Abstract
Vital stains were used in combination with fluorimetry for the elaboration of a new method to quantify Streptomyces programmed cell death, one of the key events in Streptomyces differentiation. The experimental approach described opens the possibility of designing online protocols for automatic monitoring of industrial fermentations.
Streptomyces is an extremely important bacterium for industry, since approximately two-thirds of all antibiotics are synthesized by members of this genus (4). Furthermore, streptomycetes produce large numbers of eukaryotic cell differentiation inducers and apoptosis inhibitors and inducers (19, 24, 25). Moreover, some authors consider that bacteria with complex life cycles (streptomycetes, cyanobacteria, etc.) are the evolutionary origin of some of the protein domains involved in programmed cell death (PCD) processes, including eukaryotic apoptosis: AP-ATPases (apoptotic ATPases), kinases, caspases, nucleases, etc. As such, these bacteria would constitute a simple and convenient model by which to study this important phenomenon (1, 3, 9, 12, 21, 26).
The classical Streptomyces developmental model in confluent solid cultures assumed that differentiation processes took place along the transverse axis of the cultures (bottom up): completely viable vegetative mycelia (substrate) grew on the surface and inside agar until they underwent a PCD process, after which they differentiated into a reproductive (aerial) mycelium that grew into the air (reviewed in reference 8). Although most industrial processes for secondary metabolite production are performed in liquid cultures, Streptomyces strains generally do not sporulate under these conditions (6, 18, 22), and most authors assumed that differentiation did not take place. Recently, a detailed analysis of Streptomyces differentiation in surface and submerged cultures has been performed, describing novel aspects of the differentiation processes of this bacterium (10-17). A previously unidentified compartmentalized mycelium (MI) initiates the developmental cycle and then dies following a highly ordered sequence (PCD) (10, 11, 14). Subsequently, the remaining viable segments enlarge, yielding a multinucleated mycelium (MII) that grows in successive waves that determine the characteristic complex growth curves of this microorganism. In surface cultures, two types of second mycelium were defined, based on the absence (in early development) or presence (in late development) of the hydrophobic layers characteristic of aerial hyphae (5). The traditionally denominated substrate (vegetative) mycelium corresponds, in fact, to the early second multinucleated mycelium that still lacks the hydrophobic layers coating the aerial mycelium (15). We proposed that the first compartmentalized mycelium fulfills the true vegetative role in Streptomyces development in soil (17). According to this scheme, the second early and late multinucleated mycelia should be considered jointly as part of the reproductive phase, since they are destined to sporulate (17). The second multinucleated mycelium corresponds to the antibiotic-producing structure under surface and submerged conditions (16). The knowledge of the existence of a multinucleated mycelium (MII) which differentiates from a compartmentalized mycelium (MI) after PCD opens a whole new scenario in which to study differentiation and is crucial for the analysis of differentiation in industrial fermentations (10-17).
The aim of this work was to establish a simple and reliable method to monitor and quantify cell death processes in Streptomyces fermentations. We used the vital stains SYTO 9 and propidium iodide (PI) (LIVE/DEAD BacLight bacterial viability kit; Invitrogen L-13152) previously adapted for confocal microscopic analysis of Streptomyces differentiation as described by Manteca et al. (11). SYTO 9 is a cell-permeating nucleic acid stain which labels all of the cells, i.e., both those with intact membranes and those with damaged membranes; PI penetrates only bacteria with altered membrane permeability. Thus, in the presence of both stains, bacteria with intact membranes appear fluorescent green whereas bacteria with compromised membranes appear red, given that PI causes a reduction in SYTO 9 stain fluorescence when both dyes are present (7). In this work, we go one step further in the application of these methodologies to industrial fermentations by means of the elaboration of a protocol for the quantification of Streptomyces PCD processes. To do so, we combined these stains with fluorimetric measurements (see Fig. 1 and 2). Submerged cultures of Streptomyces coelicolor M145 were performed under the conditions described by Manteca et al. in 2008 (16) (100-ml flasks with 20 ml of R5A and 107 spores per ml). The excitation and emission wavelengths were estimated using commercial calf thymus DNA (Sigma D4522) and S. coelicolor chromosomal DNA (5.5 mg/ml) stained with SYTO 9 and PI. The optimal excitation wavelengths were 480 nm for PI and 545 nm for SYTO 9 and the optimal emission wavelengths were 500 and 610 nm, respectively, coinciding with data reported in the literature (see Fig. S1a in the supplemental material). One milliliter of Streptomyces cultures was lysed by boiling in 0.5 M NaOH, and protein concentration was measured with Bradford reagent (2). Cell concentrations (expressed as mg protein/ml) for which the fluorimetric measurements were proportional to the fluorescence emissions were determined (see Fig. S1b in the supplemental material). Fluorimetric measurements in small volumes (50 μl; microtiter plates with a thin light beam) (Cary Eclipse Fluorescence spectrophotometer) were highly variable, owing to the heterogeneity of the cultures formed by relatively large pellets (around 500 μm in diameter; not shown) (16). This problem was overcome by increasing the measurement volumes (2 ml), making it possible for the light beam to include several pellets in the same measurement (Perkin-Elmer LS 50B). Two independent cultures were processed (biological replicates), and three measurements of each (methodological replicates) were performed.
