Abstract
Microbial processes are crucial for ecosystem maintenance, yet documentation of these processes in complex open field sites is challenging. Here we used a multidisciplinary strategy (site geochemistry, laboratory biodegradation assays, and field extraction of molecular biomarkers) to deduce an ongoing linkage between aromatic hydrocarbon biodegradation and nitrogen cycling in a contaminated subsurface site. Three site wells were monitored over a 10-month period, which revealed fluctuating concentrations of nitrate, ammonia, sulfate, sulfide, methane, and other constituents. Biodegradation assays performed under multiple redox conditions indicated that naphthalene metabolism was favored under aerobic conditions. To explore in situ field processes, we measured metabolites of anaerobic naphthalene metabolism and expressed mRNA transcripts selected to document aerobic and anaerobic microbial transformations of ammonia, nitrate, and methylated aromatic contaminants. Gas chromatography-mass spectrometry detection of two carboxylated naphthalene metabolites and transcribed benzylsuccinate synthase, cytochrome c nitrite reductase, and ammonia monooxygenase genes indicated that anaerobic metabolism of aromatic compounds and both dissimilatory nitrate reduction to ammonia (DNRA) and nitrification occurred in situ. These data link formation (via DNRA) and destruction (via nitrification) of ammonia to in situ cycling of nitrogen in this subsurface habitat, where metabolism of aromatic pollutants has led to accumulation of reduced metabolic end products (e.g., ammonia and methane).
Nonphotosynthetic microorganisms (particularly members of the Archaea and Bacteria) colonizing natural habitats generate metabolic energy by linking the transfer of electrons from reduced substrates (electron donors; e.g., ammonia, methane, sulfide, carbohydrates, and hydrocarbons) to oxidized substrates (electron acceptors; e.g., O2, nitrate, Fe3+, and sulfate) (18, 40, 62, 73). Whenever possible, strategies for documenting biogeochemical change involve mass balance approaches that quantitatively link materials subject to a given metabolic process (e.g., consumption of carbon substrates) to formation of metabolic by-products (e.g., CO2). However, owing to the open nature of many natural systems (including ocean water, rivers, and soils), convergent lines of evidence obtained using a variety of approaches (e.g., model incubations, analytical chemistry of metabolites, and molecular biology of genes and mRNA) are often needed to understand site biogeochemistry (40, 46, 71). Direct detection of mRNA in environmental samples has increasingly become an effective approach for documenting the in situ biogeochemical activity of microbial communities in field sites (33, 34, 42). This approach has included at least three techniques: (i) reverse transcription-PCR (RT-PCR)-based targeting of expression of specific functional genes (e.g., genes encoding naphthalene dioxygenase [74], Fe(II) uptake protein [47], RubisCo [70], or the anammox and denitrification processes [33, 34]); (ii) creation and analysis of large community cDNA libraries of expressed genes (2, 11, 51); and (iii) direct pyrosequencing of total community-derived cDNA (19, 21, 50, 67). Documenting the occurrence of, ecological impact of, and dynamic relationships between microbially mediated redox processes poses many challenges (e.g., 32, 33, 34, 37, 46, 52).
Focusing on subsurface terrestrial habitats, a variety of reports have emphasized the dynamic spatial and temporal geochemical (15, 43, 44, 69) and microbiological (9, 23, 24, 59, 79) characteristics of the habitats. Wilson et al. (76) and Anneser et al. (1) used high-resolution subsurface sampling approaches to show that gradients at the fringes of contaminated groundwater can strongly influence microbial attenuation of pollutants. Furthermore, steep redox gradients in the contaminated subsurface have been attributed to biodegradation processes (6). Although the implications of dynamic subsurface biogeochemistry have been reported for arsenic (28, 48), very little attention has been given to the biogeochemical impacts of such dynamic conditions for broader cycling of carbon, nitrogen, sulfur, and other elements. A few subsurface contaminant transport models (15, 26, 68) explicitly account for the accumulation of reduced by-products of microbial metabolism [methane, sulfide, Fe(II), Mn(II), ammonia] since these reduced substrates have the potential to foster “secondary redox reactions” that include aerobic methanotrophy, nitrification, anaerobic ammonia oxidation, and anaerobic methane oxidation. Evidence for the latter was reported by van Breukelen and Griffoen (68), who found enrichment of residual 13CH4 in the anaerobic portion of a landfill leachate groundwater plume. Much more work addressing dynamic interactions between subsurface microbial processes is warranted.
For the subsurface study site examined here there is a 16-year record of diminishing concentrations of contaminants (naphthalene, xylenes, toluene, 2-methylnaphthalene, and acenaphthylene), and the site conditions support dynamic microbial communities, including an extensive eukaryotic food chain (79). Data reported here (i) further establish that the physiological setting for contaminant biodegradation fluctuates geochemically; (ii) demonstrate that in laboratory-based incubations naphthalene biodegradation is favored under only aerobic conditions and not under five types of anaerobic, physiological conditions; and (iii) show, based on a survey of expressed mRNA and metabolites, that microbial processes anaerobically consume aromatic compounds and carry out coupled cycling of nitrogen via dissimilatory nitrate reduction to ammonia (DNRA) and nitrification.
MATERIALS AND METHODS
Site and groundwater sampling.
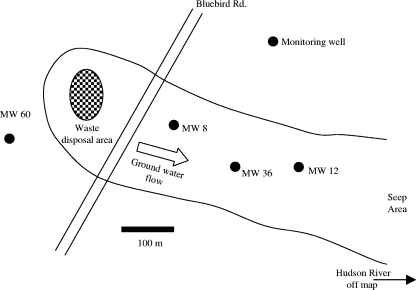
The study site is a rural wooded area in South Glens Falls, NY, where coal tar waste was buried in a shallow trench in the early 1960s. The source material was removed, and the groundwater plume is undergoing intrinsic bioremediation. This site has been described previously (5, 17) and has been used extensively for a variety of field-oriented microbiological studies (3, 4, 29, 75). A plan view of the site is shown in Fig. 1, which shows the locations of monitoring wells (MW) sampled for chemical and molecular analyses. Note that MW 60, the background control for chemical and nucleic acid assays, is not contaminated. The screening intervals (depths for accessing pumped groundwater) for all sampling wells were 5 to 7 m below the surface of the ground.
FIG. 1.
Map of field study site, showing the locations of monitoring wells (MW), the boundary of groundwater contamination, and the direction of groundwater movement. MW 60 is the site used for determining background geochemical conditions and microbiology. MW 36 is the site where there was the greatest contamination. (Adapted from reference 79.)
For DNA and mRNA extraction, groundwater samples were collected at a flow rate of 300 ml/min after 4 well volumes were purged with a peristaltic pump with new polyethylene tubing. Cells in 5 liters of groundwater (collected in ∼17 min) were concentrated on 142-mm-diameter Durapore membranes (pore size, 0.22 μm; Millipore Corp., Bedford, MA), placed in sterile Whirlpak bags, and frozen immediately by immersion in a dry ice-ethanol bath. Samples were kept on dry ice during transport to the laboratory, where they were transferred to a freezer kept at −80°C.
Geochemical analyses.
Alkalinity, the dissolved oxygen content, temperature, the total organic carbon (TOC) content, the methane content, and the concentrations of nitrate, ammonia, sulfate, sulfide, and Fe2+ were measured using previously described methods (5). The levels of precision of the assays (relative standard deviations) for groundwater constituents were as follows: nitrate, 5%; ammonia, 2%; sulfate, 2%; sulfide, 1%; methane, 1%; Fe2+, 1%; and total organic carbon, 5%.
Microcosms used to examine naphthalene biodegradation.
