Abstract
ADP-glucose pyrophosphorylase (AGPase) and glycogen synthase (GS) catalyze the first two reactions of glycogen synthesis in cyanobacteria. Mutants defective in each of these enzymes in Synechococcus elongatus PCC 7942 were constructed and characterized. Activities of the corresponding enzymes in the selected mutants were virtually undetectable, and their ability to synthesize glycogen was entirely abolished. The maximal activities of photosynthetic O2 evolution and the rates of respiration in the dark were significantly decreased in the mutants compared to those in wild-type cells. Addition of 0.2 M NaCl or 3 mM H2O2 to liquid cultures markedly inhibited the growth of the AGPase and GS mutants, while the same treatment had only marginal effects on the wild type. These results suggest a significant role for storage polysaccharides in tolerance to salt or oxidative stress.
Cyanobacteria are oxygenic photosynthetic prokaryotes and important biomass producers that are widespread in diverse environments, including freshwater, oceanic, and terrestrial habitats (39). Photosynthetic carbon assimilation in cyanobacteria results in the accumulation of polysaccharides, mostly glycogen (22), which is synthesized by the sequential actions of ADP-glucose pyrophosphorylase (AGPase, EC 2.7.7.27), glycogen synthase (GS, EC 2.4.1.21), and branching enzyme (BE, EC 2.4.1.18) (28). Although the accumulation of storage polysaccharides as intracellular inclusions has been extensively described (32, 33), its physiological significance has not been thoroughly investigated.
It is likely that glycogen in cyanobacteria has a physiological function for adaptation to an unfavorable environment (28). To study the role of storage polysaccharides, we constructed mutants of Synechococcus elongatus PCC 7942 defective in glycogen production by disrupting the structural genes coding for AGPase and GS. S. elongatus PCC 7942 is a unicellular, obligately photoautotrophic cyanobacterium, and its genomic sequence is available (accession number NC_007604) (10). The number of genes coding for enzymes in the glycogen biosynthesis pathway is variable among cyanobacterial species, but S. elongatus PCC 7942 is one of the simplest examples in that it has just one of each gene, as opposed to Synechocystis sp. PCC 6803 (accession number NC_000911) or Nostoc (Anabaena) sp. PCC 7120 (accession number NC_003272). (Both organisms have one gene for AGPase but two genes for GS.) It was therefore expected that mutants with the definite phenotype could be obtained through a single mutagenesis manipulation, avoiding possible complementary functions of paralogous genes.
The cyanobacterial glycogen biosynthesis mutants could also provide opportunities to study the mechanism of starch synthesis in plants. Based on a number of experimental observations, it is now thought that a significant proportion of the glycogen biosynthesis system in cyanobacteria is responsible for the evolution of starch biosynthesis in plants (3, 22, 26). Notable similarities have been found in the enzymatic system and its regulation between glycogen synthesis in cyanobacteria and starch synthesis in plants (2). The cyanobacterial mutants will therefore serve as hosts for the expression of heterologous AGPase and starch synthase (SS) derived from plants to examine their in vivo specificity in the absence of otherwise coexisting isozymes.
As an initial characterization, the effect of mutation on the activities of the other enzymes of the glycogen synthesis pathway was examined. Photosynthetic activities of the mutants were determined at various light intensities. Growth rates were also compared between the wild-type (WT) and mutant strains under salt and oxidative stresses due to the presence of 0.2 M NaCl and 3 mM H2O2, respectively. The role of storage polysaccharides in cyanobacteria under environmental stresses is discussed.
MATERIALS AND METHODS
Culture conditions.
Cells of S. elongatus PCC 7942 were grown in modified BG-11 medium at 30°C under continuous illumination at 50 μmol m−2 s−1, as described previously (36). Salt stress was applied by adding 0.88 g of solid NaCl to 75 ml of the liquid culture (final concentration of 0.2 M) (17, 30), followed by prompt agitation until thorough dissolution. For oxidative stress, 25 μl of 30% H2O2 was added to 75 ml of the liquid culture (final concentration of 3 mM) (27).
Gene disruption.
Total DNA was extracted from the cyanobacterial cells according to the method described by Golden et al. (6). The genomic regions flanking the AGPase gene (glgC) were amplified by PCR using two pairs of oligonucleotide primers, 5′-TGAGCGAGAAGCCTAAGCAGTGA-3′ (839B) and 5′-CTCGAGCAAGTCAAGCGGCGCTGAGA-3′ (840B) for the upstream region and 5′-CTCGAGTGGTCAAAGGGGCGGTTATT-3′ (841B) and 5′-ATCTTCATTATTGATGGTAGTTG-3′ (842B) for the downstream region (the XhoI sequence is underlined). Two DNA fragments (888 and 894 bp for the upstream and downstream regions, respectively) were independently cloned and then joined on the plasmid pGEM-T Easy (Promega). The recombinant plasmid was linearized by digestion with the unique XhoI site and ligated with the blasticidin S resistance gene (bsd) derived from pEM7/Bsd (Invitrogen). The resulting plasmid, pΔglgC::bsd, contained the genomic region in which nearly the entire coding sequence of the AGPase gene was replaced with bsd.
The GS gene (glgA) was amplified with PCR primers 5′-CATATGCGGATTCTGTTCGT-3′ (761B) and 5′-ACCAAACGGCCACCTGACTC-3′ (762B). The DNA fragment obtained (1,480 bp) was cloned into pGEM-T Easy. The recombinant plasmid was then linearized by digestion with EcoRV within the coding region and ligated with the chloramphenicol resistance gene (cat) derived from pHSG396 (Takara Bio Inc.). The resulting plasmid was designated pglgA::cat.
The DNA sequences of the plasmids were confirmed using an ABI PRISM 3100 genetic analyzer (Applied Biosystems).