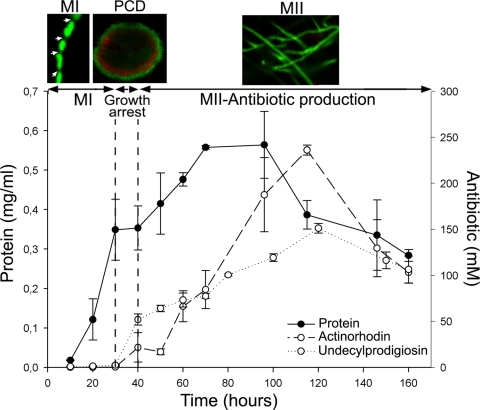
FIG. 1.
S. coelicolor growth curve and antibiotic (actinorhodin and undecylprodigiosin) production in submerged cultures. Confocal microscope images of key developmental stages stained with SYTO 9 and PI are shown at the top: individual hyphae of the first compartmentalized mycelium (MI; arrows indicate septation), second multinucleated mycelium hyphae (MII), and the mycelial pellet (240 μm in diameter) undergoing PCD processes in the center (red). The transitory growth arrest phase coinciding with PCD is indicated. See text for details.
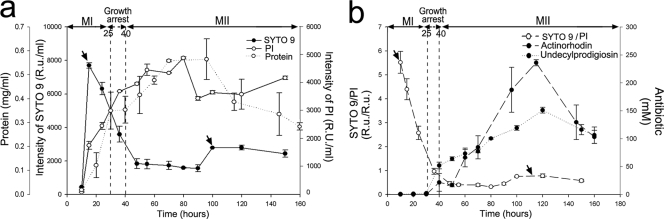
FIG. 2.
Fluorimetric measurements. (a) Intensities of SYTO 9 (live cells) and PI (dead cells) emission along the S. coelicolor developmental cycle. The cellular concentration (mg of protein per ml) is also shown. (b) Variation of the SYTO 9/PI ratio along the developmental cycle. Antibiotic (actinorhodin and undecylprodigiosin) production is indicated. MI, first compartmentalized mycelium. MII, second multinucleated mycelium. Arrows indicate maximum SYTO 9 intensities (live cells) corresponding to MI (15 h) and MII (100 h) mycelia. Data are presented as averages and standard deviations from two biological replicates measured three times each (three methodological replicates). R.U., relative units. See also Fig. S2 in the supplemental material.