Groundwater was obtained from well 36 in October 2001 and used for biodegradation assays. To prevent contact between water samples and air (25, 78), the receiving glass canning jars (2 liters for water samples and 0.5 liter for sediments) were flushed on site with nitrogen gas and, after they were filled, sealed without air bubbles. Freshly gathered, oxygen-free subsurface material (depth, 5 m) from an area adjacent to well 36 was obtained with a commercially operated Geoprobe hydraulic coring machine. The sediment was immersed in anaerobic groundwater before it was sealed in a canning jar with no headspace air. The groundwater and sediments were placed on ice and maintained at 4°C until they were dispensed into serum bottles with an N2 headspace containing a trace of H2 in an anaerobic hood (Coy Laboratory Products, Grass Lake, MI) within 2 days, as previously described (25, 78). Triplicate sterile 125-ml serum bottles received 72 ml of well 36 groundwater plus 10% (wt/vol) well 36 sediment from 2 m below the water table. Poisoned controls were prepared by adding 1 ml of poison (5% HCl, 0.25 M HgCl2) to serum bottles prior to addition of samples. After the bottles were sealed, the headspace gas in each bottle was replaced with 100% O2-free N2 gas. To promote aerobic respiration and anaerobic respiration, including nitrate reduction, sulfate reduction, manganese reduction, iron reduction, and methanogenesis, the serum bottle microcosms were amended with air or sterile anaerobic preparations so that they contained 200 mM KNO3, 200 mM NaSO4 plus 2 mM Na2S (reducing agent), 1 mg/ml MnO2 slurry, 4.6 mg/ml FeOOH slurry, and 2 mM Na2S, respectively (25, 78). The serum bottles were incubated at the ambient ground temperature (10°C) in the dark with no shaking. At various times, samples (1 ml) of the slurries were removed with a syringe. After extraction with 1 ml ethyl acetate (5), the samples were analyzed by gas chromatography-mass spectrometry (GC/MS) (5, 75) with a Hewlett-Packard model 6890 series II gas chromatograph equipped with an HP-5MS (5% phenylmethyl siloxane; Hewlett-Packard) fused silica capillary column (30 m by 0.25 mm; film thickness, 0.25 μm) connected to a Hewlett-Packard model 5973 quadrupole mass-selective detector operated with an electron energy of 70 eV and a detector voltage of 1,700 V. Splitless injection was used, with a 1-min delay before septum purge. The carrier gas was helium (linear velocity, 30 cm/s). The injector and detector temperatures were 250°C and 300°C, respectively.
Metabolite sampling and analysis.
The aerobic metabolite 1,2-dihydoxy-1,2-dihydronaphthalene (naphthalene cis-dihydrodiol) (75) and an anaerobic metabolite of naphthalenes, 2-carboxynaphthalene (20), were sought in freshly collected contaminated and uncontaminated (Fig. 1) well water using previously described procedures (20). For the cis-dihydrodiol metabolite, Envi-Chrom P solid-phase extraction tubes (6 ml; Supelco, Bellefonte, PA) were used on site to concentrate the analyte from 2 liters of well water. For carboxylated anaerobic metabolites, 2 liters of water was extracted four times with 100 ml ethyl acetate; pooled extracts were concentrated by rotary evaporation under N2 to obtain a volume of 50 ml. Extracts were concentrated under nitrogen, dehydrated, derivatized with bis(trimethylsilyl)trifluoroacetamide (BSTFA), and analyzed by GC/MS, as described above. For assays we used single-ion monitoring mode with m/z 244, 229, and 127 for the cis-dihydrodiol, and the carboxylated metabolites were analyzed in scanning mode.
Nucleic acid sampling and analysis.
For extraction of DNA and RNA from groundwater biomass, previously described procedures (4, 29, 38, 74) were used, with minor modifications. Briefly, for DNA extraction, frozen filters were crushed twice and extracted twice by boiling them for 5 min with 5 ml preheated extraction buffer (1% SDS, 0.1 M NaCl, 10 mM Tris, 1 mM EDTA; pH 8.0). Decanted extracts were added to an equal volume of phenol (pH 8.0) and mixed vigorously, which was followed by centrifugation at 4°C (15 min at 10,000 × g). The upper aqueous layer was extracted with an equal volume of phenol-chloroform (1:1), and this was followed by centrifugation as described above and then a final extraction with an equal volume of chloroform-isoamyl alcohol (24:1) and centrifugation. DNA was precipitated from the final aqueous layer at −80°C with sodium acetate (0.3 M) and an equal volume of isopropanol. After centrifugation (30 min at 15,000 × g), DNA pellets were washed in 70% ethanol and reprecipitated. Pellets were then air dried and dissolved in 50 μl 10 mM Tris-1 mM EDTA (pH 8.0).
For RNA processing, all reagents and labware were either certified RNase free, baked overnight at 200°C, treated with RNase ZAP (Ambion, Austin, TX), or treated with diethyl pyrocarbonate (DEPC) for inactivation of RNase activity as appropriate. RNA was extracted from sections of the frozen filters (see above) using a modified version of the DNA extraction protocol described above. Briefly, crushed filter pieces were processed using acid extraction buffer (pH 5.1), and acidic phenol (pH 4) was used for phenol and phenol-chloroform-isoamyl alcohol (125:25:1) extraction. RNA was precipitated from aqueous extracts with glycogen as a coprecipitant (2 μg/ml) at −80°C using sodium acetate (2.5 M) and 2 volumes of ethanol. Purified RNA pellets were recovered by centrifugation (30 min at 15,000 × g), air dried, and resuspended in 50 μl RNase-free water. DNA was removed from RNA extracts by DNase I treatment (Invitrogen), and RNA was reverse transcribed to cDNA using SuperScript III reverse transcriptase (RT) (Invitrogen) and random hexamers according to the manufacturer's instructions. Control PCRs with no RT were performed using primers and PCR conditions described below to ensure complete removal of contaminating DNA. DNA fragments associated with amoA, narG, mxaF, mcrA, nrfA, and bssA genes were PCR amplified from cDNA pools using primer pairs listed in Table 1. PCR amplification was performed using ThermoStart DNA polymerase (ABgene) and a PTC-200 DNA engine thermocycler (MJ Research). Replicate 25-μl reaction mixtures contained 0.5 to 25 ng DNA template, and the reactions were performed using previously described cycling conditions (Table 1). Pooled PCR products were resolved by agarose gel electrophoresis and were purified using a QIAquick gel extraction kit (Qiagen). Except for archaeal amoA, amplicons of the expected size were obtained for samples from well 36 and well 12 but not for samples from upgradient control well 60. Purified DNA was cloned with either a TOPO-TA cloning kit (Invitrogen) or a StrataClone PCR cloning kit (Stratagene) as described by the manufacturers. Functional biomarker gene cDNA clone libraries were generated for samples from well 36 and well 12 collected in 2006. Three bssA cDNA clone libraries were obtained for samples collected in August and November; one archaeal amoA cDNA clone library and one bacterial amoA cDNA clone library were obtained for samples collected from well 36 in August; and two nrfA clone libraries were obtained for samples collected in August. Clone libraries were screened by restriction fragment length polymorphism (RFLP) analysis, and representative inserts were sequenced. For the mxaF and narG genes, amplified fragments from DNA from well 36 (August 2006) were cloned as described above; eight and nine clones, respectively, contained inserts of the expected size. These clones were screened by RFLP analysis; one representative sequence of mxaF and three representative sequences of narG were obtained.
TABLE 1.
