Abstract
Force field parameters specifically optimized for residues important in the study of RNA catalysis are derived from density-functional calculations in a fashion consistent with the CHARMM27 all-atom empirical force field. Parameters are presented for residues that model reactive RNA intermediates and transition state analogs, thio-substituted phosphates and phosphoranes, and bound Mg2+ and di-metal bridge complexes. Target data was generated via density-functional calculations at the B3LYP/6-311++G(3df,2p)//B3LYP/6-31++G(d,p) level. Partial atomic charges were initially derived from the CHelpG electrostatic potential fitting and subsequently adjusted to be consistent with the CHARMM27 charges and Lennard-Jones parameters were determined to reproduce interaction energies with water molecules. Bond, angle and torsion parameters were derived from the density-functional calculations and renormalized to maintain compatibility with the existing CHARMM27 parameters for standard residues. The extension of the CHARMM27 force field parameters for the non-standard biological residues presented here will have considerable use in simulations of ribozymes, including the study of freeze-trapped catalytic intermediates, metal ion binding and occupation, and thio effects.
Keywords: molecular dynamics, RNA, metal ions, thio effects, phosphorane
1 Introduction
Over the last several decades a wealth of data has accumulated that demonstrates the central role RNA catalysis plays in many biological processes. Starting in the late 1970’s, it was shown that RNA could catalyze fairly complex biological reactions in ribonuclease P1,2 and Tetrahymena3,4 with an efficiency that rivaled many protein enzymes. These discoveries sparked a wave of interest in the scientific community focused on unraveling the details of how RNA enzymes (ribozymes) function. An understanding of the catalytic mechanisms of ribozymes, and their relation to sequence and tertiary structure, is opening up a variety of new frontiers. In biomedical technology, gene expression inhibitors that target viral and genetic diseases5 such as HIV6 and cancer7 are being developed, and new biotechnologies such as RNA chips8 and allosteric molecular switches in nanodevices9 are being explored.
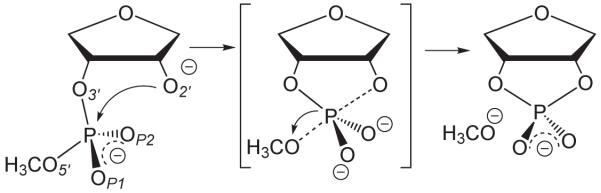
A common biological reaction catalyzed by several prototype ribozyme systems, such as the hammerhead,10,11 hairpin,12,13 and hepatitis delta virus14,15 ribozymes, involves cleavage of a phosphate group through a transesterification reaction16,17 (Figure 1). In this reaction, the 2′OH of the ribose sugar ring becomes activated via deprotonation and makes an in-line attack to the adjacent 3′-phosphate along the phosphodiester backbone. The attack produces a trigonal bipyramidal phosphorane intermediate/transition state that is accompanied by an inversion of configuration about the phosphorus center as the exocyclic P-O5′ bond is cleaved. The product of the transesterification is a 5′-OH terminus and a 2′,3′-cyclic phosphate. Additional information about this mechanism has been obtained via kinetic isotope studies18-20 and chemical modifications such as thio substitution21-23 at the scissile phosphate, although the mechanistic interpretation of these experimental studies remains a topic of discussion and some debate. Time-resolved x-ray crystallography10,24,25 has become a powerful tool to elucidate structural information at different stages along the catalytic reaction coordinate that provides valuable insight into ribozyme activity. However, this area is considerably challenging due to the difficulty of trapping a reactive intermediate and obtaining quality crystals, as well as uncertainties due to the nature of effects that arise from the crystallization conditions. For these reasons, there is considerable interest in the development of theoretical methods that can aid in the refinement and interpretation of existing experimental data, and provide structural insight into systems where such data is not yet available.
Figure 1.
Model RNA transesterification reaction.
Molecular simulation, along with experimental structural data, provides an avenue for the characterization of ribozyme dynamics in solution and refinement of key mechanistic details. Molecular simulation force fields for nucleic acids continue to improve26-31 and a variety of simulations involving ribozymes have been carried out in recent years.32-40 In order to study the structure and dynamics of different catalytic states along the reaction path of a ribozyme, however, reliable empirical force fields parameters must be developed for the transition states and reactive intermediates of these reactions. Furthermore, in order to use molecular simulation to aid in the interpretation of experimentally measured thio effects, parameters for thio-substituted phosphate and phosphorane models and their interactions with metal ions are required. In the present work, new force field parameters for residues important to the study of RNA catalysis are derived from density-functional calculations to be consistent with the CHARMM2741 all-atom empirical force field. These parameters will allow molecular dynamics (MD) simulations of ribozymes in reactive states to be performed to study the structure and dynamics that lead to catalysis.
