrated by qRT-PCR (see above), was hybridized with DNA probes targeting the genes of the ser210B locus. The results revealed a pancreatic elastase-dependent hybridization signal corresponding to an approximately 3.5-kb transcript when a probe encompassing the ser gene was used (Fig. 7a). An identical transcription pattern was obtained for one of the genes adjacent to ser210B (ORF1502), indicating that the two genes are transcribed as a polycistronic mRNA (Fig. 7a). Further proof that there was a polycistronic transcript was provided by RT-PCR experiments. cDNA templates were obtained by RT of B. breve 210B total mRNA isolated after cells were treated with pancreatic elastase. Each cDNA was used as the template in subsequent PCR with different combinations of primers spanning the ser gene and flanking genes (Fig. 7b). Notably, amplicons were obtained using the PCR primers spanning the ser210B gene (ORF1503) and the adjacent gene (ORF1502) (Fig. 7b). When the same strategy was employed to investigate cotranscription of ser210B with the other flanking genes, an RT-PCR product was obtained when primers spanning the ser210B gene and the gene predicted to be the gene encoding a LacI-type protein (ORF1504) were used, suggesting that the transcript of the ser210B operon encompasses ORF1502, ORF1503, and ORF1504 (Fig. 7b). Notably, a transcript is generally degraded from the 3′ end, which may be responsible for the fact that in microarray experiments the LacI-encoding gene was not found to be upregulated after protease treatment.
FIG. 7.
Transcriptional organization of the B. breve 210B ser operon. (a) Schematic diagram of the B. breve 210B ser locus and Northern blot analysis of B. breve 210B ser transcription. Hairpin symbols indicate predicted secondary structures. The transcript identified is indicated by a solid arrow, which points toward the 3′ end of the mRNA. RNA was isolated from a culture before and after exposure to pancreatic elastase (PPE). The molecular weights, calculated from the hybridization signal and position of 16S and 23S rRNA, are indicated. 0, 90, and 180, incubation for 0, 90, and 180 min, respectively. (b) Products generated by RT-PCR. PCR products were obtained with primers spanning the intergenic regions between ORF1502 and ser210B (primers ser1-uni and ser1-rev) (lanes 1 to 4), between ser210B and lacI (primers lacI-uni and lacI-rev) (lanes 5 to 8), and between lacI and ORF1505 (primers lac2-uni and lac2-rev) (lanes 9 to 12). The positions of the primer pairs used in RT-PCR experiments are shown in panel a. PCR products were obtained under the following conditions: lanes 1, 5, and 9, cDNA prepared from B. breve 210B RNA; lanes 3, 7, and 11, negative control in which B. breve RNA was used but reverse transcriptase was omitted; lanes 4, 8, and 12, no-template control; lanes 2, 6, and 10, positive control (B. breve DNA). Lanes MK contained a 1-kb DNA molecular marker.
Analysis of the nucleotide sequence of the ser210B operon revealed that the lacI210B gene was flanked at the 3′ end by three inverted repeats that may function as rho-independent transcriptional terminator structures (Fig. 7a).
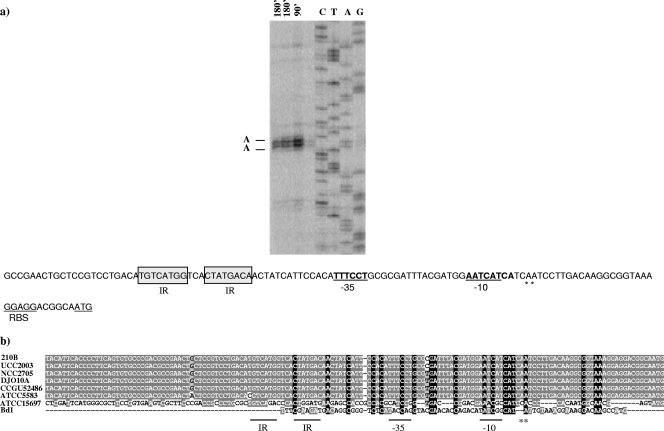
Identification of the transcriptional start site.