PCR primers used in this study
| Process | Target gene/target enzyme | Primer | Sequence (5′-3′) | Reference |
|---|---|---|---|---|
| Ammonia oxidation (Bacteria) | amoA/ammonia monooxygenase | amoA-1F* | GGGGHTTYTACTGGTGGT | 53 |
| amoA-2R | CCCCTCKGSAAAGCCTTCTTC | |||
| Ammonia oxidation (Archaea) | amoA/ammonia monooxygenase | Arch-amoAF | STAATGGTCTGGCTTAGACG | 8 |
| Arch-amoAR | GCGGCCATCCATCTGTATGT | |||
| Nitrate reduction | narG/respiratory nitrate reductase | narG1960f | TAYGTSGGCARGARAA | 60 |
| narG2659r | TTYTCRTACCABGTBGC | |||
| Methane oxidation | mxaF/methanol dehydrogenase | mxa1003f | GCGGCACCAACTGGGGCTGGT | 27 |
| mxa1561r | GGGCAGCATGAAGGGCTCCC | |||
| Methanogenesis | mcrA/methyl-coenzyme M reductase | MLf | GGTGGTGTMGGATTCACACARTAYGCWACAGC | 39 |
| MLr | TTCATTGCRTAGTTWGGRTAGTT | |||
| mcrA/methyl-coenzyme M reductase | ME1f | CGMATGCARATHGGWATGTC | 27 | |
| ME2r | TCATKGCRTAGTTDGGRTAGT | |||
| Anaerobic degradation of | bssA/benzylsuccinate synthase | 7772f | GACATGACCGACGCSATYCT | 77 |
| alkyl aromatics | 8546r | TCGTCGTCRTTGCCCCAYTT | ||
| Dissimilatory nitrate | nrfA/cytochrome c nitrite reductase | nrfAF1 | GCNTGYTGGWSNTGYAA | 60 |
| reduction to ammonia | nrfA7R1 | TWNGGCATRTGRCARTC |
Sequencing (Cornell University Life Sciences Core Laboratories Center) was conducted routinely with the M13 forward and M13 reverse primers. Sequences were manually checked for quality and edited using 4Peaks software (http://www.mekentosj.com/4peaks/). Chimeras, vector sequences, and sequences that were poor quality were excluded from further phylogenetic analyses. Sequences were compared with the GenBank nucleotide database library by performing BLAST on-line searches (http://ncbi.nlm.nih.gov/BLAST), and the closest BLAST matches were included in phylogenetic comparisons. Phylogenetic trees were constructed by performing neighbor-joining analyses (CLUSTALX alignments, ARB analysis package). Bootstrap values of ≥50% for 100 replicates were calculated for all dendrogram nodes.
Nucleotide sequence accession numbers.
The nucleotide sequences reported here have been deposited in the GenBank database under accession no. FJ810622 to FJ810667.
RESULTS
Recent site geochemistry: variability in methane and inorganic reductants with potential to fuel microbial reactions.
Data obtained for three on-site wells (wells 8, 36, and 12 [Fig. 1]) on three occasions over a 10-month period in 2006 and 2007 documented the fluctuating alkalinity and concentrations of sulfate, nitrate, and reduced substrates, including ammonia, methane, sulfide, Fe2+, and total organic carbon (Table 2). The key observation is that the values for nearly all of these analytes fluctuated over time for a given well. A previous report showed that from 1999 to 2001 the on-site concentrations of methane, sulfide, hydrogen gas, Fe2+, Mn2+, and alkalinity were elevated in the contaminated wells compared to an upgradient control well (5). The new data (Table 2) confirm the previous findings (presence of methane, sulfide, and Fe2+). However, the concentrations of sulfate, nitrate, and ammonia rose approximately 10-fold during the intervening 6 years, and the values for nearly all groundwater constituents were temporally variable (Table 2).
TABLE 2.
Time-dependent geochemical characteristics of site well water samplesa
| Characteristic | Well 12 |
Well 8 |
Well 36 |
|||||
|---|---|---|---|---|---|---|---|---|
| Sampling dates (mo/yr) |
Sampling dates (mo/yr) |
Sampling dates (mo/yr) |
||||||
| 8/2006 | 12/2006 | 5/2007 | 8/2006 | 5/2007 | 8/2006 | 12/2006 | 5/2007 | |
| Nitrate concn (mg/liter) | 2.1 | 2.7 | 1.8 | 1.8 | 1.7 | 2.3 | 1.8 | 1.6 |
| Ammonia concn (mg/liter) | 0.13 | 0.12 | 0.15 | BDb | 0.002 | 0.2 | 0.12 | 0.09 |
| Sulfate concn (mg/liter) | 100 | 99 | 480 | 110 | 590 | 150 | 51 | 67 |
| Sulfide concn (mg/liter) | 0.01 | 0.003 | 0.001 | 0.01 | 0.003 | 0.1 | 0.02 | 0.014 |
| Methane concn (μg/liter) | NDc | 2 | ND | ND | ND | ND | 21 | ND |
| Fe2+ concn (mg/liter) | 0.14 | 0.28 | 0.06 | 0.09 | 0.02 | 0.45 | 0.14 | 0.06 |
| Alkalinity (mg CaCO3) | 140 | 460 | ND | 130 | ND | 76 | 140 | 460 |
| Total organic carbon concn (mg/liter) | 5.9 | 29 | ND | ND | ND | 7.3 | 40 | ND |
The values are averages of duplicate determinations. The relative standard deviations were as follows: nitrate, 5%; ammonia, 2%; sulfate, 2%; sulfide, 1%; methane, 1%; Fe2+, 1%; and total organic carbon, 5%.
BD, below detection limit (0.001 mg/liter).
ND, not determined.
Laboratory-based biodegradation is limited to aerobic contaminant metabolism.
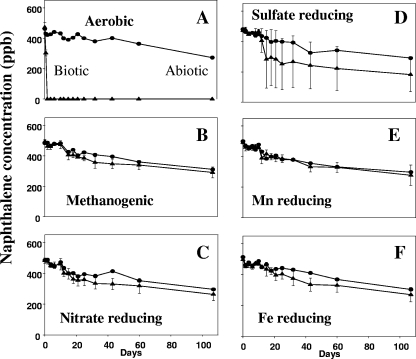
Given the prevailing oxygen-deprived conditions reported previously for the study site (5, 79), a series of laboratory-based biodegradation assays were performed to assess the propensity of the endogenous microbial community to metabolize native contaminants under a wide variety of physiological conditions. We focused on slurries of freshly gathered water and sediment from well 36 because this well had the highest levels of contamination and was anaerobic at depth (79). In the aerobic serum bottle incubations, the ambient naphthalene (480 ppb) was fully metabolized within 1 day (Fig. 2A). No significant biodegradation activity was observed over the 110-day incubation period under conditions that fostered five alternate terminal electron-accepting processes (denitrification, Fe oxide reduction, Mn oxide reduction, sulfate reduction, and methanogenesis) (Fig. 2B to F). The data indicate that oxygen-based metabolism of aromatic compounds is the most robust physiological mechanism by which the native groundwater microbial community destroys contaminants and contrast with field observations of anaerobic loss of contaminants (79). Acknowledging that field processes may depend on conditions not attained in laboratory incubations, we utilized field-based metabolite and mRNA assays to address this discrepancy.
FIG. 2.
Naphthalene biodegradation in microcosms containing well 36 groundwater and subsurface sediment from a location adjacent to well 36 in treatments (10°C) designed to favor aerobic metabolism (A), methanogenesis (B), nitrate reduction (C), sulfate reduction (D), manganese oxide reduction (E), and iron oxide reduction (F). ▴, data for viable treatments; •, data for poisoned controls. The symbols indicate the averages for triplicate microcosms; the error bars indicate one standard deviation.
Detection of metabolites.
Detection of metabolites is an insightful, independent means of assessing in situ metabolic activity (20, 22, 81). In 1995, a transient, unique metabolite of aerobic naphthalene metabolism (1,2-dihydoxy-1,2-dihydronaphthalene) was found at the study site at concentrations ranging from 0.06 ppb to 45 ppm in seven monitoring wells (75). To corroborate the previous observations and extend them to anaerobic processes, liquid- and solid-phase extractions were combined with GC/MS to concentrate and document the presence of the cis-dihydrodiol-type aerobic metabolite (see above) and key carboxylated indicators of anaerobic metabolism of both naphthalene and 2-methylnapthalene (20, 22, 81) in water samples from the three contaminated wells (wells 8, 12, and 36) and the background control (Fig. 1) in January 2001 and November 2005. The previously established assay, performed using authentic standards derivatized with BSTFA, was sensitive to ∼1 ng/liter. In single-ion monitoring mode using m/z 228, 127, and 116 and m/z 244, 229, and 127 as indicator ions for the diol and carboxylates, respectively, no evidence was found of any metabolite in samples analyzed in January 2001. However, when the assay for the carboxylated compounds was carried out in November 2005, 1-naphthoic acid and very strong evidence of 1,2,3,4-tetrahydro-1-naphthoic acid were detected in well 36 but not in the uncontaminated background control well (see Fig. S1 in the supplemental material). (The case for the presence of 1,2,3,4-tetrahydro-1-naphthoic acid rests on the fact that an authentic standard for 1,2,3,4-tetrahydro-2-naphthoic acid was available and, because the metabolite at the site had a matching mass spectrum but different retention time, it was inferred that the compound was likely the 1,2,3,4-tetrahydro-1-naphthoic acid isomer.) Benzylsuccinate-type intermediary metabolites (indicative of in situ anaerobic degradation of toluene, xylenes, etc.) were not detected in the GC/MS assays; the levels of these metabolites must have been below the limits of detection, as mRNA transcripts of the bssA gene were found (see below). Thus, while transient intermediary metabolites for both aerobic and anaerobic metabolism of naphthalenes have been found at the site, the patterns of occurrence vary temporally. This observation is fully consistent with the dynamic state of the site indicated by both geochemical (Table 2) (79) and community composition (79) data.