2 Methods
The potential energy function used for the CHARMM27 empirical force field for nucleic acids,42,43 and for the new modified RNA residues of the present work, has the general form:44
| (1) |
The first four summations are quadratic terms that give rise to energy penalties for geometrical deviations about equilibrium coordinate values. The variables b, θ, S, and φ are the bond length, bond angle, Urey-Bradley 1,3-distance, and improper torsion angle coordinates, respectively, while b0, θ0, S0, φ0 and Kb, Kθ, KS, Kφ are the corresponding force field parameters for the equilibrium geometries and force constants, respectively. The fifth summation is a trigonometric term that adjusts the periodic dihedral angle rotational barriers. The coordinate χ is the dihedral coordinate, n determines the periodicity, χ0 is a phase factor, and Kχ is the amplitude force constant. The terms involving a sum over atom pairs i, j < i (neglecting non-bonded exclusions) are the non-bonded van der Waals/Lennard-Jones (L-J) and electrostatic terms. The parameters ∊i,j and R0,ij are the van der Waals well depth and minimum distance between the ij atom pair, respectively, and are by default calculated via the Lorentz-Berthelot combining rules45 from the corresponding 1-body parameters as and R0,ij = R0,i + R0,j, respectively, although this default can be explicitly overridden using the NBFIX (non-bond fix) option if fine tuning of specific pairwise interactions is desired. The last summation is the electrostatic energy determined between atomic partial charges qi, which normally are calculated with a unit dielectric constant, ∊ = 1, as in the present work.
Parameterization of the CHARMM27 force field was based on ab initio and experimental data for small molecules,42 as well as macromolecular simulation data of DNA and RNA46 in order to more accurately capture experimental condensed phase properties. This was an important improvement to the CHARMM22 force field47 that was parameterized more heavily on the basis of small molecule target data alone. While this previous approach succeeded in capturing the target data of the selected small compounds, later simulations of duplex DNA in solution showed some disagreement with experiment in regard to the relative conformational stability of A-DNA versus B-DNA.48,49
The CHARMM27 force field was developed using a self-consistent, step-wise strategy.31,42 Partial atomic charges for the CHARMM27 force field were initially obtained from a Mulliken population analysis of the HF/6-31G(d) wave function. These were then adjusted to better reproduce scaled HF/6-31G(d) TIP3P50 water interaction energies, experimental dipole moments, and heats of sublimation where available. Equilibrium geometry parameters for the selected small molecules were initially optimized to reproduce the experimental geometries of microwave, electron diffraction and/or x-ray crystal survey data where possible. Equilibrium geometries and force constants were optimized iteratively until satisfactory fitting to the target data was achieved. Once this optimization was complete, iterative adjustment of the charges and L-J parameters was coupled with the internal parameters until overall convergence was reached. A survey of RNA and DNA crystal structures from the Nucleic Acids Database (NDB)51 was taken so that fitting to target macromolecular experimental properties, such as sugar puckering phase and dihedral angle distributions, could be made through crystal MD simulation. During this stage, in the original CHARMM27 all-atom empirical force field parameterization for nucleic acids,42 dihedral angle parameters were adjusted to lower or raise energy barriers such that target condensed phase properties were better reproduced. While this sometimes sacrificed the quality of fitting to small molecule ab initio data, it accomplished the goal of more accurately reproducing experimental condensed phase properties. This macromolecular fitting was coupled with the internal parameter optimization until satisfactory convergence was achieved.