The transcriptional start site of the ser210B locus was identified by primer extension analysis using mRNA extracted from cultures exposed to pancreatic elastase for 90 min or 180 min. Two extension products were identified 31 and 32 nucleotides upstream of the predicted translational start site of the first gene (ORF1502) of the ser210B operon (Fig. 8a). Analysis of the putative promoter region of the ser210B operon showed that there was a potential sigma-70 promoter-like sequence resembling consensus −10 and −35 sequences (Fig. 8a). Interestingly, a TG motif was found upstream of the −10 region, which suggested that there was an extended −10 region which could provide a supplementary recognition sequence for the RNA polymerase. (46).
FIG. 8.
Determination of the B. breve 210B ser transcription initiation site by primer extension analysis. (a) Primer extension results obtained using oligonucleotide SER-prom1 and mRNA isolated from exposition of pancreatic elastase at the times indicated. The deduced −10 hexamer and −35 hexamers are indicated by bold type and underlined; bold type with asterisks indicates the transcription start points; and the start codon is underlined. RBS, ribosome binding site. (b) Comparison of the promoter regions of ser genes from different bifidobacteria, including B. breve 210B, B. breve UCC2003, B. longum subsp. longum NCC2705, B. longum subsp. longum DJO10A, B. longum subsp. infantis CCUG52486, B. longum subsp. infantis ATCC 55813, B. longum subsp. infantis ATCC 15697T, and B. dentium Bd1.
The predicted promoter regions of ser loci identified in bifidobacteria, including B. breve 210B, B. breve UCC2003, B. longum subsp. longum NCC2705 (Fig. 8b), B. longum subsp. longum DJO10A, B. longum subsp. infantis ATCC 15697, B. longum subsp. infantis CCUG52486, B. longum subsp. infantis ATCC 55813, and B. dentium Bd1, were aligned in an attempt to identify putative regulatory elements. As shown in Fig. 8b, the presence of a large consensus promoter region, including the putative −10 and −35 hexamers, was deduced based on the sequences of B. breve 210B, B. breve UCC2003, B. longum subsp. longum NCC2705, B. longum subsp. longum DJO10A, B. longum subsp. infantis CCUG52486, and B. longum subsp. infantis ATCC 55813. Moreover, a number of other DNA motifs were shown to be conserved in all putative ser promoter sequences of these strains, including a nearly perfect inverted repeat (IR) (TGTCATGG-3N-CTATGACA). In contrast, the putative promoter regions of B. longum subsp. infantis ATCC 15697 and B. dentium Bd1 were highly divergent, which suggests that there is a different regulatory mechanism for ser gene expression in these bacteria (Fig. 8b).
Conclusions.
Serpin-encoding genes are widely distributed in multicellular eukaryotes and also in prokaryotes, although to a lesser extent. The association of serpin genes with bacterial and archeabacterial genomes suggests that these genes are likely to have evolved before the divergence of the main branches of life. In bacteria their presence is not universal, indicating that they either were acquired through Darwinian selection or were lost when they were not essential or important for survival. From a phylogenetic point of view, bacterial serpins are only distantly related to eukaryotic serpins, and for this reason they clustered as a separate group (5). Analysis of available bifidobacterial genomes revealed the presence of serpin-encoding genes in just a small number of species, and this may suggest that the serpin gene was originally acquired through horizontal gene transfer.