Expressed genes indicate that there was in situ microbial metabolism of aromatic hydrocarbons and ammonia.
In situ aerobic metabolism of naphthalene has previously been indicated by detection and sequencing of transcripts of the naphthalene dioxygenase gene, nahAc, in site groundwater (3, 74, 80). Substituted aromatic hydrocarbons (e.g., toluene, xylenes, and 2-methylnaphthalene) are some of the cooccurring contaminants (79). Hypothesizing that these contaminants have prominent roles in the biogeochemical repertoire of the study site, we examined six additional key processes: anaerobic catabolism of methylated aromatic hydrocarbons, aerobic oxidation of ammonia (nitrification), anaerobic reduction of nitrate (the first step in denitrification), dissimilatory reduction of nitrate to ammonia (DNRA), methanogenesis, and aerobic methane oxidation. DNA and mRNA pools extracted from water samples from the site were screened for individual functional genes (bssA for anaerobic catabolism of methylated aromatic hydrocarbons; bacterial and archaeal amoA for ammonia monooxygenase; narG for the first step in denitrification; nrfA for the second step in DNRA; mcrA for methyl-coenzyme M reductase in methanogenesis; and mxaF for the second step in aerobic methane oxidation). PCR amplification from extracted DNA was successful for all genes targeted except mcrA. narG, encoding a membrane-bound nitrate reductase, is one of nearly a dozen biomarkers widely used to assess reduction of nitrate to nitrite in anaerobic habitats (60). The three representative partial DNA sequences obtained from well 36 (Table 3) were 82 to 85% identical to genes from well-characterized nitrate reducers, including Geobacter metallireducens. Water samples from the site also harbored partial sequences that were 98% identical to the mxaF gene of Methylocystis (Table 3); mxaF is one of several established biomarkers for aerobic methylotrophy (27).
TABLE 3.
Closest relatives in the GenBank database for partial narG (652 bp) and mxaF (554 bp) DNA sequences obtained from well 36a
| Targeted gene | Sequence designation | Best BLAST match or closest match in cultured organism | Maximum % identity | Accession no. |
|---|---|---|---|---|
| narG | JMYnarG02 | Uncultured bacterium clone RT-250_16 | 84 | DQ481115 |
| Geobacter metallireducens GS-15 | 82 | CP000148 | ||
| narG | JMYnarG03 | Uncultured bacterium partial narG | 88 | AM408519 |
| Geobacter metallireducens GS-16 | 84 | CP000148 | ||
| narG | JMYnarG05 | Uncultured bacterium clone GRAMO27 | 85 | AY955194 |
| Methylobacterium sp. strain 4-46 | 81 | CP000943 | ||
| mxaF | JMYmxaF01 | Methylocystis sp. strain 5FB2 | 98 | EF212330 |
Libraries were prepared from the August 2006 samples. For narG, a library of 9 clones led to three RFLP patterns and three gene sequences. For mxaF, a library of 8 clones led to a single RFLP pattern and a single gene sequence.
Patterns of occurrence and sequences of expressed genes.
Reverse transcriptase-based detection of expressed genes was successful for bssA, nrfA, and both bacterial and archaeal amoA. In all of these assays, the reverse transcriptase-free control yielded no PCR product. Table S1 in the supplemental material shows a summary of data for site sampling, detection, and cloning of the four gene transcripts, while Table 4 shows the distribution of specific transcript sequences in cDNA libraries from the two wells from November 2005 to November 2006. Because the sampling locations and times were limited, an absence of gene amplicons in pools of DNA and mRNA did not necessarily mean that a given process did not occur at the site. Table S1 in the supplemental material shows that there were clear temporal variations in two of the transcribed genes; the data for both bacterial amoA and nrfA (November 2005, well 36) indicate that mRNA was not detected despite the presence of encoding DNA.
TABLE 4.
Summary of OTUs from cDNA clone libraries prepared from well 36 and well 12
| Gene | OTUa | Representative clone sequenceb | No. of related clones |
|||||
|---|---|---|---|---|---|---|---|---|
| Total | Well 36 |
Well 12 |
||||||
| November 2005 | August 2006 | November 2006 | August 2006 | November 2006 | ||||
| bssA | 1 | bssA-01 | 27 | 7 | 0 | 20 | 0 | 0 |
| 2 | bssA-02 | 95 | 27 | 23 | 44 | 0 | 1 | |
| 5 | bssA-05 | 11 | 3 | 0 | 2 | 6 | 0 | |
| 6 | bssA-06 | 39 | 0 | 0 | 16 | 12 | 11 | |
| 8 | bssA-08 | 2 | 0 | 0 | 0 | 2 | 0 | |
| 9 | bssA-09 | 2 | 0 | 1 | 0 | 0 | 1 | |
| 10 | bssA-10 | 13 | 0 | 0 | 0 | 8 | 5 | |
| 11 | bssA-11 | 9 | 1 | 0 | 0 | 1 | 7 | |
| 13 | bssA-13 | 7 | 4 | 0 | 0 | 0 | 3 | |
| amoA (archaeal) | 1 | amoA (arc-1) | 22 | 22 | ||||
| amoA (bacterial) | 1 | amoA-01 | 38 | 38 | ||||
| amoA-02 | ||||||||
| amoA-04 | ||||||||
| amoA-05 | ||||||||
| 3 | amoA-03 | 11 | 11 | |||||
| nrfA | 2 | nrfA-02 | 2 | 1 | 1 | |||
| 25 | nrfA-25 | 2 | 1 | 1 | ||||
| 26 | nrfA-26 | 12 | 12 | 0 | ||||
| 28 | nrfA-28 | 3 | 3 | 0 | ||||
| 3 | nrfA-03 | 18 | 0 | 18 | ||||
| 4 | nrfA-04 | 13 | 0 | 13 | ||||
| 5 | nrfA-05 | 1 | 0 | 1 | ||||
| 6 | nrfA-06 | 1 | 0 | 1 | ||||
OTUs are unique clone groups determined by RFLP screening.
Representative sequences for each OTU are shown.
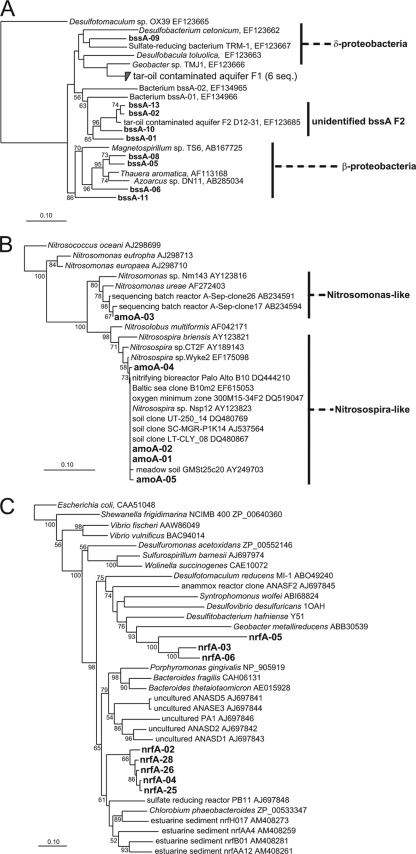
The recognition that benzylsuccinate synthase (BssA) is a pivotal enzyme in anaerobic metabolism of methylated aromatic hydrocarbons, especially toluene, has driven recent investigations of the diversity of bssA-related genes in environmental samples (7, 77). In all five attempts, bssA transcripts were obtained (a total of 207 clones in 5 libraries) for the mRNA pool from wells 12 and 36 (November 2005, August 2006, and/or November 2006) (Table 4; see Table S1 in the supplemental material). The expressed environmental bssA genes comprised 24 RFLP types, 24 sequences, and 9 distinctive operational taxonomic units (OTUs) (Table 4; see Table S1 in the supplemental material) that fell into 7 main clades (Fig. 3A), including sequences grouping with genes associated with beta- and deltaproteobacteria and a cluster associated with the unidentified bssA F2 group reported previously (77). One of the bssA alleles (bssA-05) was detected in both wells at nearly all sampling times, while other bssA alleles (e.g., bssA-09, bssA-10, bssA-11, and bssA-13) were detected rarely (Table 4).