Transformation of S. elongatus PCC 7942 was carried out using plasmids pΔglgC::bsd and pglgA::cat by the standard procedure (6). The transformants were selected on 1% agar plates of BG-11 medium containing 10 μg/ml blasticidin S (Invitrogen) or 10 μg/ml chloramphenicol (Sigma).
Insertion of the antibiotic resistance cassette at the targeted site was confirmed by PCR using total DNA extracted from the transformants as the template. Replacement of glgC with bsd was verified with PCR primers 5′-TCTCAGCGCCGCTTGACTTG-3′ (849B) and 5′-AATAACCGCCCCTTTGACCA-3′ (850B). Insertion of cat into glgA was confirmed with PCR primers 761B (see above) and 5′-AGCCCCATCTCCTTTTGGAG-3′ (764B).
Enzyme assay.
Enzyme activities were determined with crude extracts that were prepared by disrupting the cells with a French pressure cell as described previously (36).
Determination of AGPase activity was carried out according to Nakamura et al. (23). The reaction mixture consisted of 50 mM HEPES-NaOH (pH 7.5), 2 mM ADP-glucose, 2.4 mM Na pyrophosphate, 1 mM 3-phosphoglycerate, 5 mM MgCl2, 4 mM dithiothreitol (DTT), and enzyme extract (containing 50 μg protein) in a total volume of 400 μl. The reaction, initiated by the addition of the extract, was carried out at 30°C for 20 min and then stopped by heating at 100°C in a water bath for 2 min. After centrifugation, 300 μl of the supernatant was removed and mixed with an equal volume of 0.33 mM NADP+. Enzymatic activity was measured as an increase in A340 after the addition of 0.2 U of phosphoglucomutase (Roche) and 1 U of glucose 6-phosphate dehydrogenase (Roche). The extinction coefficient of NADPH at 340 nm (6.22 μmol−1 cm2) was used for the calculation.
GS activity was determined by the modified method of Nishi et al. (24). The assay was carried out at 30°C in a reaction medium that consisted of 50 mM Tris HCl (pH 8.0), 20 mM DTT, 2 mM ADP-glucose, 2 mg/ml oyster glycogen (type II; Sigma), and the crude enzyme extract in a reaction volume of 300 μl. The reaction was started by addition of the extract, and the mixture was incubated for 20 min. The reaction was stopped by heating at 100°C in a water bath for 2 min. The solution was mixed with 100 μl of a solution containing 50 mM HEPES-NaOH (pH 7.5), 10 mM phosphocreatine, 200 mM KCl, 10 mM MgCl2, and 0.5 mg/ml creatine phosphokinase (type I; Sigma) and incubated for 30 min at 30°C to convert ADP (the product of the GS reaction) to ATP. The creatine phosphokinase reaction was stopped by heating the mixture at 100°C in a water bath for 2 min, and then the reaction mixture was centrifuged at 10,000 × g for 10 min. An aliquot of the supernatant (300 μl) was mixed with 200 μl of a solution containing 125 mM HEPES-NaOH (pH 7.4), 10 mM glucose, 20 mM MgCl2, and 1 mM NADP+. The amount of ATP was measured as an increase in A340 after the addition of 1 U each of hexokinase (Roche) and glucose 6-phosphate dehydrogenase (Roche).
BE activities were detected by staining 5% (wt/vol) nondenaturing polyacrylamide gels (40) after electrophoresis (at 4°C) of the crude extracts. As a reference, crude extract of rice grains (cv. Nipponbare) in the late milking stage was run in the same gel. The gels were subsequently incubated in a reaction mixture (14 ml for a gel) consisting of 50 mM HEPES-NaOH (pH 7.0), 10% glycerol, 50 mM glucose-1-phosphate, 2.5 mM AMP, and 2.5 U/ml phosphorylase a (from rabbit muscle; Sigma) at 30°C for 15 h, rinsed with distilled water, and stained with 0.1% (wt/vol) I2/1% (wt/vol) KI solution. Band intensity was quantified using ImageJ 1.41 software (1) (http://rsb.info.nih.gov/ij/).
Protein concentration was determined using a Bio-Rad Protein Assay reagent with bovine serum albumin as the standard.
Pigment determination and spectrophotometric analyses.
Chlorophyll a (Chl a) contents were measured in a methanol solution according to Mackinney (18). Phycocyanin contents were determined according to Su et al. (34). Cells were suspended in a buffer containing 50 mM imidazole-HCl (pH 7.4), 8 mM MgCl2, 12.5% (wt/vol) glycerol, and 2% (wt/vol) Triton X-100 and disrupted by passage through a French pressure cell at 138 MPa. Unbroken cells were removed by centrifugation, and absorption spectra were determined for the supernatant preparations. The amount of phycocyanin was calculated according to the equation described by Tandeau de Marsac and Houmard (37). The percentage of cells disrupted in a given preparation was determined from the ratio of the A430 of the lysate to that of the methanol extract of the same quantity of cells, and the amount of phycocyanin was estimated after correction for the unbroken cells. Absorption spectra were determined with a Beckman DU 7400 spectrophotometer.
Composition of soluble proteins and immunological detection of D1 protein.
For visualization of the soluble proteins, crude extracts were electrophoresed on 15% sodium dodecyl sulfate (SDS)-polyacrylamide gels, followed by staining with Coomassie brilliant blue. For immunological detection of the photosystem II D1 protein, the crude extracts were resolved by 10% SDS-polyacrylamide gel electrophoresis (PAGE) and electrotransferred onto a polyvinylidenedifluoride membrane (Immobilon-P Transfer Membrane; Millipore). The D1 protein was detected using a rabbit antiserum raised against the D1 protein of spinach (12) and by an amplified alkaline phosphatase immunoblotting kit (Bio-Rad). The rabbit antiserum was kindly provided by M. Ikeuchi, University of Tokyo.