Many bifidobacteria are commensal microorganisms that naturally reside in the large intestine of humans and animals (35, 36). Recently, it has been shown that the serpin protein produced by B. longum subsp. longum NCC2705 is active against the proteolytic action of host proteases (i.e., human neutrophil elastase and pancreatic elastase) (14). In this study, we showed that the serpin genes of different bifidobacterial species and subspecies (B. breve, B. longum subsp. infantis, and B. longum subsp. longum) are highly induced when bacterial cultures are exposed to various host-derived proteases. This finding is highly relevant since many of these proteases are normally found in the human gut and thus the presence of a protease inhibitor may provide an ecological advantage to bifidobacteria since serpin activity may protect them against these host proteases. In previous reports workers have indicated that protease inhibitors secreted by bacteria such as Bacillus brevis and Prevoltella intermedia protect themselves against proteolytic attack by avoiding degradation by extracellular proteins (10, 32). In line with this, we speculate that in bifidobacteria preservation of extracellular proteins is important in order to guarantee formation of necessary extracellular structures involved in the interaction with the host. Our studies highlight the finding that some human gut-derived bifidobacterial species, such as those belonging to the B. longum phylogenetic group, possess a serpin-encoding gene, whose expression is induced by exposure to human- and plant-derived serine proteases. In contrast, other gut-associated bifidobacterial species, such as B. adolescentis, Bifidobacterium catenulatum, B. bifidum, and Bifidobacterium gallicum, do not contain a serpin gene in their genomes, suggesting that protection of these species from serine proteases depends on alternative strategies or on cross-protection offered by the (bifido)bacterial species that do produce serine proteinase inhibitors. An exception to these findings seems to be the serpin produced by B. dentium, an organism which resides in the human oral cavity. In fact, the transcription signals and control of the ser operon of B. dentium seem to be different from the transcription signals and control of the serpin-encoding loci present in human gut bifidobacteria. Such differences may be the consequence of the different repertoire of proteases to which B. dentium is exposed in its natural ecological niche compared to gut-derived bifidobacteria.
Bifidobacteria are commonly used as health-promoting bacteria in many functional or probiotic food preparations, with the aim of improving the health of the host through immunomodulatory activity, as well as reestablishing a healthy balance in the intestinal microbiota (8, 22, 23). In humans establishment of a stable intestinal microbiota is subject to tolerance of the immune system to such commensal bacteria; this is in contrast to the situation with pathogenic bacteria, which provoke a considerable immune response (29). The transcriptional activation of ser210B, ser15697, and ser15707 in response to serine proteases may represent a molecular mechanism for immunomodulation. It is well established that the release of serine proteases (e.g., neutrophile elastase) at sites of intestinal inflammation is part of the innate immunity response and that this release is caused by bacterial infection or is a consequence of pathological immune activation. The latter result is typical of inflammatory bowel disease and ulcerative colitis, which may cause breaches in the intestinal barrier (5, 24). The powerful and protease-inducible serine protease inhibitor that is synthesized by B. breve 210B as an autochthonous component of the intestinal microbiota may thus elicit anti-inflammatory activity that may reduce the negative effects of serine protease activity at sites of intestinal inflammation. This is reminiscent of the way in which exaggerated serine protease activity, which may cause pathological tissue damage, is reduced by α-antitrypsin, the physiological inhibitor present in blood plasma (14).
Further investigations should be carried out to determine the regulatory mechanisms that drive serpin gene expression in B. breve, as well as the potential role of the serpin protein in the interaction with its host.
Supplementary Material
Acknowledgments
This work was financially supported by an EMBARK postdoctoral fellowship award to A.Z. from the Italian Award for Outstanding Young Researcher scheme “Incentivazione alla mobilita' di studiosi stranieri e italiani residenti all'estero,” by Marie Curie Reintegration grant MERG-CT-2005-03080 to M.V., by Spinner 2013, by Regione Emilia Romagna, and by EFSE. This work was also financially supported by an IRCSET Embark postgraduate fellowship to F.B. D.V.S., A.Z., and M.O.M. are members of The Alimentary Pharmabiotic Centre, which is a Centre for Science and Technology (CSET) funded by Science Foundation Ireland (SFI) through the the Irish Government's National Development Plan.
We are grateful to C. Milani for technical assistance, and we thank students and coworkers for contributing data and for their enthusiasm.
Footnotes
Published ahead of print on 26 March 2010.
