Abstract
Burkholderia cenocepacia is a multidrug-resistant opportunistic pathogen that infects the airways of patients with cystic fibrosis (CF) and can survive intracellularly in macrophages and epithelial cells. The gentamicin protection assay, which relies on the poor ability of gentamicin or other aminoglycosides to permeate eukaryotic cell membranes, is traditionally employed to quantify intracellular bacteria. However, the high resistance of these bacteria to aminoglycosides hampers the use of the gentamicin protection assay to investigate intracellular infection by B. cenocepacia. Here, we report the construction of gentamicin-sensitive strains of B. cenocepacia carrying a deletion of the BCAL1674, BCAL1675, and BCAL1676 genes that form an operon encoding an AmrAB-OprA-like efflux pump. We show that bacteria carrying this deletion are hypersensitive to gentamicin and also delay phagolysosomal fusion upon infection of RAW 264.7 murine macrophages, as previously demonstrated for the parental strain. We also demonstrate for the first time that low concentrations of gentamicin can be used to effectively kill extracellular bacteria and reliably quantify the intracellular infection by B. cenocepacia, which can replicate in RAW 264.7 macrophages.
Burkholderia cenocepacia is a member of the Burkholderia cepacia complex (Bcc), a group of phenotypically similar Gram-negative bacteria that are found ubiquitously in nature (14). Over the past 20 years, species of this complex have emerged as important opportunistic pathogens of immunocompromised individuals, especially in patients suffering from cystic fibrosis (CF) and chronic granulomatous disease (33, 53). Infections in CF patients by B. cenocepacia are particularly alarming since bacteria are transmissible between CF patients, and a proportion of infected individuals rapidly deteriorate and develop cepacia syndrome, a necrotizing pneumonia that is virtually always fatal (23, 25, 27, 34). Furthermore, treatment of B. cenocepacia infections is often difficult because these bacteria, along with other members of the Bcc, are highly resistant to most clinically useful antibiotics (1, 16).
B. cenocepacia can survive intracellularly within a variety of eukaryotic cells such as amoebae, epithelial cells, and macrophages (10, 30, 31, 39, 40, 47, 50, 53, 56). However, the capacity for intracellular replication of B. cenocepacia in eukaryotic cells and the factors involved in bacterial survival remain unclear. Some studies have reported that B. cenocepacia and other Bcc strains can replicate inside eukaryotic cells (6, 31, 36, 40, 46, 47, 59) while others have shown minimal to no replication (29, 30, 39, 45) (Table 1 summarizes intracellular survival/replication assays with Burkholderia species in phagocytic cells of mammalian origin). These discrepancies could be due to variations in the experimental procedure, including differences in eukaryotic cell types and Bcc strains, and the method for quantification of intracellular replication. Also, depending on the time of the invasion assay, phagocytic cells can display signs of toxicity and various forms of cell death (36, 45; D. Hynes and M. A. Valvano unpublished). Therefore, it is difficult to conclusively demonstrate intracellular replication in the presence of cell death since intracellular bacteria may be able to replicate better in dying cells (36). Traditionally, intracellular survival and replication of bacteria in eukaryotic cells have been studied and quantified using the gentamicin protection assay (20). In this assay, gentamicin is added to kill extracellular bacteria while intracellular bacteria remain protected from gentamicin killing due to the poor ability of this antibiotic to permeate eukaryotic cells. The survival of intracellular bacteria can then be determined by lysing infected eukaryotic cells with mild, nonionic detergents, followed by bacterial quantification through serial dilution and colony counts. Unfortunately, the gentamicin protection assay is not suitable for the study of intracellular B. cenocepacia since this bacterium is highly resistant to gentamicin and other aminoglycosides. To overcome this limitation, several laboratories have modified this assay and utilized a combination of ceftazidime and gentamicin at very high concentrations ranging from 500 to 1,000 μg ml−1 for ceftazidime and 250 to 500 μg ml−1 for gentamicin to eliminate extracellular bacteria (6, 47). These modifications are not ideal since aminoglycosides can penetrate eukaryotic cells by pinocytosis and may accumulate in phagolysosomes, causing bacterial cell death (19) or rapidly affecting the normal physiology of intracellular bacteria by causing bacterial stress (41). These effects depend on the vacuolar pH and the aminoglycoside concentration as the bacterial sensitivity to aminoglycosides decreases markedly at acidic pH (41). These observations are relevant to the intracellular survival of B. cenocepacia in macrophages, which occurs in a bacteria-containing vacuole that delays acidification and maintains a relatively high pH for several hours postinfection (29). Aminoglycosides can also escape to the cytosol (52), promoting mistranslation of eukaryotic proteins (4, 18) and thereby compromising the physiology of the host cell. Also, high concentrations of ceftazidime can cause toxicity in macrophages, as determined by the release of lactic acid dehydrogenase (28).
TABLE 1.
Intracellular replication/survival assays in phagocytic cells infected with Burkholderia species
| Species and strain(s) (description) | Host cell type | Infection assay conditionsa | Intracellular survival |
Reference | |
|---|---|---|---|---|---|
| No. of CFU (log10 units) | Time (h) of detection of maximum no. of CFU | ||||
| B. ambifaria | |||||
| Several strains | THP-1 human monocytes | MOI, 10; 125 μg ml−1 Tet and 1 mg ml−1 Cfz for 8 h | 0.5 | 8 | 59 |
| B. cenocepacia | |||||
| J2315 | U937 human monocytes | MOI, 10; 1 mg ml−1 Cfz and 1 mg ml−1 Ak for 2 h; no antibiotic thereafter | 1 | 24 | 40 |
| K56-2 | U937 human monocytes | MOI, 10; 500 μg ml−1 Cfz and 250 μg ml−1 Gm for 2 h; 100 μg ml−1 Gm thereafter | 1 | 20 | 46 |
| C6433 | Human dendritic cells | MOI, 0.3-1; 8 μg ml−1 Mer for 24 h | 2 | 24 | 36 |
| B. multivorans | |||||
| C5568 | Human dendritic cells | MOI, 0.3-1; 8 μg ml−1 Mer for 24 h | 0.5 | 24 | 36 |
| B. vietnamiensis | |||||
| CEP040 | PU5-1.8 murine macrophages | MOI, 1-5; 1 mg ml−1 Cfz and 500 μg ml−1 Kn for 2 h; 25 μg ml−1 Kn thereafter | 1b | 24 | 45 |
| B. mallei | |||||
| SR1A (ATCC 23344 with a deletion of type VI secretion cluster) | RAW 264.7 murine macrophages | MOI, 1; 200 μg ml−1 Gm for 24 h | 24 | 9 | |
| ATCC 23344 | J774.2 murine macrophages | MOI, 10; 250 μg ml−1 Kn for 18 h | 2 | 18 | 44 |
| B. pseudomallei | |||||
| Clinical isolates 0035, 0037, and 0038 | Human monocytes | MOI, 10; no use of antibiotics reported | 2-3 | 18 | 43 |
| 316c | Rat alveolar macrophages | MOI, 10; 100 μg ml−1 Kn for 2 h; 20 μg ml−1 Kn thereafter | 2 | 16 | 26 |
| 10276 | J774.2 murine macrophages | MOI, 1; 250 μg ml−1 Kn for 12 h | 2 | 12 | 55 |
| Various isogenic morphotypes of strain 153 | J774A.1 murine macrophages | MOI, 25-50; 250 μg ml−1 Kn for 2 h; 20 μg ml−1 Kn for 8 h | 1.5 | 8 | 11 |
| DD503 (1026b; ΔamrR-oprA) | RAW 264.7 murine macrophages | MOI, 10; 250 μg ml−1 Kn for 18 h | 2 | 18 | 8 |
| 1026b | RAW 264.7 murine macrophages | MOI, 2; antibiotics used but not specified | 1.5 | 8 | 13 |
| KHW | C57BL/6 mouse bone-marrow derived monocytes | MOI, 3, 100 μg ml−1 Kn | >1c | 6 | 5 |
Ak, amikacin; Cfz, ceftazidime; Gm, gentamicin; Kn, kanamycin; Mer, meropemen; Tet, tetracycline.