A growth curve of S. coelicolor cultivated under the conditions described above is shown in Fig. 1. The growth arrest phase and the two waves of cell growth (MI and MII) are readily visible. The SYTO 9 and PI emissions correlate well with this growth curve and the differentiation processes (Fig. 2a): at early time points, there is an initial exponential growth phase of the MI reflected in a rapid increase in SYTO 9 fluorescence; PI intensity increases slowly as a result of the hyphae that begin to die in the center of the mycelial pellets at early time points (16). Subsequently, SYTO 9 fluorescence decreases quickly in the phases preceding transitory growth arrest, indicating that the fluorescence derived from the MI growing cells cannot offset the loss of the MI cells which are dying in the center of the mycelial pellets. Finally, MI differentiates to MII and undergoes a new exponential growth phase, which is also visible as a stabilization and posterior increase in SYTO 9 fluorescence. Despite the strong correlation between Streptomyces development and SYTO 9 and PI emissions, these values were not terribly informative; for instance, a SYTO 9 intensity of 4,000 relative units could be observed during the MI or MII stage. A value of 3,500 PI relative units (Fig. 2a) was likewise visible during both stages. However, when the data were normalized as a SYTO 9/PI ratio (Fig. 2b), we were able to obtain a reliable marker of differentiation: antibiotic production occurred when these ratios reached values between 0.5 and 1. This overlaps with the transitory growth arrest and MII phases (Fig. 2b). The reproducibility of these measurements was excellent, with average coefficients of variation between biological replicates of about 0.09 and 50% for methodological replicates (see Fig. S2 in the supplemental material), demonstrating that the SYTO 9/PI index is a powerful indicator of Streptomyces differentiation. In the case reported here, antibiotic production began only at values of less than 1 (Fig. 2b).
Streptomyces differentiation in submerged cultures has been largely ignored (16, 20, 22, 23), since the optimization of industrial fermentations is an essentially empirical process. A keen understanding of antibiotic production and differentiation that relies on an autophagic process (PCD) is crucial to manipulating fermentation parameters and inducing the formation of the antibiotic-producing mycelium (MII). The methodology developed here constitutes a straightforward experimental means by which to do so, opening the possibility of automatic online protocols for their implementation on an industrial scale.
Supplementary Material
Acknowledgments
This research was funded by a grant from the DGI, Subdireccion General de Proyectos de Investigacion, MEC, Spain (BIO2007-66313). P. Yagüe was supported by a predoctoral grant from the Fundacion para el Fomento en Asturias de la Investigacion Científica Aplicada y la Tecnologia (FICYT), and A. Manteca was supported by a postdoctoral grant from the Ministerio de Ciencia e Innovacion, Spain.
We thank Priscilla A. Chase for proofreading the text.
Footnotes
Published ahead of print on 26 March 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Aravind, L., V. M. Dixit, and E. V. Koonin. 1999. The domains of death: evolution of the apoptosis machinery. Trends Biochem. Sci. 24:47-53. [DOI] [PubMed] [Google Scholar]
- 2.Bradford, M. M. 1976. A rapid and sensitive for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 3.Cal, S., J. F. Aparicio, C. G. de los Reyes-Gavilan, R. G. Nicieza, and J. Sanchez. 1995. A novel exocytoplasmic endonuclease from Streptomyces antibioticus. Biochem. J. 306:93-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Champness, W. C. 2000. Prokaryotic development, p. 11-31. In Y. V. Brun and L. J. Skimkets (ed.), Actinomycete development, antibiotic production, and phylogeny: questions and challenges. American Society for Microbiology, Washington, DC.
- 5.Claessen, D., H. A. Wosten, G. van Keulen, O. G. Faber, A. M. Alves, W. G. Meijer, and L. Dijkhuizen. 2002. Two novel homologous proteins of Streptomyces coelicolor and Streptomyces lividans are involved in the formation of the rodlet layer and mediate attachment to a hydrophobic surface. Mol. Microbiol. 44:1483-1492. [DOI] [PubMed] [Google Scholar]
- 6.Daza, A., J. F. Martin, A. Dominguez, and J. A. Gil. 1989. Sporulation of several species of Streptomyces in submerged cultures after nutritional downshift. J. Gen. Microbiol. 135:2483-2491. [DOI] [PubMed] [Google Scholar]
- 7.Haugland, R. P. 2002. Nucleic acid detection and genomics technology, p. 269-287. In J. Gregory (ed.), Handbook of fluorescent probes and research chemicals, ninth edition. Molecular Probes, Inc., Eugene, OR.
- 8.Hodgson, D. A. 1992. Differentiation in actinomycetes, p. 407-440. In S. Mohan, C. Dow, and J. A. Cole (ed.), Prokaryotic structure and function: a new perspective. Society for General Microbiology symposium. Cambridge University Press, Cambridge, England.