REFERENCES
- 1.Barrangou, R., E. P. Briczinski, L. L. Traeger, J. R. Loquasto, M. Richards, P. Horvath, A. C. Coute-Monvoisin, G. Leyer, S. Rendulic, J. L. Steele, J. R. Broadbent, T. Oberg, E. G. Dudley, S. Schuster, D. A. Romero, and R. F. Roberts. 2009. Comparison of the complete genome sequences of Bifidobacterium animalis subsp. lactis DSM 10140 and Bl-04. J. Bacteriol. 191:4144-4151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Benjamini, Y., and Y. Hochberg. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. 57:289-300. [Google Scholar]
- 3.Bilban, M., L. K. Buehler, S. Head, G. Desoye, and V. Quaranta. 2002. Defining signal thresholds in DNA microarrays: exemplary application for invasive cancer. BMC Genomics 3:19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolstad, B. M., R. A. Irizarry, M. Astrand, and T. P. Speed. 2003. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19:185-193. [DOI] [PubMed] [Google Scholar]
- 5.Burg, N. D., and M. H. Pillinger. 2001. The neutrophil: function and regulation in innate and humoral immunity. Clin. Immunol. 99:7-17. [DOI] [PubMed] [Google Scholar]
- 6.Elliott, P. R., J. P. Abrahams, and D. A. Lomas. 1998. Wild-type alpha 1-antitrypsin is in the canonical inhibitory conformation. J. Mol. Biol. 275:419-425. [DOI] [PubMed] [Google Scholar]
- 7.Felsenstein, J. 1997. An alternating least squares approach to inferring phylogenies from pairwise distances. Syst. Biol. 46:101-111. [DOI] [PubMed] [Google Scholar]
- 8.Furrie, E., S. Macfarlane, A. Kennedy, J. H. Cummings, S. V. Walsh, D. A. O'Neil, and G. T. Macfarlane. 2005. Synbiotic therapy (Bifidobacterium longum/Synergy 1) initiates resolution of inflammation in patients with active ulcerative colitis: a randomised controlled pilot trial. Gut 54:242-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gettins, P. G. 2002. Serpin structure, mechanism, and function. Chem. Rev. 102:4751-4804. [DOI] [PubMed] [Google Scholar]
- 10.Grenier, D. 1994. Characteristics of a protease inhibitor produced by Prevotella intermedia. FEMS Microbiol. Lett. 119:13-18. [DOI] [PubMed] [Google Scholar]
- 11.Huntington, J. A., R. J. Read, and R. W. Carrell. 2000. Structure of a serpin-protease complex shows inhibition by deformation. Nature 407:923-926. [DOI] [PubMed] [Google Scholar]
- 12.Irving, J. A., R. N. Pike, A. M. Lesk, and J. C. Whisstock. 2000. Phylogeny of the serpin superfamily: implications of patterns of amino acid conservation for structure and function. Genome Res. 10:1845-1864. [DOI] [PubMed] [Google Scholar]
- 13.Irving, J. A., P. J. Steenbakkers, A. M. Lesk, H. J. Op den Camp, R. N. Pike, and J. C. Whisstock. 2002. Serpins in prokaryotes. Mol. Biol. Evol. 19:1881-1890. [DOI] [PubMed] [Google Scholar]
- 14.Ivanov, D., C. Emonet, F. Foata, M. Affolter, M. Delley, M. Fisseha, S. Blum-Sperisen, S. Kochhar, and F. Arigoni. 2006. A serpin from the gut bacterium Bifidobacterium longum inhibits eukaryotic elastase-like serine proteases. J. Biol. Chem. 281:17246-17252. [DOI] [PubMed] [Google Scholar]
- 15.Kimura, M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 16.Leahy, S. C., D. G. Higgins, G. F. Fitzgerald, and D. van Sinderen. 2005. Getting better with bifidobacteria. J. Appl. Microbiol. 98:1303-1315. [DOI] [PubMed] [Google Scholar]
- 17.Lee, J. H., V. N. Karamychev, S. A. Kozyavkin, D. Mills, A. R. Pavlov, N. V. Pavlova, N. N. Polouchine, P. M. Richardson, V. V. Shakhova, A. I. Slesarev, B. Weimer, and D. J. O'Sullivan. 2008. Comparative genomic analysis of the gut bacterium Bifidobacterium longum reveals loci susceptible to deletion during pure culture growth. BMC Genomics 9:247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marco, M. L., S. Pavan, and M. Kleerebezem. 2006. Towards understanding molecular modes of probiotic action. Curr. Opin. Biotechnol. 17:204-210. [DOI] [PubMed] [Google Scholar]
- 19.Mènard, R., and A. C. Storer. 1998. Papain. Academic Press, London, United Kingdom.