Growth rates and activities of photosynthesis and respiration in the dark.
Growth rates of S. elongatus PCC 7942 cells in liquid cultures were determined by measuring the A730 of the cell suspension. Activities of photosynthetic oxygen evolution in the cyanobacterium were determined by a Clark-type oxygen electrode (Rank Brothers Ltd., Bottisham, UK). The cells were suspended in 50 mM Tricine-KOH (pH 7.5) at a concentration of 5 μg Chl ml−1 in the presence of 10 mM NaHCO3 and kept at 30°C. Light was provided by a halogen lamp (Iwasaki Electric Co., Ltd., Tokyo, Japan) at various intensities. A measurement at a constant light intensity was carried out for at least 5 min to ensure the linearity of the slope. Respiration in the dark was measured under the same conditions, except that the cell suspension was placed in complete darkness.
Hydrogen evolution.
The activities of light-dependent hydrogen evolution were measured by the method of Gutthann et al. (7) using a Clark-type oxygen electrode which was inversely connected so that the platinum electrode was polarized at +600 mV relative to the Ag/AgCl electrode. A cell suspension containing 100 μg ml−1 Chl was incubated with 40 U of glucose oxidase-50 U of catalase-1 mM glucose for 15 min at 30°C in darkness. After the completion of fermentative hydrogen production, the suspension was illuminated at 800 μmol m−2 s−1 to induce photohydrogen production. For comparison, Synechocystis sp. PCC 6803 was grown and subjected to the assay under the same conditions.
Carbohydrate extraction and analysis.
S. elongatus PCC 7942 cells were grown in liquid culture to an A730 of approximately 2.0, and aliquots (1.5 ml) were removed at intervals for the determination of glycogen and sucrose accumulation in the cells. Cells were collected by centrifugation at 10,000 × g for 5 min, resuspended with 1.5 ml of absolute methanol, and kept at −20°C for 24 h. After centrifugation, the pellet and 1 ml of the supernatant were brought to dryness in a centrifugal vacuum evaporator for the determination of glycogen and sucrose, respectively.
The dried pellet was resuspended in 1 ml of distilled water and incubated at 100°C for 40 min. A portion (200 μl) of the suspension was mixed with 100 μl of 2.5 mM Na-acetate (pH 5) containing 0.5 mg/ml glucoamylase (from Rhizopus niveus; Seikagaku Kogyo) and incubated at 40°C for 1 h. After centrifugation at 10,000 × g for 5 min, 150 μl of the supernatant was mixed with 375 μl of distilled water and 105 μl of a reaction medium (S1 solution) containing 400 mM HEPES-NaOH (pH 7.5), 10 mM MgSO4, 3 mM NADP+, 10 mM ATP, 3 U of hexokinase (from yeast; Roche), and 2 U of glucose 6-phosphate dehydrogenase (from yeast; Roche). The amount of glucose moieties derived from glycogen was determined as the increase in A340.
The dried supernatant was dissolved in 667 μl of distilled water and heated at 100°C for 40 min. A portion (100 μl) of the suspension was mixed with 25 μl of a solution (S2) containing 50 mM Na-acetate (pH 5) and 4 mg/ml β-fructosidase (from yeast; Roche) and incubated at 30°C for 30 min. The solution was then mixed with 175 μl of distilled water and 200 μl of S1 solution. The sucrose content was determined from the amount of glucose produced as described above.
RESULTS AND DISCUSSION
Construction of mutants.
Genes coding for AGPase (glgC, Synpcc7942_0603) and GS (glgA, Synpcc7942_2518) in S. elongatus PCC 7942 were cloned by PCR based on the genomic sequences of this organism (accession number NC_007604) (10) and S. elongatus PCC 6301 (NC_006576) (35), which were nearly identical to each other. Unlike Escherichia coli, in which the AGPase and GS genes are in the same operon (25), these genes were distantly located on the chromosome of S. elongatus PCC 7942 and were therefore inactivated independently. We used different antibiotic resistance markers (bsd and cat) for disruption of the two genes, leaving the opportunity to construct double mutants in future studies.
We also adopted different procedures for the disruption of the two genes; nearly the entire coding region of AGPase gene was removed and replaced with bsd, while cat was simply inserted at an EcoRV restriction site in the GS gene. We chose a much more tedious procedure for mutation of the AGPase gene, intending to make a host for heterologous expression of AGPase from plants (in future work). As the conservation of the sequence of the AGPase gene between different organisms is much more substantial than that of the GS/SS gene, deletion of the coding sequence would prevent any possible recombination event (through sequence similarity) between intrinsic and exogenous genes from occurring.
On the chromosome of S. elongatus PCC 7942, both the AGPase and GS genes are immediately followed by another open reading frame of unknown function, apparently constituting polycistronic units. To preserve the activity of the downstream genes, we used antibiotic markers without a transcriptional terminator or Ω element.
Antibiotic-resistant colonies were grown, and total DNA was extracted and examined by PCR for gene replacement. When the PCR primers described in Materials and Methods were used, a DNA fragment of 1.3 kb was amplified from the AGPase gene of the WT strain. In contrast, the DNA fragment found in the WT was replaced with a 0.6-kb DNA fragment from a blasticidin S-resistant transformant (AGP5). A portion of the GS gene was amplified as a 0.8-kb DNA fragment from the WT strain, while it was replaced with a 1.8-kb fragment in two independent chloramphenicol-resistant transformants (GS1 and GS2). We concluded that the WT alleles of these genes were completely removed from the transformants. After the establishment of the gene disruption, the mutant strains were maintained without the selective antibiotics.
Enzymatic activities for glycogen synthesis.