Only in the absence of 25 μg ml−1 kanamycin.
For untreated cells; no replication in IFN-γ-treated cells.
We sought to overcome these shortcomings by constructing a gentamicin-sensitive strain of B. cenocepacia that can be used to study intracellular infections using gentamicin protection assays. In this study, we report the construction of such a mutant and show not only that it can be effectively killed using low concentrations of gentamicin but also that it displays the same trafficking properties in macrophages that we have previously reported for the parental isolate (29, 38, 48). Using macrophage infections and gentamicin protection assays, we demonstrate that B. cenocepacia gentamicin-sensitive mutants replicate intracellularly in RAW 264.7 macrophages.
MATERIALS AND METHODS
Bacterial strains, cell lines, growth conditions, and reagents.
B. cenocepacia strains J2315 and K56-2 are clinical isolates from CF patients and belong to the highly transmissible epidemic ET12 clone (37). Escherichia coli strains DH5α [λ− φ 80dlacZΔ M15 Δ(lacZYA-argF)U169 recA1 endA hsdR17(rK− mK−) supE44 thi-1 gyrA relA1] (laboratory stock) and GT115 [F− mcrA Δ(mrr-hsdRMS-mcrBC) φ80ΔlacZΔM15 ΔlacX74 recA1 rpsL (StrA) endA1 Δdcm uidA(ΔMluI)::pir-116 ΔsbcC-sbcD] (Invivogen, San Diego, CA) were used for cloning experiments. E. coli and B. cenocepacia were grown in Luria-Bertani (LB) broth with shaking or on LB agar plates and were incubated at 37°C. When necessary, E. coli cultures were supplemented with tetracycline (20 μg ml−1), kanamycin (40 μg ml−1), trimethoprim (50 μg ml−1), and chloramphenicol (30 μg ml−1). RAW 264.7 murine macrophages were obtained from the American Type Culture Collection (Manassas, VA) and were maintained in antibiotic-free Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS). RAW 264.7 cells were incubated at 37°C in a humidified atmosphere containing 5% carbon dioxide. Antibiotics and chemicals were purchased from Sigma Chemical (St. Louis, MO). LysoTracker Red DND-99 was purchased from Invitrogen (Eugene, OR).
General molecular techniques and reagents.
DNA manipulations and cloning were performed as described previously (51). Restriction endonuclease and T4 DNA ligase were purchased from Roche (Roche Diagnostics, Laval, Quebec, Canada) and were used as recommended by the manufacturer. DNA amplification was performed by PCR using Taq DNA polymerase or Proof Start polymerase (Qiagen Inc., Mississauga, Ontario, Canada). Plasmid DNA was isolated using a QiaPrep Spin kit (Qiagen). PCR products were purified using a QIAquick PCR purification kit or a QIAquick gel extraction kit (Qiagen). Transformation of E. coli was done by the calcium chloride method (15). Mobilization of plasmids into B. cenocepacia was achieved by triparental mating (17) using the helper plasmid pRK2013 (21).
Antimicrobial susceptibility testing.
Growth rates and MICs were determined as previously described (35). Overnight cultures, grown in LB broth, were diluted to an optical density at 600 nm (OD600) of 0.002 in LB broth, and antibiotics were added to samples at 2-fold dilutions. Samples were then aliquoted into 100-well plates, which were incubated at 37°C with constant shaking for 24 h in a Bioscreen C automated growth curve analyzer (MTX Lab Systems, Inc., Vienna, VA). Growth rate was determined by measuring OD600 readings taken every hour. The MIC was determined as the lowest concentration of antibiotic that inhibited visible growth after 24 h of incubation.
Construction of gentamicin-sensitive deletion strains.
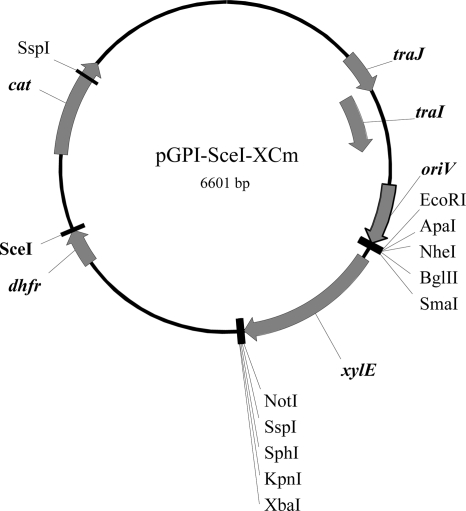
Deletion of BCAL1674-BCAL1676 was achieved with a homing endonuclease mutagenesis system described previously (22). This system relies on a suicide vector that contains an I-SceI restriction site (pGPI-SceI) and a replicative but unstable plasmid that encodes the I-SceI endonuclease (pDAI-SceI). To improve genetic manipulations in B. cenocepacia J2315 (see below), we modified pGPI-SceI by introducing a chloramphenicol resistance determinant and the xylE reporter gene. The chloramphenicol acetyltransferase (cat) gene from pAp2 (32) was PCR amplified using primers 3994 (5′-TTACTATCTAGACTGCAGATCGATAAGTATAGGAACTTCGGCGC-3′; PstI site is underlined) and 3995 (5′-GTAATCTAGAGACTGCAGATCGATTCATCGCAGTACTGTTG-3′; PstI site is underlined) and with pAp2 DNA as a template. The resulting amplicon was digested with PstI and cloned into PstI-linearized pGPI-SceI to create pGPI-SceI-Cm. The xylE gene from pMo130 (24) was amplified using primers 3925 (5′-TGGGAATTCGGGCCCCGCTAGCCAGATCTTCCCGGGAAG-3′; EcoRI site is underlined) and 3929 (5′-CTATAGTCTAGACGGTACCGCATGCAATATTGGCGGCCGCTCAGGTCAGGTCAGCAC-3′; XbaI site is underlined) and pMo130 DNA as a template (24). This PCR product was digested with EcoRI and XbaI and cloned into pGPI-SceI-Cm linearized with the same enzymes, resulting in pGPI-SceI-XCm (Fig. 1).