FIG. 3.
Phylogenetic analysis of mRNA transcripts found in water from the site for three key site metabolic processes: anaerobic degradation of methylated aromatic compounds (bssA) (A), aerobic metabolism of ammonia (bacterial amoA) (B), and anaerobic dissimilatory reduction of nitrate to ammonia (nrfA) (C). The alignments are based on data for 250, 137, and 209 deduced amino acid sites, respectively. Neighbor-joining analyses were performed with CLUSTALX alignments using the ARB analysis package. Sequences obtained in this study are indicated by bold type, while reference sequences are indicated by light type. The distribution of clones in wells 36 and 12 is shown in Table 4. Bootstrap values of ≥50% for 100 replicates are indicated at the nodes. The GenBank accession numbers for reference sequences are indicated after the clone and organism designations. Scale bars = 0.10 change per nucleotide position.
Members of the Bacteria and Archaea both carry out nitrification reactions, although the catalytic subunits (AmoA) of the ammonia monooxygenase enzymes that they utilize are distinct (8). Expression of both types of amoA was detected in water from well 36 (August 2006). In all three attempts, transcripts of archaeal amoA genes were amplified; only a single RFLP-OTU type was detected in a single 22-member clone library (Table 4; see Table S1 in the supplemental material), and its sequence matched (99% sequence identity) that of uncultured archaeal amoA environmental clones (e.g., GenBank accession no. EU852688). In two of three attempts, transcripts of bacterial amoA were detected; from a pool of 49 clones in one library, two RFLP types and four distinct representative clone sequences were obtained (Table 4; see Table S1 in the supplemental material). Phylogenetically, the pool of bacterial AmoA mRNA sequences fell into two main clades (Fig. 3B), and these were associated with established nitrifying genera (e.g., Nitrosospira and Nitrosomonas).
nrfA encodes an enzyme (cytochrome c nitrite reductase) that is responsible for a 6-electron transfer that reduces nitrite to ammonia in the DNRA process (59, 60). In two of three detection attempts, nrfA was found to be expressed (in a total of 52 clones in two libraries) (Table 4; see Table S1 in the supplemental material) and occurred as 8 RFLP types and 8 representative sequences or OTUs in water from wells 12 and 36 in August 2006 (Table 4; see Table S1 in the supplemental material). The environmental transcripts, shown as 7 main clades in Fig. 3C, were related to nrfA genes previously associated with Chlorobium phaeobacteroides and Geobacter (60). Two of the nrfA alleles (nrfA2, and nrfA25) were detected in both wells, while the other alleles occurred in single wells (Table 4).
In contrast to the results described above for the contaminated wells (wells 36 and 12), the results of searches for mRNA transcripts in samples from the background control (well 60) were negative in nearly all cases. There was one exception, however; an amplicon for archaeal amoA was found. This suggests that aerobic ammonia oxidation may be an ambient process at the site, probably fed by mineralization of native organic matter.
DISCUSSION
Spatial variability and temporal variability of geochemical conditions at the groundwater site examined, as documented in this study and previously (79), largely preclude construction of high-resolution maps for microbial processes. In order to decipher causation in subsurface biogeochemical processes, it is logical to give high weight to the presence of genes, transcripts, and metabolites and low weight to their absence. For example, the inability in this investigation to detect the biomarker of aerobic naphthalene metabolism (1,2-dihydoxy-1,2-dihydronaphthalene) does not negate the prior detection of this biomarker (75) documenting aerobic naphthalene biodegradation at the site. In contrast, the expression of bssA transcripts (Fig. 3A) and the discovery of anaerobic metabolites characteristic of naphthalene and 2-methylnaphthalene (see Fig. S1 in the supplemental material) in the groundwater establish that the microbial community native to the study site metabolizes aromatic contaminant compounds in situ via anaerobic pathways.
The potential physiological electron donors fueling microbial reactions occurring at the site include coal tar-derived organic compounds, native organic compounds in the dissolved organic carbon pool, methane, ammonia, and sulfide. The complementary set of potential physiological electron acceptors includes O2, nitrate, sulfate, oxides of Fe and Mn (on aquifer solids), and CO2. Depletion of O2 and nitrate inside but not outside the zone of contamination has provided evidence of in situ aerobic and anaerobic metabolism of coal tar waste constituents (5, 79). The constant slow delivery of ambient electron acceptors (e.g., oxygen, sulfate, and nitrate) to the site via groundwater flow (on-site velocity, ∼12 m/year [17]) has the potential to drive (govern) the rates and types of electron-accepting processes and lead to accumulation of reduced metabolic by-products (see below).
This study resulted in a variety of insights about a suite of biogeochemical processes important to N cycling at the site. The pools of ammonia and nitrate fluctuate (Table 2). Detection of archaeal amoA transcripts in the upgradient background well indicated that nitrification is among the myriad biogeochemical processes that predictably occur in the absence of contamination. However, the prevalence of anaerobic conditions and the transcripts of the nrfA gene in groundwater (Fig. 3C) indicate that dissimilatory reduction of nitrate to ammonia (DNRA) (59, 60) is likely a key mechanism of in situ ammonia production. Furthermore, the presence of mRNA transcripts of both bacterial and archaeal amoA genes (Fig. 3B) indicates that there is in situ ammonia oxidation. Therefore, redox cycling of N appears to occur at the site, and it likely stems indirectly from coal tar waste, whose oxidation creates elevated concentrations of ammonia (via DNRA) in water at the site. The presence in 16S rRNA clone libraries of sequences representing microbial taxa known to carry out ammonia oxidation (e.g., taxa related to Nitrosopumilus [32, 79]) provides additional support for the hypothesis that there is redox cycling of nitrogen. The conclusion that emerges from the observations described above is that the dynamic geochemistry of this subsurface site fosters the development of microbial populations that catalyze biogeochemical redox reactions, separated spatially and/or temporally, that are complementary to one another. In the present study we simultaneously used both empirical field geochemical measurements and the results of a molecular (mRNA)-based strategy to argue that nitrification can cooccur with DNRA and thus that there is coupled cycling of N in the subsurface.
Dissimilatory reduction of nitrate to ammonia is the 8-electron-accepting conversion of NO3− to NH3 (59, 64). This process has long been considered a physiological peculiarity restricted to enteric bacteria (16) or sulfur reducers (55), and its ecological significance has been underexplored and underemphasized for decades (13, 14, 64, 65, 66). However, DNRA has been reported to occur in a variety of habitats, including anaerobic bioreactors (41), landfills (10), freshwater sediment (35), a glacial meltwater stream (45), marine sediments (36, 61, 72), and anaerobic river sediments (12, 31). Furthermore, there has been a resurgence of efforts aimed at documenting and understanding the occurrence of DNRA in tropical forest soils (49, 56, 57, 63) and in African coastal waters (30) by using measurements of the physiological conversion of 15NO3− to 15NH3 by environmental samples. In the oxygen minimum zone off the coast of Peru, DNRA has emerged as a crucial N-cycling process, generating the ammonia pool that fosters the anammox reaction (33).