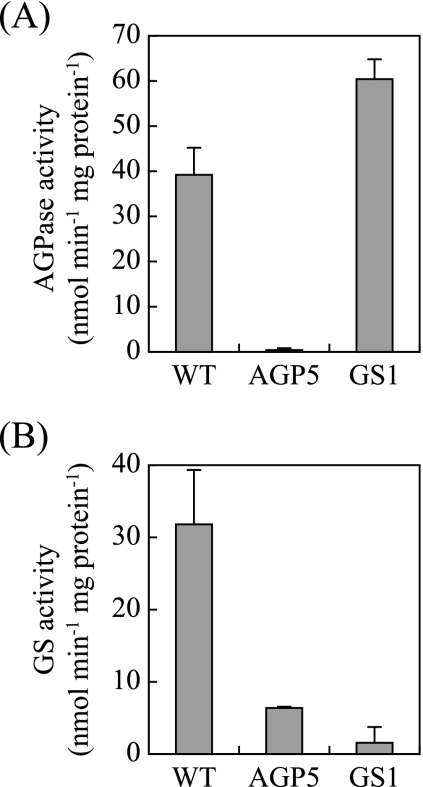
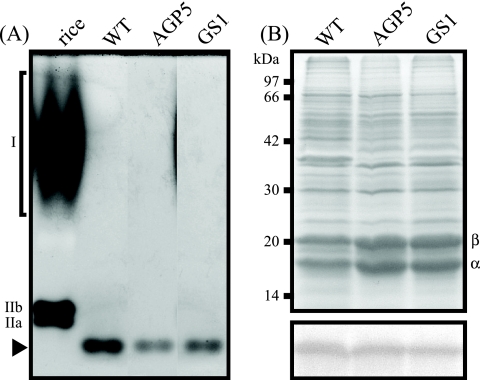
The activities of enzymes involved in glycogen biosynthesis were measured in the crude extracts of WT and mutant S. elongatus strain PCC 7942. Figure 1A shows that the AGPase activity in the crude extract of the WT was approximately 40 (nmol min−1 mg protein−1), while it was undetectable in the AGPase mutant. In the GS mutant, the AGPase activity was 1.5 times as high as that in the WT. Figure 1B shows that the GS activity in the WT was 32 (nmol min−1 mg protein−1), while it was within the background level in GS mutant. In the AGPase mutant, the GS activity was decreased to 1/5 of that in the WT. The BE activities in the crude extracts of the WT and the mutants were visualized by nondenaturing PAGE and activity staining (Fig. 2A). The activity in the WT was detected on the gel as a single staining band whose mobility was greater than those of three BE isozymes in rice endosperm. The intensities of the staining bands of the mutants were decreased to 1/2 of that in the WT when an equal amount of total protein was loaded onto the lane (Fig. 2B). These results indicated that mutation of the AGPase or GS gene resulted in total loss of the corresponding enzyme activity. The altered activities of other enzymes in the glycogen biosynthesis pathway would be secondary effects caused by the mutation. When the enzymes responsible for later steps in the metabolic pathway were synthesized in the mutants, they would not be supplied with the substrate. Without substrate, these enzymes are nonfunctional and therefore may be subject to turnover at elevated rates. As shown below (see Fig. 4), the ability to accumulate glycogen was entirely abolished in these mutants. This result strongly suggests that AGPase and GS are solely responsible for storage polysaccharide synthesis in S. elongatus PCC 7942.
FIG. 1.
Enzymatic activities in crude extracts of WT and mutant S. elongatus strain PCC 7942. (A) AGPase activity. (B) GS activity. Extracts containing 50 μg of protein were used for the assay. The data shown for each strain are averages of three independent measurements, and standard deviation bars are shown.
FIG. 2.
Enzymatic activity of BE and protein composition in crude extracts of WT and mutant S. elongatus strain PCC 7942. (A) The extracts of the cyanobacterial strains (containing 6.7 μg of protein) were run on a nondenaturing polyacrylamide gel along with an extract of immature rice grains (containing 2.2 μg of protein) as a reference. The gel was then subjected to BE activity staining as described in the text. An arrowhead indicates the mobility of the band corresponding to the BE activity of S. elongatus PCC 7942. The migration of BE isoforms in rice endosperm (BEI, BEIIa, and BEIIb) is also indicated. The activity of the BEI isoform is visible as a characteristic smeary band (40). (B) Composition of soluble proteins (upper panel) and immunological detection of the photosystem II D1 protein (lower panel) in crude extracts of S. elongatus PCC 7942. Crude extract containing 40 μg of protein was loaded onto each lane. The bands indicated by the characters α and β are α- and β-phycocyanin, respectively. The D1 protein was detected using an antiserum raised against the D1 protein of spinach as described in the text.
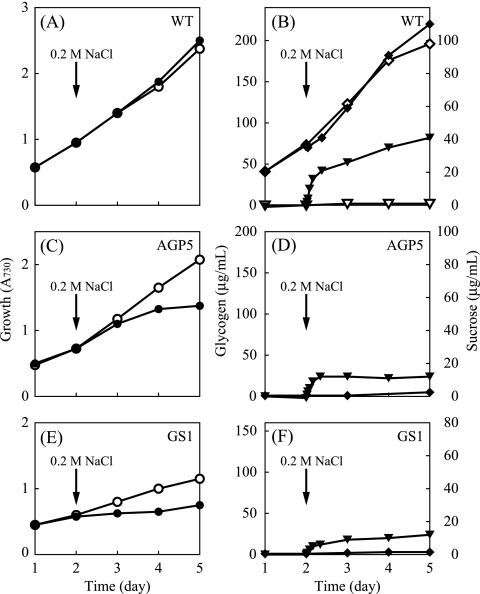
FIG. 4.