FIG. 1.
Schematic map indicating the salient features and restriction sites of pGPI-SceI-XCm. cat, chloramphenicol acetyltransferase gene; dhfr, dihydrofolic acid reductase gene; traJ-I, plasmid mobilization region; oriV, replication region; xylE, 2,3-catechol-dioxygenase gene.
To facilitate detection of cured colonies carrying the desired gene deletion, we added a modified sacB gene into pDAISce-I. The sacB gene was amplified from pMo130 DNA using primers 4160 (5′-TAGCGACTCGAGCTAGCGCCTTCTTGACGAGTTCTTCTG-3′; NheI site is underlined) and 4161 (5′-ATCGACGCTAGCTCGAGCAAGCTGCAGTTATTTGTTAAC-3′; NheI site is underlined). The PCR product was digested with NheI and cloned into NheI linearized pDAISce-I to create pDAISce-I-SacB.
The mutagenesis plasmid for the BCAL1674-BCAL1676 deletion was constructed by PCR amplification of 700-bp DNA fragments flanking this gene cluster, which were cloned into pGPI-SceI-XCm. The upstream fragment was amplified using primer 3928 (5′-ATCGATAATATTCGGCGGCGAGCTCGCCGGCGGCGATCG-3′; SspI site is underlined) and 3855 (5′-GTCGATGGTACCGCCCTTTCCGCATCCGGCCACGGCCAG-3′; KpnI site is underlined). The downstream fragment was amplified using primer 3856 (5′-GATCGAGGTACCCTCGAACTGCTCGACGCGCAGCGCAGC-3′; KpnI site is underlined) and 3927 (5′-GTCGCATCTAGAACATTGTCCTGCGCCGCCATCG-3′; XbaI site is underlined). The upstream fragment was digested with SspI and KpnI, the downstream fragment was digested with XbaI and KpnI, and both fragments were ligated into pGPI-SceI-XCm to create pMH304. The mutagenic pMH304 plasmid was mobilized into B. cenocepacia J2315 and K56-2 by triparental mating. Trimethoprim at 100 μg ml−1 was used to select for cointegrants in strain K56-2, and both trimethoprim and chloramphenicol at 200 μg ml−1 and 400 μg ml−1, respectively, were used to select for cointegrants in J2315. To distinguish true cointegrants from colonies that spontaneously became resistant to chloramphenicol and/or trimethoprim, plates were sprayed with catechol since in the presence of this compound colonies expressing 2,3-catechol-dioxygenase encoded by xylE turn a bright yellow (24). To prevent the potential isolation of B. cenocepacia gentamicin-resistant suppressor mutants, ampicillin (200 μg ml−1) and polymyxin B (25 μg ml−1) instead of gentamicin were used for selection against the E. coli donor and helper strains after triple mating. For the final mutagenesis stage, pDAISce-I-SacB was mobilized into B. cenocepacia cointegrants, and exconjugants were selected with 100 μg ml−1 and 250 μg ml−1 tetracycline for K56-2 and J2315 derivatives, respectively. Tetracycline-resistant colonies were screened by PCR to confirm the deletion using the primers 4017 (5′-CCCTTGCCGGCATTGGCTCATGC-3′) and 4018 (5′-CGGCGTCGAGCATCGCCTCGGCGTCGTTC-3′) that anneal to sequences outside the deleted region. Detection of deletion mutants cured from the plasmid pDAISce-I-SacB was achieved by growing B. cenocepacia on LB plates without salt and supplemented with 5% (wt/vol) sucrose and then screening the resulting colonies for loss of tetracycline resistance.
Microscopy.
Macrophage infections and microscopic analysis were performed as previously described (30, 48). RAW 264.7 macrophages were grown on glass coverslips in six-well plates and maintained in DMEM with 10% FBS at 37°C in a humidified atmosphere with 5% carbon dioxide. Overnight cultures of B. cenocepacia were washed three times with DMEM-FBS and used to infect macrophages at a multiplicity of infection (MOI) of 50. Upon infection, plates were centrifuged for 1 min at 300 × g to allow for bacterial contact with macrophages. Infection was allowed to proceed for 4 h at 37°C under 5% carbon dioxide. Infected macrophages were washed with phosphate-buffered saline (PBS), and 0.5 μM LysoTracker Red DND-99 was added for 1 min to stain acidic vacuoles. Coverslips were then visualized using an Axioscope 2 (Carl Zeiss) microscope with a 100× oil immersion objective.
Kill curves.
Efflux pump mutant strains MH1K and MH1J were grown overnight in LB broth at 37°C with shaking. Cultures were washed three times with DMEM-FBS, diluted to 106 CFU ml−1 in the same medium, and statically incubated in a six-well plate at 37°C in a humidified atmosphere with 5% carbon dioxide for 2 h. After this time, gentamicin at 10 μg ml−1 or distilled water (vehicle control) was added to the wells. Samples from each well were taken, serially diluted, and plated onto LB agar to enumerate viable bacteria.
Gentamicin protection assay.