- 9.Koonin, E. V., and L. Aravind. 2002. Origin and evolution of eukaryotic apoptosis: the bacterial connection. Cell Death Differ. 9:394-404. [DOI] [PubMed] [Google Scholar]
- 10.Manteca, A., M. Fernandez, and J. Sanchez. 2005. A death round affecting a young compartmentalized mycelium precedes aerial mycelium dismantling in confluent surface cultures of Streptomyces antibioticus. Microbiology 151:3689-3697. [DOI] [PubMed] [Google Scholar]
- 11.Manteca, A., M. Fernandez, and J. Sanchez. 2005. Mycelium development in Streptomyces antibioticus ATCC11891 occurs in an orderly pattern which determines multiphase growth curves. BMC Microbiol. 5:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Manteca, A., A. I. Pelaez, R. Zardoya, and J. Sanchez. 2006. Actinobacteria cyclophilins: phylogenetic relationships and description of new class- and order-specific paralogues. J. Mol. Evol. 63:719-732. [DOI] [PubMed] [Google Scholar]
- 13.Manteca, A., M. Fernandez, and J. Sanchez. 2006. Cytological and biochemical evidence for an early cell dismantling event in surface cultures of Streptomyces antibioticus. Res. Microbiol. 157:143-152. [DOI] [PubMed] [Google Scholar]
- 14.Manteca, A., U. Mäder, B. A. Connolly, and J. Sanchez. 2006. A proteomic analysis of Streptomyces coelicolor programmed cell death. Proteomics 6:6008-6022. [DOI] [PubMed] [Google Scholar]
- 15.Manteca, A., D. Claessen, C. Lopez-Iglesias, and J. Sanchez. 2007. Aerial hyphae in surface cultures of Streptomyces lividans and Streptomyces coelicolor originate from viable segments surviving an early programmed cell death event. FEMS Microbiol. Lett. 274:118-125. [DOI] [PubMed] [Google Scholar]
- 16.Manteca, A., R. Alvarez, N. Salazar, P. Yagüe, and J. Sanchez. 2008. Mycelium differentiation and antibiotic production in submerged cultures of Streptomyces coelicolor. Appl. Environ. Microbiol. 74:3877-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manteca, A., and J. Sanchez. 2009. Streptomyces development in colonies and soils. Appl. Environ. Microbiol. 75:2920-2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Novella, I. S., C. Barbes, and J. Sanchez. 1992. Sporulation of Streptomyces antibioticus ETHZ 7451 in submerged culture. Can. J. Microbiol. 38:769-773. [DOI] [PubMed] [Google Scholar]
- 19.Omura, S. 1992. The expanded horizon for microbial metabolites—a review. Gene 115:141-149. [DOI] [PubMed] [Google Scholar]
- 20.Pamboukian, C. R. D., L. M. Guimarães, and M. C. R. Facciotti. 2002. Applications of image analysis in the characterization of Streptomyces olindensis in submerged culture. Braz. J. Microbiol. 33:17-21. [Google Scholar]
- 21.Petricková, K., and M. Petricek. 2003. Eukaryotic-type protein kinases in Streptomyces coelicolor: variations on a common theme. Microbiology 149:1609-1621. [DOI] [PubMed] [Google Scholar]
- 22.Rueda, B., E. M. Miguelez, C. Hardisson, and M. B. Manzanal. 2001. Mycelial differentiation and spore formation by Streptomyces brasiliensis in submerged culture. Can. J. Microbiol. 47:1042-1047. [PubMed] [Google Scholar]
- 23.Stocks, S. M., and C. R. Thomas. 2001. Viability, strength, and fragmentation of Saccharopolyspora erythraea in submerged fermentation. Biotechnol. Bioeng. 75:702-709. [DOI] [PubMed] [Google Scholar]
- 24.Tamaoki, T., and H. Nakano. 1990. Potent and specific inhibitors of protein kinase C of microbial origin. Biotechnology (N.Y.) 8:732-735. [DOI] [PubMed] [Google Scholar]
- 25.Umezawa, K. 1997. Induction of cellular differentiation and apoptosis by signal transduction inhibitors. Adv. Enzyme Regul. 37:393-401. [DOI] [PubMed] [Google Scholar]
- 26.Zhang, C. C. 1996. Bacterial signalling involving eukaryotic-type protein kinases. Mol. Microbiol. 20:9-15. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.