This study reports apparent contradictions between two types of information pertinent to aromatic hydrocarbon biodegradation. Laboratory incubations (Fig. 2) provided evidence only for the commonly observed aerobic metabolism of naphthalene (3, 80). Yet data for two independent field-derived biomarkers (metabolites and mRNA transcripts) (Fig. 3; see Fig. S1 in the supplemental material) clearly indicated that anaerobic microbial metabolism of methylated and nonmethylated aromatic pollutants occurs in situ at the study site. We reconcile the apparent contradictions by pointing to the fact that metabolic relationships within microbial consortia (necessary for expression of many anaerobic processes) may be difficult to assemble in laboratory settings (54). The field-derived information trumps the laboratory information. It is possible that trace amounts of oxygen entered our sampling bottles; this may have led to elimination of populations essential for anaerobic naphthalene metabolism. Also, the concentrations of final electron acceptors and/or reducing agents may have been so high that they inhibited the physiological processes examined. The recovery of both metabolites and transcripts indicative of anaerobic aromatic hydrocarbon metabolism provides impetus for redoubling our efforts to cultivate and isolate microbes catalyzing these processes.
Supplementary Material
Acknowledgments
Funding was provided by National Institute of Environmental Health Sciences grant 1-R21-ES012834 and NSF grant DEB-0841999 (to E.L.M.). J.M.Y. was funded in part by the NSF IGERT Program and by NIEHS training grant 5-T32-ES00752-28 (to S. Bloom and A. Yen, Cornell University).
We appreciate constructive criticisms from reviewers.
Footnotes
Published ahead of print on 26 March 2010.
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Anneser, B., F. Einsiedl, R. U. Meckenstock, L. Richters, F. Wisotzky, and C. Griebler. 2008. High-resolution monitoring of biogeochemical gradients in a tar oil-contaminated aquifer. Appl. Geochem. 23:1715-1730. [Google Scholar]
- 2.Bailly, J., L. Fraissinet-Tachet, J.-C. Verner, J.-C. Debaud, M. Lemaire, M. Wesolowski-Louvel, and R. Marmeisse. 2007. Soil eukaryotic functional diversity, a metatranscriptomic approach. ISME J. 1:632-642. [DOI] [PubMed] [Google Scholar]
- 3.Bakermans, C., and E. L. Madsen. 2002. Detection in coal tar waste-contaminated groundwater of mRNA transcripts related to naphthalene dioxygenase by fluorescent in situ hybridization with tyramide signal amplification. J. Microbiol. Methods 50:75-84. [DOI] [PubMed] [Google Scholar]
- 4.Bakermans, C., and E. L. Madsen. 2002. Diversity of 16S rDNA and naphthalene dioxygenase genes from coal-tar-waste-contaminated aquifer waters. Microb. Ecol. 44:95-106. [DOI] [PubMed] [Google Scholar]
- 5.Bakermans, C., A. M. Hohnstock-Ashe, S. Padmanabhan, P. Padmanabhan, and E. L. Madsen. 2002. Geochemical and physiological evidence for mixed aerobic and anaerobic field biodegradation of coal tar waste by subsurface microbial communities. Microb. Ecol. 44:107-117. [DOI] [PubMed] [Google Scholar]
- 6.Bauer, R. D., M. Rolle, S. Bauer, C. Eberhardt, P. Grathwohl, O. Kolditz, R. U. Meckenstock, and C. Griebler. 2009. Enhanced biodegradation by hydraulic heterogeneities in petroleum hydrocarbon plumes. J. Contam. Hydrol. 105:56-68. [DOI] [PubMed] [Google Scholar]
- 7.Beller, H., S. Kane, T. Legler, and P. Alvarez. 2002. A real-time polymerase chain reaction method for monitoring anaerobic, hydrocarbon-degrading bacteria based on a catabolic gene. Environ. Sci. Technol. 36:3977-3984. [DOI] [PubMed] [Google Scholar]
- 8.Beman, J. M., and C. A. Francis. 2006. Diversity of ammonia-oxidizing Archaea and Bacteria in the sediments of a hypernutrified subtropical estuary: Bahía del Tóbari, Mexico. Appl. Environ. Microbiol. 72:7767-7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bennett, P. C., F. K. Hiebert, and J. R. Rogers. 2000. Microbial control of mineral-groundwater equilibria: macroscale to microscale. Hydrogeol. J. 8:47-62. [Google Scholar]
- 10.Berge, N. D., D. R. Reinhart, and T. G. Townsend. 2005. The fate of nitrogen in bioreactor landfills. Crit. Rev. Environ. Sci. Technol. 35:365-399. [Google Scholar]
- 11.Botero, L. M., S. D'Imperio, M. Burr, T. R. McDermott, M. Young, and D. J. Hassett. 2005. Poly(A) polymerase modification and reverse transcriptase PCR amplification of environmental RNA. Appl. Environ. Microbiol. 71:1267-1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brunet, C. R., and L. J. Garcia-Gil. 1996. Sulfide-induced dissimilatory nitrate reduction to ammonia in anaerobic freshwater sediments. FEMS Microbiol. Ecol. 21:131-138. [Google Scholar]
- 13.Burgin, A. J., and S. K. Hamilton. 2007. Have we overemphasized the role of denitrification in aquatic systems? A review of nitrate removal pathways. Front. Ecol. Environ. 5:89-96. [Google Scholar]
- 14.Caskey, W. H., and J. M. Tiedje. 1979. Evidence for Clostridia as agents of dissimilatory reduction of nitrate to ammonium in soils. Soil Sci. Soc. Am. J. 43:931-936. [Google Scholar]
- 15.Christensen, T. H., P. L. Bjerg, S. A. Banwart, R. Jakobsen, G. Heron, and H.-J. Albrechtsen. 2000. Characterization of redox conditions in groundwater contaminant plumes. J. Contam. Hydrol. 45:165-241. [Google Scholar]
- 16.Cole, J. 1996. Nitrate reduction to ammonia by enteric bacteria: redundancy or a strategy for survival during oxygen starvation? FEMS Microbiol. Lett. 136:1-11. [DOI] [PubMed] [Google Scholar]
- 17.Electric Power Research Institute. 1996. Characterization and monitoring before and after source removal at a former manufactured gas plant (MGP) disposal site. Report TR-105921. Electric Power Research Institute, Pleasant Hill, CA.