Gentamicin protection assays were performed using a modified protocol described for Salmonella enterica (54). RAW 264.7 macrophages were grown in six-well plates at 1 × 106 cells per well and incubated under conditions described above. Overnight bacterial cultures were grown in LB broth at 37°C with shaking. Bacterial cultures were washed three times with DMEM plus FBS and were used to infect macrophages at an MOI of 1. Upon infection, plates were centrifuged for 1 min at 300 × g and were incubated for 2 h at 37°C under 5% carbon dioxide. Infected macrophages were washed three times with PBS to remove nonadherent bacteria. To kill extracellular bacteria, DMEM plus FBS containing 50 μg ml−1 gentamicin was added for 30 min, removed by aspiration, and replaced with fresh medium containing 10 μg ml−1 gentamicin for the remainder of the experiment. To enumerate intracellular bacteria, infected macrophages were washed once with PBS and then lysed with 1% Triton X-100 in PBS at the appropriate times. Recovered intracellular bacteria were quantified by plating serial dilutions on LB agar plates and enumerating colony counts.
Statistical analyses.
The statistical significance of differences in the data was determined by a Student's t test using the GraphPad Prism software package, version 4.0 (GraphPad Software, La Jolla, CA).
RESULTS AND DISCUSSION
BCAL1674-BCAL1676 encode a putative efflux pump that confers resistance to aminoglycoside antibiotics.
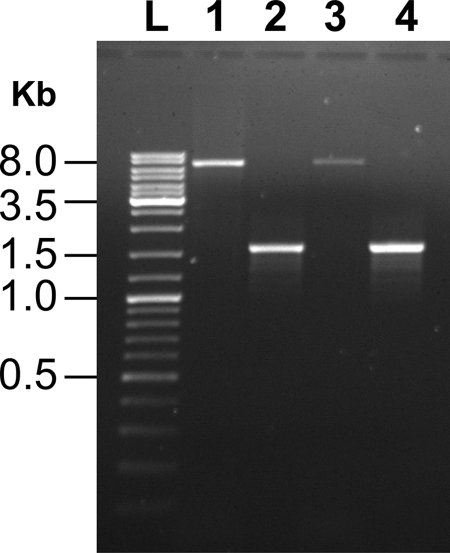
In Burkholderia pseudomallei, a multidrug efflux pump encoded by the three-gene cluster amrAB-oprA mediates resistance to aminoglycoside antibiotics (42, 57). We scanned the genome of B. cenocepacia strain J2315 using position-specific iterated (PSI) BLAST (2) for genes encoding putative AmrAB-OprA-like efflux pumps. Our scan revealed a three-gene cluster (annotated as BCAL1674, BCAL1675, and BCAL1676) encoding proteins that share 73%, 79%, and 86% amino acid sequence identity to B. pseudomallei 1106a OprA, AmrA, and AmrB proteins, respectively. To assess whether this putative efflux pump mediates aminoglycoside resistance in B. cenocepacia, we constructed a markerless deletion of the genes BCAL1674-BCAL1676 in B. cenocepacia strains J2315 and K56-2, resulting in the strains MH1J and MH1K, respectively. PCR amplification using genomic DNA from the parental strains and primers flanking the deletion endpoints (as described in Materials and Methods) yielded an amplicon of ∼7.1 kb in both cases (Fig. 2, lanes 1 and 3). In contrast, an ∼1.6-kb fragment was amplified from the genomes of the deletion mutants (Fig. 2, lanes 2 and 4), demonstrating that a deletion of the expected size (∼5.5 kb) had occurred.
FIG. 2.
Confirmation that MH1K and MH1J strains carry a 5.5-kb deletion of BCAL1674-BCAL1676. Agarose gel showing PCR products that were amplified using primers that anneal outside the deletion endpoints, as indicated in Materials and Methods. Lane L, DNA ladder; lanes 1 and 3, 7.1-kb PCR product amplified from genomic DNA of parental K56-2 and J2315 strains, respectively; lanes 2 and 4, 1.6-kb PCR product amplified from genomic DNA of MH1K and MH1J mutant strains, respectively.
The susceptibilities of parental and mutant strains to various aminoglycoside and nonaminoglycoside antibiotics were tested. The MICs of MH1J and MH1K to gentamicin were 8 μg ml−1 compared to MIC values greater than 1,024 μg ml−1 in the parental strains (Table 2). Similarly, the MIC values of MH1J and MH1K to tobramycin decreased to 8 μg ml−1 compared to MIC values of 512 μg ml−1 for the parental strains (Table 2). The kanamycin MIC for MH1J and MH1K was 64 μg ml−1 versus MIC values greater than 1,024 μg ml−1 for the respective parental strains (Table 2). The sensitivities of our deletion mutants to aminoglycosides are similar to those observed in a ΔamrAB-oprA B. pseudomallei mutant (12, 42), suggesting that the function of this pump in both species is conserved. In addition, a 4-fold reduction in MIC values to tetracycline was observed in MH1J and MH1K compared to their respective parental strains, while chloramphenicol MIC values remained unchanged in mutant and parental bacteria (Table 2). These results suggest that this efflux pump may also contribute to resistance to tetracycline but not chloramphenicol. Taken together, our results demonstrate that the efflux pump encoded by BCAL1674-BCAL1676 was critical for the resistance of B. cenocepacia J2315 and K56-2 against aminoglycoside antibiotics. Furthermore, growth rates of MH1J and MH1K were similar to those of the corresponding parental strains, indicating that the deletion of these genes has no major effects on the normal physiology of B. cenocepacia (data not shown).
TABLE 2.
MIC values for parental and mutant strains of B. cenocepacia
| Antibiotic | MIC for the strain (μg/ml)a |
|||
|---|---|---|---|---|
| K56-2 | MH1K | J2315 | MH1J | |
| Gentamicin | >1,024 | 8 | >1,024 | 8 |
| Tobramicin | 512 | 8 | 512 | 8 |
| Kanamycin | >1,024 | 64 | >1,024 | 64 |
| Tetracycline | 32 | 8 | 64 | 16 |
| Chloramphenicol | 64 | 64 | 64 | 64 |
K56-2 and J2315 are the parental B. cenocepacia strains, and MH1K and MH1J are their respective efflux pump deletion mutants. MICs were recorded as the minimal concentration of antibiotic to inhibit visible growth in LB broth after 24 h. Results represent triplicate readings from two independent experiments.
The MH1J and MH1K mutants are suitable for gentamicin protection assays.