- 20.Mottonen, J., A. Strand, J. Symersky, R. M. Sweet, D. E. Danley, K. F. Geoghegan, R. D. Gerard, and E. J. Goldsmith. 1992. Structural basis of latency in plasminogen activator inhibitor-1. Nature 355:270-273. [DOI] [PubMed] [Google Scholar]
- 21.O'Hara, A. M., and F. Shanahan. 2007. Mechanisms of action of probiotics in intestinal diseases. Sci. World J. 7:31-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Mahony, L., J. McCarthy, P. Kelly, G. Hurley, F. Luo, K. Chen, G. C. O'Sullivan, B. Kiely, J. K. Collins, F. Shanahan, and E. M. Quigley. 2005. Lactobacillus and bifidobacterium in irritable bowel syndrome: symptom responses and relationship to cytokine profiles. Gastroenterology 128:541-551. [DOI] [PubMed] [Google Scholar]
- 23.Ouwehand, A. C., S. Salminen, and E. Isolauri. 2002. Probiotics: an overview of beneficial effects. Antonie Van Leeuwenhoek 82:279-289. [PubMed] [Google Scholar]
- 24.Reeves, E. P., H. Lu, H. L. Jacobs, C. G. Messina, S. Bolsover, G. Gabella, E. O. Potma, A. Warley, J. Roes, and A. W. Segal. 2002. Killing activity of neutrophils is mediated through activation of proteases by K+ flux. Nature 416:291-297. [DOI] [PubMed] [Google Scholar]
- 25.Roberts, T. H., J. Hejgaard, N. F. Saunders, R. Cavicchioli, and P. M. Curmi. 2004. Serpins in unicellular Eukarya, Archaea, and Bacteria: sequence analysis and evolution. J. Mol. Evol. 59:437-447. [DOI] [PubMed] [Google Scholar]
- 26.Sali, A., and T. L. Blundell. 1993. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 234:779-815. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook, J., E. E. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 28.Sanchez, B., M. C. Champomier-Verges, P. Anglade, F. Baraige, C. G. de Los Reyes-Gavilan, A. Margolles, and M. Zagorec. 2005. Proteomic analysis of global changes in protein expression during bile salt exposure of Bifidobacterium longum NCIMB 8809. J. Bacteriol. 187:5799-5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sansonetti, P. J. 2004. War and peace at mucosal surfaces. Nat. Rev. Immunol. 4:953-964. [DOI] [PubMed] [Google Scholar]
- 30.Schell, M. A., M. Karmirantzou, B. Snel, D. Vilanova, B. Berger, G. Pessi, M. C. Zwahlen, F. Desiere, P. Bork, M. Delley, R. D. Pridmore, and F. Arigoni. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. U. S. A. 99:14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sela, D. A., J. Chapman, A. Adeuya, J. H. Kim, F. Chen, T. R. Whitehead, A. Lapidus, D. S. Rokhsar, C. B. Lebrilla, J. B. German, N. P. Price, P. M. Richardson, and D. A. Mills. 2008. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc. Natl. Acad. Sci. U. S. A. 105:18964-18969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shiga, Y., H. Yamagata, N. Tsukagoshi, and S. Udaka. 1995. BbrPI, an extracellular proteinase inhibitor of Bacillus brevis, protects cells from the attack of exogenous proteinase. Biosci. Biotechnol. Biochem. 59:2348-2350. [DOI] [PubMed] [Google Scholar]
- 33.Silverman, G. A., P. I. Bird, R. W. Carrell, F. C. Church, P. B. Coughlin, P. G. Gettins, J. A. Irving, D. A. Lomas, C. J. Luke, R. W. Moyer, P. A. Pemberton, E. Remold-O'Donnell, G. S. Salvesen, J. Travis, and J. C. Whisstock. 2001. The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Evolution, mechanism of inhibition, novel functions, and a revised nomenclature. J. Biol. Chem. 276:33293-33296. [DOI] [PubMed] [Google Scholar]
- 34.Smyth, G. K. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3:Article3. [DOI] [PubMed]