- 18.Falkowski, P. G., T. Fenchel, and E. F. Delong. 2008. The microbial engines that drive Earth's biogeochemical cycles. Science 320:1034-1039. [DOI] [PubMed] [Google Scholar]
- 19.Frias-Lopez, J., Y. Shi, G. W. Tyson, M. L. Coleman, S. C. Schuster, S. W. Chisholm, and E. F. Delong. 2008. Microbial community gene expression in ocean surface waters. Proc. Natl. Acad. Sci. U. S. A. 105:3805-3810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gieg, L., and J. Suflita. 2002. Detection of anaerobic metabolites of saturated and aromatic hydrocarbons in petroleum-contaminated aquifers. Environ. Sci. Technol. 36:3755-3762. [DOI] [PubMed] [Google Scholar]
- 21.Gilbert, J. A., D. Field, Y. Huang, R. Edwards, W. Li, P. Gilna, and I. Joint. 2008. Detection of large numbers of novel sequences in the metatranscriptomes of complex marine microbial communities. PLoS One 3:e3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Griebler, C., M. Safinowski, A. Vieth, H. H. Richnow, and R. U. Meckenstock. 2004. Combined application of stable carbon isotope analysis and specific metabolites determination for assessing in situ degradation of aromatic hydrocarbons in a tar oil contaminated aquifer. Environ. Sci. Technol. 38:617-631. [DOI] [PubMed] [Google Scholar]
- 23.Haack, S. K., L. R. Fogarty, T. G. West, E. W. Alm, J. T. McGuire, D. T. Long, D. W. Hyndman, and L. R. Forney. 2004. Spatial and temporal changes in microbial community structure associated with recharge-influenced chemical gradients in a contaminated aquifer. Environ. Microbiol. 6:438-448. [DOI] [PubMed] [Google Scholar]
- 24.Hendrickx, B., W. Dejonghe, W. Boënne, M. Brennerova, M. Cernik, T. Lederer, M. Bucheli-Witshcel, L. Bastiaens, W. Verstraete, E. M. Top, L. Diels, and D. Springael. 2005. Dynamics of an oligotrophic bacterial aquifer community during contact with a groundwater plume contaminated with benzene, toluene, ethylbenzene, and xylenes: an in situ mesocosm study. Appl. Environ. Microbiol. 71:3815-3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hohnstock-Ashe, A. M., S. E. Bilotta, and E. L. Madsen. 2001. Further biogeochemical characterization of a trichloroethene-contaminated fractured dolomite aquifer: electron source and microbial communities involved in reductive dechlorination. Environ. Sci. Technol. 35:4449-4456. [DOI] [PubMed] [Google Scholar]
- 26.Hunter, K. S., Y. Wang, and P. Van Cappellen. 1998. Kinetic modeling of microbially-driven redox chemistry of subsurface environments: coupling transport, microbial metabolism and geochemistry. J. Hydrol. 209:53-80. [DOI] [PubMed] [Google Scholar]
- 27.Inagaki, F., U. Tsunogai, M. Suzuki, A. Kosaka, H. Machiyama, K. Takai, T. Nunoura, K. Nealson, and K. Horikoshi. 2004. Characterization of C1-metabolizing prokaryotic communities in methane seep habitats at the Kuroshima Knoll, Southern Ryukyu Arc, by analyzing pmoA, mmoX, mxaF, mcrA, and 16S rRNA genes. Appl. Environ. Microbiol. 70:7445-7455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Islam, F. S., A. G. Gault, C. Boothman, D. A. Polya, J. M. Charnock, D. Chatterjee, and J. R. Lloyd. 2004. Role of metal-reducing bacteria in arsenic release from Bengal delta sediments. Nature 430:68-71. [DOI] [PubMed] [Google Scholar]
- 29.Jeon, C. O., P. Padmanabhan, C. M. DeRito, J. R. Snape, and E. L. Madsen. 2003. Discovery of a bacterium, with distinctive dioxygenase, that is responsible for in situ biodegradation in contaminated sediment. Proc. Natl. Acad. Sci. U. S. A. 100:13591-13596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kartal, B., M. M. M. Kuypers, G. Lavik, J. Schalk, H. J. M. Op den Camp, M. S. M. Jetten, and M. Strous. 2007. Anammox bacteria disguised as denitrifiers: nitrate reduction to dinitrogen gas via nitrite and ammonium. Environ. Microbiol. 9:635-642. [DOI] [PubMed] [Google Scholar]
- 31.Kelso, B. H. L., R. V. Smith, R. J. Laughlin, and D. S. Lennox. 1997. Dissimilatory nitrate reduction in anaerobic sediments leading to river nitrite accumulation. Appl. Environ. Microbiol. 63:4679-4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Konneke, M., A. E. Bernhard, J. R. de la Torre, C. B. Walker, J. B. Waterbury, and D. A. Stahl. 2005. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature 437:543-546. [DOI] [PubMed] [Google Scholar]
- 33.Lam, P., G. Lavik, M. M. Jensen, J. van de Vossenberg, M. Schmid, D. Woebken, D. Gutiérrez, D. Amann, M. S. M. Jetten, and M. M. M. Kuypers. 2009. Revising the nitrogen cycle in the Peruvian oxygen minimum zone. Proc. Natl. Acad. Sci. U. S. A. 106:4752-4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lam, P., M. M. Jensen, B. Lavik, D. F. McGinnis, B. Mueller, C. J. Schubert, R. Amann, et al. 2007. Linking crenarchaeal and bacterial nitrification to anammox in the Black Sea. Proc. Natl. Acad. Sci. U. S. A. 104:7104-7109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laverman, A. M., R. W. Canavan, C. P. Slomp, and P. Van Cappellen. 2007. Potential nitrate removal in a coastal freshwater sediment (Haringvliet Lake, the Netherlands) and response to salinization. Water Res. 41:3061-3068. [DOI] [PubMed] [Google Scholar]
- 36.Laverman, A. M., P. Van Cappellen, D. Van Rotterdam-Los, C. Pallud, and J. Abell. 2006. Potential rates and pathways of microbial nitrate reduction in coastal sediments. FEMS Microbiol. Ecol. 58:179-192. [DOI] [PubMed] [Google Scholar]
- 37.Lavik, G., T. Stuehrmann, V. Bruechert, A. Van der Plas, V. Mohrholz, P. Lam, M. Mussmann, B. M. Fuchs, R. Amann, U. Lass, and M. M. M. Kuypers. 2009. Detoxification of sulphidic African shelf waters by blooming chemolithotrophs. Nature 457:581-584. [DOI] [PubMed] [Google Scholar]
- 38.Liou, J. S.-C., C. M. DeRito, and E. L. Madsen. 2008. Field-based and laboratory stable isotope probing surveys of the identities of both aerobic and anaerobic benzene-metabolizing microorganisms in freshwater sediment. Environ. Microbiol. 10:1964-1977. [DOI] [PubMed] [Google Scholar]
- 39.Luton, P. E., J. M. Wayne, R. J. Sharp, and P. W. Riley. 2002. The mcrA gene as an alternative to 16S rRNA in the phylogenetic analysis of methanogen populations in landfill. Microbiology 148:3521-3530. [DOI] [PubMed] [Google Scholar]
- 40.Madsen, E. L. 2008. Environmental microbiology: from genomes to biogeochemistry. Blackwell, Malden, MA.
- 41.Mazeas, L., V. Vigneron, and K. Le-Menach. 2008. Elucidation of nitrate reduction pathways in anaerobic bioreactors using a stable isotope approach. Rapid Commun. Mass Spectrom. 22:1746-1750. [DOI] [PubMed] [Google Scholar]
- 42.McGrath, K. C., S. R. Thomas-Hall, C. T. Cheng, L. Leo, A. Alexa, and S. Schmidt. 2008. Isolation and analysis of mRNA from environmental microbial communities. J. Microbiol. Methods 75:172-176. [DOI] [PubMed] [Google Scholar]
- 43.McGuire, J. T., E. W. Smith, D. T. Long, D. W. Hyndman, S. K. Haack, M. J. Klug, and M. A. Velbel. 2000. Temporal variations in parameters reflecting terminal-electron-accepting-processes in an aquifer contaminated with waste fuel and chlorinated solvents. Chem. Geol. 169:471-485. [Google Scholar]
- 44.McGuire, J. T., D. T. Long, M. J. Klug, S. K. Haack, and D. W. Hyndman. 2002. Evaluating behavior of oxygen, nitrate and sulfate during recharge and quantifying reaction rates in a contaminated aquifer. Environ. Sci. Technol. 36:2693-2700. [DOI] [PubMed] [Google Scholar]
- 45.McKnight, D. M., R. L. Runkel, C. M. Tate, J. H. Duff, and D. L. Moorhead. 2004. Inorganic N and P dynamics of Antarctic glacial meltwater streams as controlled by hyporheic exchange and benthic autotrophic communities. J. N. Am. Benthol. Soc. 23:171-188. [Google Scholar]
- 46.Michaelis, W., R. Seifert, K. Nauhaus, T. Treude, V. Thiel, M. Blumenberg, et al. 2002. Microbial reefs in the black sea fueled by anaerobic oxidation of methane. Science 297:1013-1015. [DOI] [PubMed] [Google Scholar]