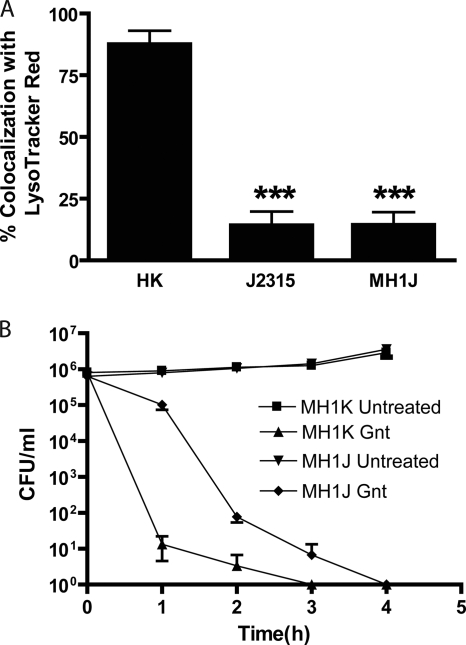
The primary objective of this work was to obtain a gentamicin-sensitive strain of B. cenocepacia that could be used in gentamicin protection assays to reliably quantify intracellular bacteria in eukaryotic cells. Therefore, we determined whether the BCAL1674-BCAL1676 deletion affected the trafficking of B. cenocepacia in macrophages compared to the parental strains. Infected macrophages were treated with LysoTracker Red, which is an acidotropic red fluorescent dye that accumulates in lysosomes and acidic cell organelles (3, 58). We have previously used this methodology to demonstrate that in contrast to heat-killed bacteria, which reach acidic compartments rapidly after internalization, live B. cenocepacia cells delay phagosomal-lysosomal fusion and do not colocalize with LysoTracker Red-rich vacuoles for at least 4 h postinfection (29, 48). RAW 264.7 macrophages were infected with live and heat-inactivated B. cenocepacia strains J2315 and MH1J, and colocalization of bacteria-containing vacuoles with LysoTracker Red was determined by fluorescence microscopy. At 4 h postinfection, 87.5% ± 9.5% of heat-killed bacteria colocalized with LysoTracker Red while the colocalization of LysoTracker Red with live J2315 and MH1J cells was 14.2% ± 9.6% and 14.3% ± 8.9%, respectively (Fig. 3A). These results demonstrate that B. cenocepacia strain J2315 and its ΔBCAL1674-BCAL1676 derivative display the same trafficking properties in macrophages.
FIG. 3.
Characterization of the gentamicin-sensitive B. cenocepacia strains. (A) Percentage of LysoTracker Red colocalization with bacterium-containing vacuoles. The values represent the average and standard error of three experiments in which 21 fields of view were examined. Significant differences were determined using the unpaired t test. ***, P < 0.001 between heat-killed cells (HK) and J2315 or MH1J. (B) Gentamicin killing of efflux pump deletion mutants. Efflux pump mutant strains of B. cenocepacia MH1K and MH1J, grown in DMEM plus FBS, were treated with 10 μg ml−1 gentamicin (Gnt) or H2O (untreated), and viability was monitored through plating onto LB agar plates. Results represent the average and standard error of three independent experiments.
Next, the ability of gentamicin to effectively kill MH1J and MH1K was evaluated. MH1J and MH1K strains were grown in macrophage cell culture medium consisting of DMEM with 10% FBS and challenged with 10 μg ml−1 gentamicin. Bacterial viability was determined by serial dilution and colony counts. After 1 h of gentamicin treatment, MH1K cells displayed a rapid decline in viability (reduction of 5.8 log10 units), and the culture was completely sterilized within 3 h of antibiotic treatment. The viability of MH1J bacteria was reduced by 3.9 log10 units 2 h after gentamicin treatment, and the culture was completely sterilized by 4 h (Fig. 3B). No growth was observed for either mutant strain beyond 4 h after the addition of gentamicin (data not shown). In contrast, untreated MH1K and MH1J cultures continued to grow in DMEM-FBS, indicating that the killing observed in treated samples is specific to the antibacterial activity of gentamicin (Fig. 3B). These results demonstrate that the deletion of the amrAB-oprA-like efflux pump gene cluster in B. cenocepacia leads to effective bacterial killing by low concentrations of gentamicin. Therefore, we concluded that MH1K and MH1J mutants are suitable for gentamicin protection assays.
B. cenocepacia gentamicin-sensitive mutants replicate intracellularly in RAW 264.7 macrophages.
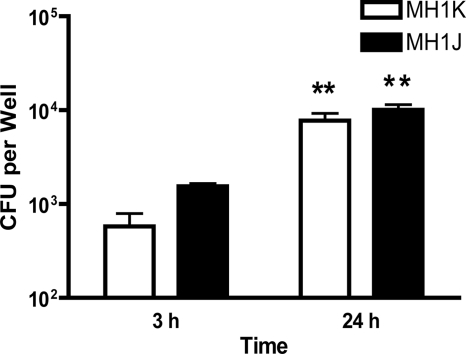
The efflux pump mutants MH1K and MH1J were tested for their ability to survive and replicate in RAW 264.7 macrophages, which were infected for 2 h at an MOI of 1. Gentamicin was added for 30 min at 50 μg ml−1 to kill extracellular bacteria. Infected cells were further incubated for 3 or 24 h in 10 μg ml−1 gentamicin, as indicated in Materials and Methods. Even though gentamicin effectively kills the B. cenocepacia mutants at 10 μg ml−1, we chose to use a higher concentration of antibiotic at the beginning of the experiments to completely ensure rapid sterilization of the extracellular medium. To enumerate intracellular bacteria, infected macrophages were washed and lysed, and the lysates were serially diluted and plated to determine bacterial counts at 3 and 24 h postinfection. The average number of bacteria in the wash after 24 h was 62 bacteria per well for MH1K and 31 bacteria per well for MH1J, which corresponds to 0.7% and 0.3% of the average number of bacterial cells in each well at 24 h. The presence of a relatively small number of bacteria in the wash is likely due to detachment of some infected macrophages during the wash steps. This was confirmed by observing under the microscope the presence of macrophages containing bacteria after low-speed centrifugation of the wash (data not shown). From these experiments we concluded that the presence of residual bacteria in the wash was negligible, demonstrating the effectiveness of gentamicin in killing extracellular bacteria. At 3 h postinfection, the MH1J strain was internalized 2.6-fold more than the MH1K strain (Fig. 4), a result consistent with our previous finding regarding the antiphagocytic activity of the B. cenocepacia O antigen (49), which is expressed on the surface of K56-2 (the parental strain for MH1K) but absent from J2315. Upon further incubation to 24 h, the number of intracellular bacteria increased by 13.5-fold (1.1 log10 units) for MH1K and by 6.5-fold (0.8 log10 units) for MH1J compared to the number of intracellular bacteria at 3 h postinfection (Fig. 4). The difference in the intracellular replication rates from MH1K and MH1J reflected the difference of the doubling times between these strains since the generation time of K56-2 and MH1K is approximately half that for the J2315 and MH1J strains (data not shown). These results demonstrate that the gentamicin-sensitive mutants of B. cenocepacia J2315 and K56-2 are capable of intracellular replication within RAW 264.7 macrophages.