Growth and carbohydrate content of S. elongatus PCC 7942 during liquid culture in the presence of NaCl. (A, C, and E) Changes in cell density (○, •) measured by A730. (B, D, and F) Glycogen (⋄, ♦) and sucrose (▿, ▾) contents (left and right y axes, respectively). The name of the strain is indicated in each panel. For the culture indicated by the closed symbols, 0.2 M NaCl was added at the time indicated. For clarity, the carbohydrate content of the mutant strains in the absence of NaCl is not shown; these values remained below detectable levels throughout the experiment.
Composition of pigments and proteins.
Both the AGPase and GS mutants exhibited a pale blue-green color, suggesting an altered composition of their pigments. The cellular contents of phycocyanin and Chl a, the major photosynthetic pigments in this organism, were therefore compared between the WT and mutant strains (Table 1). In both of the mutants, the phycocyanin contents were increased, compared to those in the WT, while the Chl a contents were comparable. Consequently, the phycocyanin/Chl a ratio was consistently higher in the mutants. An increased level of phycocyanin was visualized when the soluble protein was resolved by SDS-PAGE (Fig. 2B). The change in phycocyanin content could disturb the equality of the amount of the other proteins (e.g., BE shown in Fig. 2A) loaded on each lane. However, the substantial reduction in the band intensity of BE could not be explained solely as a consequence of increased phycocyanin content. We therefore concluded that the activity of BE was specifically decreased as an enzyme involved in glycogen metabolism.
TABLE 1.
Phycocyanin and Chl a contents in WT and mutants of S. elongatus PCC 7942
| Strain | Avg amt (μg unit of A730−1) ± SDa |
Phycocyanin/Chl a ratio | |
|---|---|---|---|
| Phycocyanin | Chl a | ||
| WT | 36.5 ± 2.7 | 6.5 ± 0.0 | 5.6 ± 0.4 |
| AGP5 | 44.0 ± 0.4 | 6.6 ± 0.2 | 6.7 ± 0.3 |
| GS1 | 41.7 ± 0.9 | 6.4 ± 0.4 | 6.5 ± 0.5 |
The values shown are from three independent measurements.
Light energy absorbed by phycobilisomes is transferred predominantly to photosystem II. To determine whether the level of photosystem II is altered in the mutants, we carried out immunoblot detection analysis of the D1 protein, a core subunit of photosystem II. As shown in Fig. 2B, the amount of D1 protein was not significantly different between the WT and mutant strains.
Activities of photosynthesis and respiration.
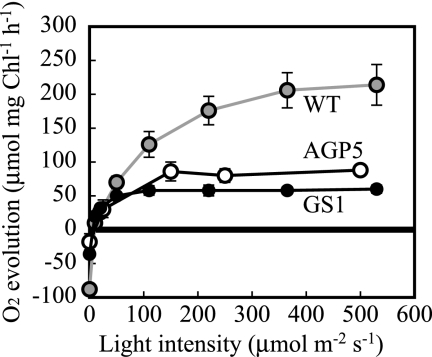
Photosynthetic activities in the WT and mutant strains were measured as bicarbonate-dependent O2 evolution with an oxygen electrode at various light intensities. The activity in the WT showed a hyperbolic curve, increasing gradually at high light intensities of up to 500 μmol m−2 s−1, and it exceeded 200 μmol mg Chl−1 h−1 at a saturating light intensity (Fig. 3). In contrast, the activities in the mutants were saturated at 100 μmol m−2 s−1 and the maximum activities in AGPase and GS mutants were approximately 1/3 and 1/4, respectively, of that observed in the WT.
FIG. 3.
Oxygen evolution by the cells of WT and mutant S. elongatus strain PCC 7942 under illumination at various light intensities. Negative values observed in darkness indicate oxygen consumption. The data shown for each strain are averages of three independent measurements, and standard deviation bars are shown.
Figure 3 also shows that the activities of respiration in the mutants placed in darkness were reduced to 1/2 of that in the WT. This result suggests that glycogen serves as a major form of respiratory substrate in darkness.
The decreased activity of photosynthetic O2 evolution would be related to a limited capacity to consume reducing equivalents due to the defect in glycogen synthesis. We have shown that the level of photosystem II protein was not altered (Fig. 2B) while that of antenna pigments (phycocyanin) for photosystem II was increased (Table 1; Fig. 2B) in the mutants. A possible explanation for these observations is that the efficiency of energy transfer from the antenna pigments to the reaction center is impaired in the mutants.
In the absence of adequate consumption of reducing equivalents through glycogen synthesis, one of the candidates for alternative electron acceptors is H+, leading to H2 production. The capacity for light-dependent, anaerobic H2 evolution was therefore measured according to the method described by Gutthann et al. (7) with the WT and mutant forms of S. elongatus PCC 7942. The activity in Synechocystis PCC 6803 was also determined for comparison (7). While a limited but definite amount of H2 evolution was observed with WT Synechocystis PCC 6803, no appreciable activity was detectable with S. elongatus strain PCC 7942 under the same conditions (data not shown). The relative activity of hydrogenase in S. elongatus PCC 7942 may be much lower, and/or other compound(s) may be primarily responsible for the consumption of reducing equivalents.
Carbohydrate contents and responses to salt and oxidative stresses.
The carbohydrate contents of the WT and mutant strains were compared during their growth in liquid culture under continuous light. Growth rates were determined by measuring the A730 of cell suspensions. A cell suspension with an A730 of 1.0 contained 1.1 × 108 ± 0.1 × 108 cells ml−1 (average ± standard deviation; n = 10), and no appreciable difference was observed between the values of WT and mutant cells. In WT cells, the amount of glycogen increased steadily as the cell density increased (Fig. 4A and B), indicating that a constant amount of polysaccharide was accumulated during growth under continuous illumination. Figure 4C and E show that the growth rates of AGPase and GS mutants were much lower than that of the WT. In these mutants, glycogen was hardly detectable throughout the experiment (Fig. 4D and F).