- 35.Turroni, F., E. Foroni, P. Pizzetti, V. Giubellini, A. Ribbera, P. Merusi, P. Cagnasso, B. Bizzarri, G. L. de'Angelis, F. Shanahan, D. van Sinderen, and M. Ventura. 2009. Exploring the diversity of the bifidobacterial population in the human intestinal tract. Appl. Environ. Microbiol. 75:1534-1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Turroni, F., J. R. Marchesi, E. Foroni, M. Gueimonde, F. Shanahan, A. Margolles, D. van Sinderen, and M. Ventura. 2009. Microbiomic analysis of the bifidobacterial population in the human distal gut. ISME J. 3:745-751. [DOI] [PubMed] [Google Scholar]
- 37.Ventura, M., C. Canchaya, A. Del Casale, F. Dellaglio, E. Neviani, G. F. Fitzgerald, and D. van Sinderen. 2006. Analysis of bifidobacterial evolution using a multilocus approach. Int. J. Syst. Evol. Microbiol. 56:2783-2792. [DOI] [PubMed] [Google Scholar]
- 38.Ventura, M., C. Canchaya, G. F. Fitzgerald, R. S. Gupta, and D. van Sinderen. 2007. Genomics as a means to understand bacterial phylogeny and ecological adaptation: the case of bifidobacteria. Antonie Van Leeuwenhoek 91:351-372. [DOI] [PubMed] [Google Scholar]
- 39.Ventura, M., C. Canchaya, A. Tauch, G. Chandra, G. F. Fitzgerald, K. F. Chater, and D. van Sinderen. 2007. Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiol. Mol. Biol. Rev. 71:495-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ventura, M., J. G. Kenny, Z. Zhang, G. F. Fitzgerald, and D. van Sinderen. 2005. The clpB gene of Bifidobacterium breve UCC 2003: transcriptional analysis and first insights into stress induction. Microbiology 151:2861-2872. [DOI] [PubMed] [Google Scholar]
- 41.Ventura, M., S. O'Flaherty, M. J. Claesson, F. Turroni, T. R. Klaenhammer, D. van Sinderen, and P. O'Toole. 2009. Genome-scale analyses of health-promoting bacteria: probiogenomics. Nat. Rev. Microbiol. 7:61-71. [DOI] [PubMed] [Google Scholar]
- 42.Ventura, M., R. Reniero, and R. Zink. 2001. Specific identification and targeted characterization of Bifidobacterium lactis from different environmental isolates by a combined multiplex-PCR approach. Appl. Environ. Microbiol. 67:2760-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ventura, M., F. Turroni, A. Zomer, E. Foroni, V. Giubellini, F. Bottacini, C. Canchaya, M. J. Claesson, F. He, M. Mantzourani, L. Mulas, A. Ferrarini, B. Gao, M. Delledonne, B. Henrissat, P. Coutinho, M. Oggioni, R. S. Gupta, Z. Zhang, D. Beighton, G. F. Fitzgerald, P. W. O'Toole, and D. van Sinderen. 2009. The Bifidobacterium dentium Bd1 genome sequence reflects its genetic adaptation to the human oral cavity. PLoS Genet. 5:e1000785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ventura, M., D. van Sinderen, G. F. Fitzgerald, and R. Zink. 2004. Insights into the taxonomy, genetics and physiology of bifidobacteria. Antonie Van Leeuwenhoek 86:205-223. [DOI] [PubMed] [Google Scholar]
- 45.Ventura, M., and R. Zink. 2003. Comparative sequence analysis of the tuf and recA genes and restriction fragment length polymorphism of the internal transcribed spacer region sequences supply additional tools for discriminating Bifidobacterium lactis from Bifidobacterium animalis. Appl. Environ. Microbiol. 69:7517-7522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wagner, R. 2000. Transcription regulation in prokaryotes. Oxford University Press, New York, NY.
- 47.Zhang, Q., A. M. Buckle, R. H. Law, M. C. Pearce, L. D. Cabrita, G. J. Lloyd, J. A. Irving, A. I. Smith, K. Ruzyla, J. Rossjohn, S. P. Bottomley, and J. C. Whisstock. 2007. The N terminus of the serpin, tengpin, functions to trap the metastable native state. EMBO Rep. 8:658-663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zoetendal, E. G., C. C. Booijink, E. S. Klaassens, H. G. Heilig, M. Kleerebezem, H. Smidt, and W. M. de Vos. 2006. Isolation of RNA from bacterial samples of the human gastrointestinal tract. Nat. Protoc. 1:954-959. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.