FIG. 4.
B. cenocepacia replicates intracellularly in RAW 264.7 macrophages. RAW 264.7 macrophages were infected at an MOI of 1 with either MH1K or MH1J for 2 h. B. cenocepacia cultures were then washed, gentamicin was added for 30 min at 50 μg ml−1 to kill extracellular bacteria, and infected cells were further incubated for 3 or 24 h in 10 μg ml−1 gentamicin, as described in Materials and Methods. Infected cells were lysed with 1% Triton X-100 in PBS at 3 h or 24 h after infection, and intracellular bacteria were enumerated by serial dilution and colony counts on LB agar plates. Results represent the average and standard error of three independent experiments. Significant differences were determined using an unpaired t test. **, P < 0.01 between bacterial counts at 3 h and 24 h.
Concluding remarks.
We have identified the efflux pump encoded by BCAL1674-BCAL1676 as a critical factor for aminoglycoside resistance in B. cenocepacia. A similar deletion in B. cenocepacia J2315 was previously shown to have no phenotype regarding sensitivity to aminoglycosides (7). We do not know the exact reason for this discrepancy, but it is possible that differences in the antibiotic selection strategy to generate the mutant may be important. In the study by Buroni et al. (7), gentamicin was used to kill donor E. coli during the triparental mating procedure required for construction of the B. cenocepacia mutant, which could have resulted in the selection of gentamicin-resistant suppressor mutants in the B. cenocepacia deletion strain. In this study, we anticipated this possibility and therefore used ampicillin and polymyxin B to kill donor E. coli cells.
This work demonstrates that gentamicin protection assays with the gentamicin-sensitive mutants MH1K and MH1J can be effectively used to reliably quantify the intracellular replication of B. cenocepacia in eukaryotic cells. Indeed, we could show that both mutants undergo intracellular replication within macrophages at levels similar to those previously found in other Bcc strains, including B. cenocepacia J2315 and K56-2 (Table 1). In these studies (Table 1), depending on the strains and the host cell type, intracellular replication ranged from 0.5 log10 units (B. ambifaria in THP-1 human monocytes) to 2 log10 units (B. cenocepacia in human dendritic cells). These values are slightly lower than those reported for B. pseudomallei and B. mallei (Table 1), which ranged from <1 log10 units (for B. pseudomallei in gamma interferon [IFN-γ]-treated macrophages) to 2 to 3 log10 units; these organisms are recognized human pathogens with the ability of intracellular survival and replication in phagocytic cells. Therefore, it would appear that B. cenocepacia and other Bcc bacteria can also undergo intracellular replication, albeit at reduced levels compared to B. mallei and B. pseudomallei. The gentamicin-sensitive mutants constructed in this work do not show differences in growth rates in vitro, and they also exhibit trafficking properties in macrophages similar to those of their respective parental bacteria. Therefore, since these mutants behave similarly to the parental strains, we conclude that they will be useful tools to investigate intracellular adaptations and virulence mechanisms employed by B. cenocepacia upon infection of human cells without the potentially confounding effects of using large amounts of antibiotics in the assay system. Furthermore, the strategy employed here could be generally applicable to other nonfermentative opportunistic Gram-negative bacteria, which are also highly resistant to aminoglycosides.
Acknowledgments
We thank Slade Loutet for critical reading of the manuscript.
This work was supported by a grant from the Canadian Cystic Fibrosis Foundation. A.M.S. was supported by a Graduate Award from the Canadian Institutes of Health Research. M.A.V. holds a Canada Research Chair in Infectious Diseases and Microbial Pathogenesis.
Footnotes
Published ahead of print on 26 March 2010.
REFERENCES
- 1.Aaron, S. D., W. Ferris, D. A. Henry, D. P. Speert, and N. E. Macdonald. 2000. Multiple combination bactericidal antibiotic testing for patients with cystic fibrosis infected with Burkholderia cepacia. Am. J. Respir. Crit. Care Med. 161:1206-1212. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., and D. J. Lipman. 1990. Protein database searches for multiple alignments. Proc. Natl. Acad. Sci. U. S. A. 87:5509-5513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Al-Younes, H. M., T. Rudel, and T. F. Meyer. 1999. Characterization and intracellular trafficking pattern of vacuoles containing Chlamydia pneumoniae in human epithelial cells. Cell. Microbiol. 1:237-247. [DOI] [PubMed] [Google Scholar]
- 4.Böttger, E. C., B. Springer, T. Prammananan, Y. Kidan, and P. Sander. 2001. Structural basis for selectivity and toxicity of ribosomal antibiotics. EMBO Rep. 2:318-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breitbach, K., G. W. Sun, J. Köhler, K. Eske, P. Wongprompitak, G. Tan, Y. Liu, Y.-H. Gan, and I. Steinmetz. 2009. Caspase-1 mediates resistance in murine melioidosis. Infect. Immun. 77:1589-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burns, J. L., M. Jonas, E. Y. Chi, D. K. Clark, A. Berger, and A. Griffith. 1996. Invasion of respiratory epithelial cells by Burkholderia (Pseudomonas) cepacia. Infect. Immun. 64:4054-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buroni, S., M. R. Pasca, R. S. Flannagan, S. Bazzini, A. Milano, I. Bertani, V. Venturi, M. A. Valvano, and G. Riccardi. 2009. Assessment of three resistance-nodulation-cell division drug efflux transporters of Burkholderia cenocepacia in intrinsic antibiotic resistance. BMC Microbiol. 9:200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Burtnick, M. N., P. J. Brett, V. Nair, J. M. Warawa, D. E. Woods, and F. C. Gherardini. 2008. Burkholderia pseudomallei type III secretion system mutants exhibit delayed vacuolar escape phenotypes in RAW 264.7 murine macrophages. Infect. Immun. 76:2991-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burtnick, M. N., D. Deshazer, V. Nair, F. C. Gherardini, and P. J. Brett. 2010. Burkholderia mallei cluster 1 type VI secretion mutants exhibit growth and actin polymerization defects in RAW 264.7 murine macrophages. Infect. Immun. 78:88-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caraher, E., C. Duff, T. Mullen, S. Mc Keon, P. Murphy, M. Callaghan, and S. McClean. 2007. Invasion and biofilm formation of Burkholderia dolosa is comparable with Burkholderia cenocepacia and Burkholderia multivorans. J. Cyst. Fibros. 6:49-56. [DOI] [PubMed] [Google Scholar]