The effect of salt stress (addition of 0.2 M NaCl) on the WT and mutant strains was examined next. Although S. elongatus is not tolerant to high salt concentrations, it has been reported that sucrose synthesis is induced in moderate salinity (17, 30). It was therefore possible that a significant modulation of carbohydrate metabolism takes place under salt stress. In the presence of 0.2 M NaCl, the A730 of the culture of WT cells increased at a rate comparable to that in the culture without NaCl (Fig. 4A). Addition of 0.2 M NaCl to the culture did not cause a significant change in the glycogen content of WT cells (Fig. 4B). When cells were grown in standard medium without NaCl, sucrose was undetectable. After the addition of NaCl, sucrose rapidly accumulated in the cells in 8 h. Intracellular sucrose attained a steady level (18 μg ml−1 A730−1) and remained constant for at least 2 days. The amount of sucrose was expressed conventionally on a culture volume basis. Assuming that the cell volume to Chl a ratio of this organism is 60 μl mg−1 Chl (29), the intracellular sucrose concentration is estimated to be approximately 0.28 M. If accumulation of sucrose is confined to the cytoplasmic space (excluding the thylakoid lumen), the value would be much higher but still within a physiologically plausible range. Since the fluctuation of the glycogen content upon the addition of NaCl was rather small, a major fraction of the carbon used to synthesize sucrose should be derived from de novo CO2 fixation through the Calvin-Benson cycle. Compared to the WT, the most notable effect of NaCl treatment on the mutants was the substantial reduction of the growth rates (Fig. 4C and E). Addition of NaCl led to the synthesis of sucrose in these cells, as in the WT (Fig. 4D and F). As the ability to synthesize glycogen was lost in these mutants, sucrose should be synthesized solely through de novo assimilation of CO2 instead of conversion from polysaccharide.
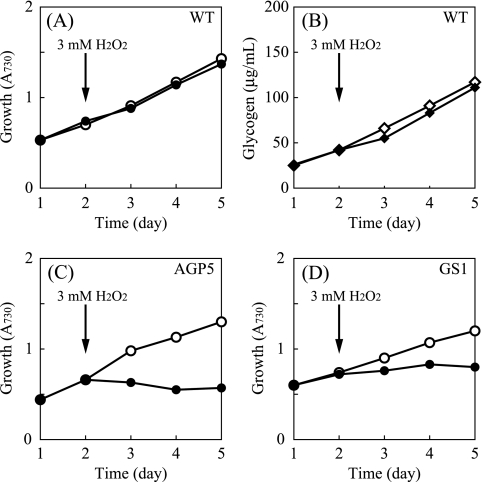
The marked inhibition of the growth of the mutants by NaCl raised the possibility that these mutants are susceptible to environmental stress. To see if the sensitivity (growth inhibition) of the mutants is a general effect under environmental stresses, growth rates were measured in the presence of 3 mM H2O2. Figure 5A shows that the addition of hydrogen peroxide to a liquid culture hardly affected the growth of the WT. The glycogen content did not show a significant change after the addition of H2O2 (Fig. 5B). In contrast to the WT, a severe inhibition of the growth of AGPase and GS mutants during oxidative stress was observed (Fig. 5C and D).
FIG. 5.
Growth and carbohydrate content of S. elongatus PCC 7942 during liquid culture in the presence of H2O2. (A, C, and D) Change in cell density as determined by A730. The name of the strain is indicated in each panel. (B) Glycogen content of WT cells. For the culture indicated by the closed symbols, 3 mM H2O2 was added at the time indicated. The glycogen content of the mutant strains remained below detectable levels throughout the experiment (not shown).
Under salt and osmotic stresses, sucrose is responsible for the protective functions as a compatible solute (9). In addition to the synthesis of the compatible solute(s), other responsive and adaptive processes are also induced under the salt and osmotic stresses. These include extrusion of Na+ ion by Na+/H+ antiporter (4, 13, 31, 38) and synthesis of stress-responsive proteins, including enzymes for the production of compatible solutes, heat shock proteins, and enzymes acting on reactive oxygen species (5, 15, 19). Salt and oxidative stresses are therefore causally related in the cells of cyanobacteria, as supported by much experimental evidence (16). The stress response processes indicated above induce a high demand for ATP synthesis. A considerable proportion of the glycogen that was transiently degraded in the WT upon the addition of NaCl or H2O2 would be responsible for the production of ATP. As supporting evidence, it has been reported that the activities and expression of photosystem I and cytochrome c oxidase were enhanced during salt stress in Synechocystis (14). It is plausible that glycogen serves as a substrate for respiration through cytochrome c oxidase. Because of the deficiency of glycogen synthesis and accumulation, AGPase and GS mutants would be unable to synthesize a sufficient amount of ATP to fulfill their cellular needs. These mutants were thus incapable of adapting to salt and oxidative stresses and consequently showed growth inhibition.
Concluding remarks.
Mutants defective in glycogen/starch biosynthesis due to the lesion in AGPase or GS (SS) have been characterized in a number of photosynthetic organisms. An AGPase mutant of the cyanobacterium Synechocystis PCC 6803 has been isolated (20, 21). In contrast to AGPases in plants and eukaryotic algae, the enzyme in cyanobacteria is a homotetramer (11) and is encoded by a single gene. In Synechocystis PCC 6803, however, ADP-glucose serves as the precursor for both glycogen and the primary compatible solute glucosylglycerol (8). The ability to synthesize both of these compounds was simultaneously abolished by the lesion in the AGPase gene in Synechocystis PCC 6803 (20). In the AGPase mutant of Synechocystis exposed to salt stress, sucrose was accumulated in the place of glucosylglycerol. The carbohydrate metabolism of S. elongatus PCC 7942 is thus much simpler than that of Synechocystis PCC 6803 and would be suitable for studying the role of sucrose during salt stress. The homologues of GS in plants and algae are SSs, which also show diversification into multiple isoforms. To date, the complete elimination of starch (polysaccharide) synthesis due to the lesion in SS activities has not been reported for any plant species.