- 47.O'Neil, R. A., D. E. Holmes, M. V. Coppi, L. A. Adams, M. J. Larrahondo, J. E. Ward, K. P. Nevin, T. L. Woodard, H. A. Vrionis, A. L. N′Guessan, and D. R. Lovley. 2008. Gene transcript analysis of assimilatory iron limitation in Geobacteraceae during groundwater bioremediation. Environ. Microbiol. 10:1218-1230. [DOI] [PubMed] [Google Scholar]
- 48.Oremland, R. S., and J. F. Stolz. 2003. The ecology of arsenic. Science 300:939-944. [DOI] [PubMed] [Google Scholar]
- 49.Pett-Ridge, J., W. L. Silver, and M. K. Firestone. 2006. Redox fluctuations frame microbial community impacts on N-cycling rates in a humid tropical forest soil. Biogeochemistry 81:95-110. [Google Scholar]
- 50.Poretsky, R. S., I. Hewson, S. Sun, A. E. Allen, J. P. Zehr, and M. A. Moran. 2009. Comparative day/night metatranscriptomic analysis of microbial communities in the North Pacific subtropical gyre. Environ. Microbiol. 6:1358-1375. [DOI] [PubMed] [Google Scholar]
- 51.Poretsky, R. S., N. Bano, A. Buchan, G. LeCleir, J. Kleikemper, M. Pickering, W. M. Pate, M. A. Moran, and J. T. Hollibaugh. 2005. Analysis of microbial gene transcripts in environmental samples. Appl. Environ. Microbiol. 71:4121-4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Raghoebarsing, A. A., A. Pol, K. T. van de Pas-Schoonen, A. J. P. Smolders, K. F. Ettwig, W. I. C. Rijpstra, et al. 2006. A microbial consortium couples anaerobic methane oxidation to denitrification. Nature 440:918-921. [DOI] [PubMed] [Google Scholar]
- 53.Rotthauwe, J.-H., K. P. Witzel, and W. Liesack. 1997. The ammonia monooxygenase structural gene amoA as a functional marker: molecular fine-scale analysis of natural ammonia-oxidizing population. Appl. Environ. Microbiol. 63:4704-4712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schink, B. 2002. Synergistic interactions in the microbial world. Antonie Van Leeuwenhoek 81:257-261. [DOI] [PubMed] [Google Scholar]
- 55.Schumacher, W., and P. M. H. Kroneck. 1992. Anaerobic energy-metabolism of the sulfur-reducing bacterium spirillum 5175 during dissimilatory nitrate reduction to ammonia. Arch. Microbiol. 157:464-470. [Google Scholar]
- 56.Silver, W. L., D. J. Herman, and M. K. Firestone. 2001. Dissimilatory nitrate reduction to ammonium in upland tropical forest soils. Ecology 82:2410-2416. [Google Scholar]
- 57.Silver, W. L., A. W. Thompson, A. Reich, J. J. Ewel, and M. K. Firestone. 2005. Nitrogen cycling in tropical plantation forest: potential controls on nitrogen retention. Ecol. Appl. 15:1604-1614. [Google Scholar]
- 58.Simon, J. 2002. Enzymology and bioenergetics of respiratory nitrite ammonification. FEMS Microbiol. Rev. 26:285-309. [DOI] [PubMed] [Google Scholar]
- 59.Simon, K. S., J. Gibert, P. Petiot, and R. Laurent. 2001. Spatial and temporal patterns of bacterial density and metabolic activity in a Karst aquifer. Arch. Hydrobiol. 151:67-82. [Google Scholar]
- 60.Smith, C. J., D. B. Nedwell, L. F. Dong, and A. M. Osborn. 2007. Diversity and abundance of nitrate reductase genes (narG and napA), nitrite reductase genes (nirS and nrfA), and their transcripts in estuarine sediments. Appl. Environ. Microbiol. 73:3612-3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sorensen, J. 1978. Capacity for denitrification and reduction of nitrate to ammonia in a coastal marine sediment. Appl. Environ. Microbiol. 35:301-305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stumm, W., and J. J. Morgan. 1996. Aquatic chemistry: chemical equilibria and rates in natural waters, 3rd ed. Wiley, New York, NY.
- 63.Templer, P. H., W. L. Silver, and J. Pett-Ridge. 2008. Plant and microbial controls on nitrogen retention and loss in a humic tropical forest. J. Ecol. 89:3030-3040. [DOI] [PubMed] [Google Scholar]
- 64.Tiedje, J. M. 1988. Ecology of denitrification and dissimilatory nitrate reduction to ammonium, p. 179-244. In A. J. B. Zehnder (ed.), Biology of anaerobic microorganisms. Wiley & Sons, New York, NY.
- 65.Tiedje, J. M., A. J. Sextone, D. D. Myrold, and J. A. Robinson. 1982. Denitrification: ecological niches, competition and survival. Antonie Van Leeuwenhoek 48:569-583. [DOI] [PubMed] [Google Scholar]
- 66.Tiedje, J. M., J. Sørensen, and Y.-Y. L. Chang. 1981. Assimilatory and dissimilatory nitrate reduction: perspectives and methodology of simultaneous measurements of several nitrogen cycle processes. Terr. Nitr. Cycles Ecol. Bull. 33:331-342. [Google Scholar]
- 67.Urich, T., A. Lanzén, J. Qi, D. H. Huson, C. Schleper, and S. C. Schuster. 2008. Simultaneous assessment of soil microbial community structure and function through analysis of the meta-transcriptome. PLoS One 3:e2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.van Breukelen, B. M., and J. Griffoen. 2004. Biogeochemical processes at the fringe of a landfill leachate pollution plume: potential for dissolved organic carbon, Fe(II), Mn(II), NH4, and CH4 oxidation. J. Contam. Hydrol. 73:181-205. [DOI] [PubMed] [Google Scholar]
- 69.Vroblesky, D. A., and F. H. Chapelle. 1994. Temporal and spatial changes of terminal electron accepting processes in a petroleum hydrocarbon contaminated aquifer and the significance for contaminant biodegradation. Water Resour. Res. 30:1561-1570. [Google Scholar]
- 70.Wawrik, B., J. H. Paul, and F. R. Tabita. 2002. Real-time PCR quantification of rbcL (ribulose-1,5-bisphosphate carboxylase/oxygenase) mRNA in diatoms and pelagophytes. Appl. Environ. Microbiol. 68:3771-3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Weiss, J. V., and I. M. Cozzarelli. 2008. Biodegradation in contaminated aquifers: incorporating microbial/molecular methods. Groundwater 46:305-322. [DOI] [PubMed] [Google Scholar]
- 72.Welsh, D. T., G. Castadelli, M. Bartoli, D. Poli, M. Careri, R. deWit, and P. Viaroli. 2001. Denitrification in an intertidal seagrass meadow, a comparison of 15N-isotope and acetylene-block techniques: dissimilatory nitrate reduction to ammonia as a source of N2O? Mar. Biol. 139:1029-1036. [Google Scholar]
- 73.White, D. 2000. The physiology and biochemistry of prokaryotes, 2nd ed. Oxford University Press, New York, NY.
- 74.Wilson, M. S., C. Bakermans, and E. L. Madsen. 1999. In situ, real-time catabolic gene expression: extraction and characterization of naphthalene dioxygenase mRNA transcripts from groundwater. Appl. Environ. Microbiol. 65:80-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wilson, M. S., and E. L. Madsen. 1996. Field extraction of a transient intermediary metabolite indicative of real time in situ naphthalene biodegradation. Environ. Sci. Technol. 30:2099-2103. [Google Scholar]
- 76.Wilson, R. D., S. F. Thornton, and D. M. Mackay. 2004. Challenges in monitoring the natural attenuation of spatially variable plumes. Biodegradation 15:359-369. [DOI] [PubMed] [Google Scholar]
- 77.Winderl, C., S. Schaefer, and T. Lueders. 2007. Detection of anaerobic toluene and hydrocarbon degraders in contaminated aquifers using benzylsuccinate synthase (bssA) genes as a functional marker. Environ. Microbiol. 9:1035-1046. [DOI] [PubMed] [Google Scholar]
- 78.Yager, R. M., S. E. Bilotta, C. L. Mann, and E. L. Madsen. 1997. Metabolic adaptation and in situ attenuation of chlorinated ethenes by naturally occurring microorganisms in a fractured dolomite aquifer near Niagara Falls, New York. Environ. Sci. Technol. 31:3138-3147. [Google Scholar]
- 79.Yagi, J. M., E. F. Neuhauser, J. A. Ripp, D. M. Mauro, and E. L. Madsen. 2010. Subsurface ecosystem resilience: long-term attenuation of subsurface contaminants supports an elaborate eukaryotic food chain and a dynamic microbial community. ISME J. 4:131-143. [DOI] [PubMed] [Google Scholar]
- 80.Yagi, J. M., and E. L. Madsen. 2009. Diversity, abundance, and consistency of microbial oxygenase expression and biodegradation in a shallow contaminated aquifer. Appl. Environ. Microbiol. 75:6478-6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Young, L. Y., and C. D. Phelps. 2005. Metabolic biomarkers for monitoring in situ anaerobic hydrocarbon degradation. Environ. Health Perspect. 113:62-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.