- 11.Chantratita, N., V. Wuthiekanun, K. Boonbumrung, R. Tiyawisutsri, M. Vesaratchavest, D. Limmathurotsakul, W. Chierakul, S. Wongratanacheewin, S. Pukritiyakamee, N. J. White, N. P. J. Day, and S. J. Peacock. 2007. Biological relevance of colony morphology and phenotypic switching by Burkholderia pseudomallei. J. Bacteriol. 189:807-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi, K. H., T. Mima, Y. Casart, D. Rholl, A. Kumar, I. R. Beacham, and H. P. Schweizer. 2008. Genetic tools for select-agent-compliant manipulation of Burkholderia pseudomallei. Appl. Environ. Microbiol. 74:1064-1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chuaygud, T., S. Tungpradabkul, S. Sirisinha, K. L. Chua, and P. Utaisincharoen. 2008. A role of Burkholderia pseudomallei flagella as a virulent factor. Trans. R. Soc. Trop. Med. Hyg. 102(Suppl. 1):S140-S144. [DOI] [PubMed] [Google Scholar]
- 14.Coenye, T., and P. Vandamme. 2003. Diversity and significance of Burkholderia species occupying diverse ecological niches. Environ. Microbiol. 5:719-729. [DOI] [PubMed] [Google Scholar]
- 15.Cohen, S. N., A. C. Chang, and L. Hsu. 1972. Nonchromosomal antibiotic resistance in bacteria: genetic transformation of Escherichia coli by R-factor DNA. Proc. Natl. Acad. Sci. U. S. A. 69:2110-2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Conway, S. P., K. G. Brownlee, M. Denton, and D. G. Peckham. 2003. Antibiotic treatment of multidrug-resistant organisms in cystic fibrosis. Am. J. Respir. Med. 2:321-332. [DOI] [PubMed] [Google Scholar]
- 17.Craig, F. F., J. G. Coote, R. Parton, J. H. Freer, and N. J. Gilmour. 1989. A plasmid which can be transferred between Escherichia coli and Pasteurella haemolytica by electroporation and conjugation. J. Gen. Microbiol. 135:2885-2890. [DOI] [PubMed] [Google Scholar]
- 18.Diop, D., C. Chauvin, and O. Jean-Jean. 2007. Aminoglycosides and other factors promoting stop codon readthrough in human cells. C. R. Biol. 330:71-79. [DOI] [PubMed] [Google Scholar]
- 19.Drevets, D. A., B. P. Canono, P. J. Leenen, and P. A. Campbell. 1994. Gentamicin kills intracellular Listeria monocytogenes. Infect. Immun. 62:2222-2228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Elsinghorst, E. A. 1994. Measurement of invasion by gentamicin resistance. Methods Enzymol. 236:405-420. [DOI] [PubMed] [Google Scholar]
- 21.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U. S. A. 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flannagan, R. S., T. Linn, and M. A. Valvano. 2008. A system for the construction of targeted unmarked gene deletions in the genus Burkholderia. Environ. Microbiol. 10:1652-1660. [DOI] [PubMed] [Google Scholar]
- 23.Govan, J. R., P. H. Brown, J. Maddison, C. J. Doherty, J. W. Nelson, M. Dodd, A. P. Greening, and A. K. Webb. 1993. Evidence for transmission of Pseudomonas cepacia by social contact in cystic fibrosis. Lancet 342:15-19. [DOI] [PubMed] [Google Scholar]
- 24.Hamad, M. A., S. L. Zajdowicz, R. K. Holmes, and M. I. Voskuil. 2009. An allelic exchange system for compliant genetic manipulation of the select agents Burkholderia pseudomallei and Burkholderia mallei. Gene 430:123-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Isles, A., I. Maclusky, M. Corey, R. Gold, C. Prober, P. Fleming, and H. Levison. 1984. Pseudomonas cepacia infection in cystic fibrosis: an emerging problem. J. Pediatr. 104:206-210. [DOI] [PubMed] [Google Scholar]
- 26.Jones, A. L., T. J. Beveridge, and D. E. Woods. 1996. Intracellular survival of Burkholderia pseudomallei. Infect. Immun. 64:782-790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones, A. M., M. E. Dodd, and A. K. Webb. 2001. Burkholderia cepacia: current clinical issues, environmental controversies and ethical dilemmas. Eur. Respir. J. 17:295-301. [DOI] [PubMed] [Google Scholar]
- 28.Judy, B. M., G. C. Whitlock, A. G. Torres, and D. M. Estes. 2009. Comparison of the in vitro and in vivo susceptibilities of Burkholderia mallei to ceftazidime and levofloxacin. BMC Microbiol. 9:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lamothe, J., K. K. Huynh, S. Grinstein, and M. A. Valvano. 2007. Intracellular survival of Burkholderia cenocepacia in macrophages is associated with a delay in the maturation of bacteria-containing vacuoles. Cell. Microbiol. 9:40-53. [DOI] [PubMed] [Google Scholar]
- 30.Lamothe, J., S. Thyssen, and M. A. Valvano. 2004. Burkholderia cepacia complex isolates survive intracellularly without replication within acidic vacuoles of Acanthamoeba polyphaga. Cell. Microbiol. 6:1127-1138. [DOI] [PubMed] [Google Scholar]
- 31.Landers, P., K. G. Kerr, T. J. Rowbotham, J. L. Tipper, P. M. Keig, E. Ingham, and M. Denton. 2000. Survival and growth of Burkholderia cepacia within the free-living amoeba Acanthamoeba polyphaga. Eur. J. Clin. Microbiol. Infect. Dis. 19:121-123. [DOI] [PubMed] [Google Scholar]
- 32.Law, R. J., J. N. Hamlin, A. Sivro, S. J. McCorrister, G. A. Cardama, and S. T. Cardona. 2008. A functional phenylacetic acid catabolic pathway is required for full pathogenicity of Burkholderia cenocepacia in the Caenorhabditis elegans host model. J. Bacteriol. 190:7209-7218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lipuma, J. J. 2003. Burkholderia cepacia complex as human pathogens. J. Nematol. 35:212-217. [PMC free article] [PubMed] [Google Scholar]
- 34.LiPuma, J. J., S. E. Dasen, D. W. Nielson, R. C. Stern, and T. L. Stull. 1990. Person-to-person transmission of Pseudomonas cepacia between patients with cystic fibrosis. Lancet 336:1094-1096. [DOI] [PubMed] [Google Scholar]