In conclusion, in this study, we have demonstrated the previously unexplored significance of storage polysaccharides in cyanobacteria. Glycogen metabolism is thus physiologically important for these organisms to cope with the ever-changing environment.
Acknowledgments
We thank Chieko Sugita and Mamoru Sugita of Nagoya University for providing the genomic sequence of S. elongatus PCC 6301 prior to publication. We are also grateful to Masahiko Ikeuchi of the University of Tokyo for providing the antiserum raised against the D1 protein. Technical support by the members of the Biotechnology Center, Akita Prefectural University, is greatly appreciated.
Footnotes
Published ahead of print on 2 April 2010.
REFERENCES
- 1.Abramoff, M. D., P. J. Magelhaes, and S. J. Ram. 2004. Image processing with ImageJ. Biophotonics Int. 11:36-42. [Google Scholar]
- 2.Ball, S. G., and K. M. Morell. 2003. From bacterial glycogen to starch: understanding the biogenesis of the plant starch granule. Annu. Rev. Plant Biol. 54:207-233. [DOI] [PubMed] [Google Scholar]
- 3.Deschamps, P., C. Colleoni, Y. Nakamura, E. Suzuki, J. L. Putaux, A. Buléon, S. Haebel, G. Ritte, M. Steup, L. I. Falcón, D. Moreira, W. Loffelhardt, J. N. Raj, C. Plancke, C. d'Hulst, D. Dauvillée, and S. Ball. 2008. Metabolic symbiosis and the birth of the plant kingdom. Mol. Biol. Evol. 25:536-548. [DOI] [PubMed] [Google Scholar]
- 4.Elanskaya, I. V., I. V. Karandashova, A. V. Bogachev, and M. Hagemann. 2002. Functional analysis of the Na+/H+ antiporter encoding genes of the cyanobacterium Synechocystis PCC 6803. Biochemistry (Moscow) 67:432-440. [DOI] [PubMed] [Google Scholar]
- 5.Fulda, S., S. Mikkat, F. Huang, J. Huckauf, K. Marin, B. Norling, and M. Hagemann. 2006. Proteome analysis of salt stress response in the cyanobacterium Synechocystis sp. strain PCC 6803. Proteomics 6:2733-2745. [DOI] [PubMed] [Google Scholar]
- 6.Golden, S. S., J. Brusslan, and R. Haselkorn. 1987. Genetic engineering of the cyanobacterial chromosome. Methods Enzymol. 153:215-231. [DOI] [PubMed] [Google Scholar]
- 7.Gutthann, F., M. Egert, A. Marques, and J. Appel. 2007. Inhibition of respiration and nitrate assimilation enhances photohydrogen evolution under low oxygen concentrations in Synechocystis sp. PCC 6803. Biochim. Biophys. Acta 1767:161-169. [DOI] [PubMed] [Google Scholar]
- 8.Hagemann, M., and N. Erdmann. 1994. Activation and pathway of glucosylglycerol synthesis in the cyanobacterium Synechocystis sp. PCC 6803. Microbiology 140:1427-1431. [Google Scholar]
- 9.Hincha, D. K., and M. Hagemann. 2004. Stabilization of model membranes during drying by compatible solutes involved in the stress tolerance of plants and microorganisms. Biochem. J. 383:277-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holtman, C. K., Y. Chen, P. Sandoval, A. Gonzales, M. S. Nalty, T. L. Thomas, P. Youderian, and S. S. Golden. 2005. High-throughput functional analysis of the Synechococcus elongatus PCC 7942 genome. DNA Res. 12:103-115. [DOI] [PubMed] [Google Scholar]
- 11.Iglesias, A. A., G. Kakefuda, and J. Preiss. 1991. Regulatory and structural properties of the cyanobacterial ADPglucose pyrophosphorylases. Plant Physiol. 97:1187-1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ikeuchi, M., and Y. Inoue. 1987. Specific 125I labeling of D1 (herbicide-binding protein). An indication that D1 functions on both the donor and acceptor sides of photosystem II. FEBS Lett. 210:71-76. [Google Scholar]
- 13.Inaba, M., A. Sakamoto, and N. Murata. 2001. Functional expression in Escherichia coli of low-affinity and high-affinity Na+ (Li+)/H+ antiporters of Synechocystis. J. Bacteriol. 183:1376-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jeanjean, R., H. C. P. Matthijs, B. Onana, M. Havaux, and F. Joset. 1993. Exposure of the cyanobacterium Synechocystis PCC6803 to salt stress induces concerted changes in respiration and photosynthesis. Plant Cell Physiol. 34:1073-1079. [Google Scholar]
- 15.Kanesaki, Y., I. Suzuki, S. I. Allakhverdiev, K. Mikami, and N. Murata. 2002. Salt stress and hyperosmotic stress regulate the expression of different sets of genes in Synechocystis sp. PCC 6803. Biochem. Biophys. Res. Commun. 290:339-348. [DOI] [PubMed] [Google Scholar]
- 16.Latifi, A., M. Ruiz, and C.-C. Zhang. 2009. Oxidative stress in cyanobacteria. FEMS Microbiol. Rev. 33:258-278. [DOI] [PubMed] [Google Scholar]
- 17.Mackay, M. A., R. S. Norton, and L. J. Borowitzka. 1984. Organic osmoregulatory solutes in cyanobacteria. J. Gen. Microbiol. 130:2177-2191. [Google Scholar]
- 18.Mackinney, G. 1941. Absorption of light by chlorophyll solutions. J. Biol. Chem. 140:315-322. [Google Scholar]