- 35.Loutet, S. A., S. J. Bartholdson, J. R. Govan, D. J. Campopiano, and M. A. Valvano. 2009. Contributions of two UDP-glucose dehydrogenases to viability and polymyxin B resistance of Burkholderia cenocepacia. Microbiology 155:2029-2039. [DOI] [PubMed] [Google Scholar]
- 36.MacDonald, K. L., and D. P. Speert. 2008. Differential modulation of innate immune cell functions by the Burkholderia cepacia complex: Burkholderia cenocepacia but not Burkholderia multivorans disrupts maturation and induces necrosis in human dendritic cells. Cell. Microbiol. 10:2138-2149. [DOI] [PubMed] [Google Scholar]
- 37.Mahenthiralingam, E., T. Coenye, J. W. Chung, D. P. Speert, J. R. Govan, P. Taylor, and P. Vandamme. 2000. Diagnostically and experimentally useful panel of strains from the Burkholderia cepacia complex. J. Clin. Microbiol. 38:910-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maloney, K. E., and M. A. Valvano. 2006. The mgtC gene of Burkholderia cenocepacia is required for growth under magnesium limitation conditions and intracellular survival in macrophages. Infect. Immun. 74:5477-5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marolda, C. L., B. Hauröder, M. A. John, R. Michel, and M. A. Valvano. 1999. Intracellular survival and saprophytic growth of isolates from the Burkholderia cepacia complex in free-living amoebae. Microbiology 145:1509-1517. [DOI] [PubMed] [Google Scholar]
- 40.Martin, D. W., and C. D. Mohr. 2000. Invasion and intracellular survival of Burkholderia cepacia. Infect. Immun. 68:24-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Menashe, O., E. Kaganskaya, T. Baasov, and S. Yaron. 2008. Aminoglycosides affect intracellular Salmonella enterica serovars Typhimurium and Virchow. Antimicrob. Agents Chemother. 52:920-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moore, R. A., D. DeShazer, S. Reckseidler, A. Weissman, and D. E. Woods. 1999. Efflux-mediated aminoglycoside and macrolide resistance in Burkholderia pseudomallei. Antimicrob. Agents Chemother. 43:465-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pruksachartvuthi, S., N. Aswapokee, and K. Thankerngpol. 1990. Survival of Pseudomonas pseudomallei in human phagocytes. J. Med. Microbiol. 31:109-114. [DOI] [PubMed] [Google Scholar]
- 44.Ribot, W. J., and R. L. Ulrich. 2006. The animal pathogen-like type III secretion system is required for the intracellular survival of Burkholderia mallei within J774.2 macrophages. Infect. Immun. 74:4349-4353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saini, L. S., S. B. Galsworthy, M. A. John, and M. A. Valvano. 1999. Intracellular survival of Burkholderia cepacia complex isolates in the presence of macrophage cell activation. Microbiology 145:3465-3475. [DOI] [PubMed] [Google Scholar]
- 46.Sajjan, S. U., L. A. Carmody, C. F. Gonzalez, and J. J. LiPuma. 2008. A type IV secretion system contributes to intracellular survival and replication of Burkholderia cenocepacia. Infect. Immun. 76:5447-5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sajjan, U. S., J. H. Yang, M. B. Hershenson, and J. J. LiPuma. 2006. Intracellular trafficking and replication of Burkholderia cenocepacia in human cystic fibrosis airway epithelial cells. Cell. Microbiol. 8:1456-1466. [DOI] [PubMed] [Google Scholar]
- 48.Saldías, M. S., J. Lamothe, R. Wu, and M. A. Valvano. 2008. Burkholderia cenocepacia requires the RpoN sigma factor for biofilm formation and intracellular trafficking within macrophages. Infect. Immun. 76:1059-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Saldías, M. S., X. Ortega, and M. A. Valvano. 2009. Burkholderia cenocepacia O antigen lipopolysaccharide prevents phagocytosis by macrophages and adhesion to epithelial cells. J. Med. Microbiol. 58:1542-1548. [DOI] [PubMed] [Google Scholar]
- 50.Saldías, M. S., and M. A. Valvano. 2009. Interactions of Burkholderia cenocepacia and other Burkholderia cepacia complex bacteria with epithelial and phagocytic cells. Microbiology 155:2809-2817. [DOI] [PubMed] [Google Scholar]
- 51.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 52.Sandoval, R. M., and B. A. Molitoris. 2004. Gentamicin traffics retrograde through the secretory pathway and is released in the cytosol via the endoplasmic reticulum. Am. J. Physiol. Renal. Physiol. 286:F617-F624. [DOI] [PubMed] [Google Scholar]
- 53.Speert, D. P. 2002. Advances in Burkholderia cepacia complex. Paediatr. Respir. Rev. 3:230-235. [DOI] [PubMed] [Google Scholar]
- 54.Steele-Mortimer, O. 2008. Infection of epithelial cells with Salmonella enterica. Methods Mol. Biol. 431:201-211. [DOI] [PubMed] [Google Scholar]
- 55.Stevens, M. P., M. W. Wood, L. A. Taylor, P. Monaghan, P. Hawes, P. W. Jones, T. S. Wallis, and E. E. Galyov. 2002. An Inv/Mxi-Spa-like type III protein secretion system in Burkholderia pseudomallei modulates intracellular behaviour of the pathogen. Mol. Microbiol. 46:649-659. [DOI] [PubMed] [Google Scholar]
- 56.Taylor, J. B., L. A. Hogue, M. J. Walter, S. L. Brody, and C. L. Cannon. 2010. Entry of Burkholderia organisms into respiratory epithelium: CFTR, microfilament and microtubule dependence. J. Cyst. Fibros. 9:36-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Trunck, L. A., K. L. Propst, V. Wuthiekanun, A. Tuanyok, S. M. Beckstrom-Sternberg, J. S. Beckstrom-Sternberg, S. J. Peacock, P. Keim, S. W. Dow, and H. P. Schweizer. 2009. Molecular basis of rare aminoglycoside susceptibility and pathogenesis of Burkholderia pseudomallei clinical Isolates from Thailand. PLoS Negl. Trop. Dis. 3:e519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Via, L. E., R. A. Fratti, M. McFalone, E. Pagan-Ramos, D. Deretic, and V. Deretic. 1998. Effects of cytokines on mycobacterial phagosome maturation. J. Cell Sci. 111:897-905. [DOI] [PubMed] [Google Scholar]
- 59.Vial, L., M. Groleau, M. Lamarche, G. Filion, J. Castonguay-Vanier, V. Dekimpe, F. Daigle, S. Charette, and E. Déziel. 2010. Phase variation has a role in Burkholderia ambifaria niche adaptation. ISME J. 4:49-60. [DOI] [PubMed] [Google Scholar]