- 19.Marin, K., Y. Kanesaki, D. A. Los, N. Murata, I. Suzuki, and M. Hagemann. 2004. Gene expression profiling reflects physiological processes in salt acclimation of Synechocystis sp. strain PCC 6803. Plant Physiol. 136:3290-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miao, X., Q. Wu, G. Wu, and N. Zhao. 2003. Sucrose accumulation in salt-stressed cells of agp gene deletion-mutant in cyanobacterium Synechocystis sp. PCC 6803. FEMS Microbiol. Lett. 218:71-77. [DOI] [PubMed] [Google Scholar]
- 21.Miao, X., Q. Wu, G. Wu, and N. Zhao. 2003. Changes in photosynthesis and pigmentation in an agp deletion mutant of the cyanobacterium Synechocystis sp. Biotech. Lett. 25:391-396. [DOI] [PubMed] [Google Scholar]
- 22.Nakamura, Y., J. Takahashi, A. Sakurai, Y. Inaba, E. Suzuki, S. Nihei, S. Fujiwara, M. Tsuzuki, H. Miyashita, H. Ikemoto, M. Kawachi, H. Sekiguchi, and N. Kurano. 2005. Some cyanobacteria synthesize semi-amylopectin type α-polyglucans instead of glycogen. Plant Cell Physiol. 46:539-545. [DOI] [PubMed] [Google Scholar]
- 23.Nakamura, Y., K. Yuki, S.-Y. Park, and T. Ohya. 1989. Carbohydrate metabolism in the developing endosperm of rice grains. Plant Cell Physiol. 30:833-839. [Google Scholar]
- 24.Nishi, A., Y. Nakamura, N. Tanaka, and H. Satoh. 2001. Biochemical and genetic analysis of the effects of amylose-extender mutation in rice endosperm. Plant Physiol. 127:459-472. [PMC free article] [PubMed] [Google Scholar]
- 25.Okita, T. W., R. L. Rodriguez, and J. Preiss. 1981. Biosynthesis of bacterial glycogen. Cloning of the glycogen biosynthetic enzyme structural genes of Escherichia coli. J. Biol. Chem. 256:6944-6952. [PubMed] [Google Scholar]
- 26.Patron, N. J., and P. J. Keeling. 2005. Common evolutionary origin of starch biosynthetic enzymes in green and red algae. J. Phycol. 41:1131-1141. [Google Scholar]
- 27.Perelman, A., A. Uzan, D. Hacohen, and R. Schwarz. 2003. Oxidative stress in Synechococcus sp. strain PCC 7942: various mechanisms for H2O2 detoxification with different physiological roles. J. Bacteriol. 185:3654-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Preiss, J. 1984. Bacterial glycogen synthesis and its regulation. Annu. Rev. Microbiol. 38:419-458. [DOI] [PubMed] [Google Scholar]
- 29.Price, G. D., and M. R. Badger. 1989. Ethoxyzolamide inhibition of CO2 uptake in the cyanobacterium Synechococcus PCC7942 without apparent inhibition of internal carbonic anhydrase activity. Plant Physiol. 89:37-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reed, R. H., D. L. Richardson, S. R. C. Warr, and W. D. P. Stewart. 1984. Carbohydrate accumulation and osmotic stress in cyanobacteria. J. Gen. Microbiol. 130:1-4. [Google Scholar]
- 31.Serrano, R., and A. Rodriguez-Navarro. 2001. Ion homeostasis during salt stress in plants. Curr. Opin. Cell Biol. 13:399-404. [DOI] [PubMed] [Google Scholar]
- 32.Shively, J. M. 1988. Inclusions: granules of polyglucose, polyphosphate, and poly-β-hydroxybutyrate. Methods Enzymol. 167:195-203. [Google Scholar]
- 33.Stanier, G. 1988. Fine structure of cyanobacteria. Methods Enzymol. 167:157-172. [Google Scholar]
- 34.Su, X., P. G. Fraenkel, and L. Bogorad. 1992. Excitation energy transfer from phycocyanin to chlorophyll in an apcA-defective mutant of Synechocystis sp. PCC 6803. J. Biol. Chem. 267:22944-22950. [PubMed] [Google Scholar]
- 35.Sugita, C., K. Ogata, M. Shikata, H. Jikuya, J. Takano, M. Furumichi, M. Kanehisa, T. Omata, M. Sugiura, and M. Sugita. 2007. Complete nucleotide sequence of the freshwater unicellular cyanobacterium Synechococcus elongatus PCC 6301 chromosome: gene content and organization. Photosynth. Res. 93:55-67. [DOI] [PubMed] [Google Scholar]
- 36.Suzuki, E., K. Umeda, S. Nihei, K. Moriya, H. Ohkawa, S. Fujiwara, M. Tsuzuki, and Y. Nakamura. 2007. Role of the GlgX protein in glycogen metabolism of the cyanobacterium, Synechococcus elongatus PCC 7942. Biochim. Biophys. Acta 1770:763-773. [DOI] [PubMed] [Google Scholar]
- 37.Tandeau de Marsac, N., and J. Houmard. 1988. Complementary chromatic adaptation: physiological conditions and action spectra. Methods Enzymol. 167:318-328. [Google Scholar]
- 38.Waditee, R., T. Hibino, T. Nakamura, A. Incharoensakdi, and T. Takabe. 2002. Overexpression of a Na+/H+ antiporter confers salt tolerance on a freshwater cyanobacterium, making it capable of growth in sea water. Proc. Natl. Acad. Sci. U. S. A. 99:4109-4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Whitton, B. A., and M. Potts. 2000. The ecology of cyanobacteria. Their diversity in time and space, p 669. Kluwer Academic Publishers, Dordrecht, Netherlands.
- 40.Yamanouchi, H., and Y. Nakamura. 1992. Organ specificity of isoforms of starch branching enzyme (Q-enzyme) in rice. Plant Cell Physiol. 33:985-991. [Google Scholar]