Due to the lack of high-resolution experimental data for RNA reactive intermediates and chemically modified nucleic acids, the parameterization of the present study is based solely on ab initio data, the optimized strucutures for which are shown in Figures 2, 3, 4, and 5. Parameter optimization for modified CHARMM27 RNA, Mg2+, and OH− residues was based on density functional theory (DFT) calculations for training sets of small molecules that represented the desired target systems. Density-functional calculations were performed using the Becke three-parameter hybrid functional combined with the Lee-Yang-Parr exchange-correlation functional (B3LYP)52,53 with the 6–31++G(d,p) basis set for geometry and frequency calculations followed by single-point electronic structure refinement with the 6–311++G(3df,2p) basis set in a manner analogous to recent studies of biological phosphates.54-58 The B3LYP model employed here neglects proper treatment of long-range dispersion interactions; however, these weak dispersion interactions are small in comparison with the highly polar (e.g., hydrogen bonded) and ionic interactions investigated here, for which B3LYP has been demonstrated previously to be generally reliable. All DFT calculations were performed using the GAUSSIAN0359 package. Partial atomic charges were based on the CHelpG60,61 method (CHarges from ELectrostatic Potentials using a Grid) which fits atomic centered point charges to the molecular electrostatic potential. The CHelpG charges were also constrained to reproduce molecular dipole moments. Similar approaches to force field charge determination have been applied and validated previously.60,62 These calculations were performed with the 6–311++G(3df,2p) basis set, and the CHelpG charges were subsequently modified to be consistent with the original force field by taking into account only the charge differences from standard CHARMM residues, and making further adjustments for polar and non-polar hydrogens and integer charged groups in accord with the original parameterization procedure31,42 (see Section 3.2 for complete details). The CHelpG charges used in the present work to obtain the charge differences with respect to similar standard CHARMM residues are different than the Mulliken charges that were used as a starting point for optimization in the original CHARMM27 force field, and in somees these charge modelsdiffer significantly. However, it should be emphasized that the Mulliken charges As in the original CHARMM27 force field, transferring charges from the small model compounds to the nucleic acid fragments was accomplished by adding the charge of the removed hydrogen atom to the heavy atom from which it was deleted. Once charges were obtained, Lennard-Jones (L-J) parameters were determined by fitting CHARMM residue-TIP3P water interaction energies and distances to the density-functional target values. This was accomplished through scaling of the interaction energies and shifting of the binding distances such that the DFT values matched those of the original CHARMM27 force field for unsubstituted dimethyl phosphate (DMP−). Although the original CHARMM27 force field parameterization fixed waters to the TIP3P geometry in ab initio calculations, it was found in this work that the relative differences in geometries predicted by the DFT level of theory used compared better with the HF/6-31G(d) used in the original force field when the waters remained unconstrained (see Table S2 in Supporting Information). Internal parameters were fit to the density-functional geometries and energies for the model compounds.
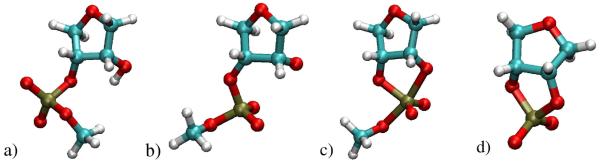
Figure 2.
a) CHARMM27 standard and modified nucleotide residues: b) A standard CHARMM27 ribonucleotide analog (no base) with the 2′ position protonated; c) 2′ deprotonated ribose phosphate (i.e., activated at the 2′ position); d) ribose phosphorane (i.e., intermediate/transition state model); 2′,3′-cyclic ribose phosphate (i.e., transesterification product). Color coding of atoms is as follows: carbon=turquoise, oxygen=red, phosphorous=brown, hydrogen=white.
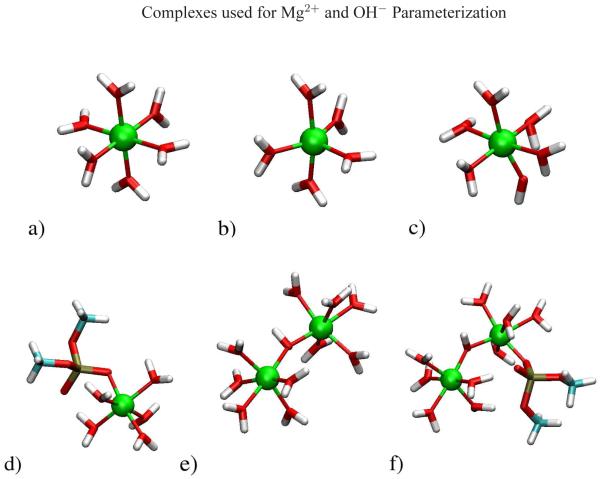
Figure 3.
a), b), c)Mg(H2O)5(OH−)1+, d)Mg(H2O)5:DMP1+, e), f)Mg(H2O)5· (OH−) Mg(H2O)4:DMP2+. Color coding of atoms is as follows: magnesium=green, oxygen=red, carbon=turquoise, hydrogen=white.
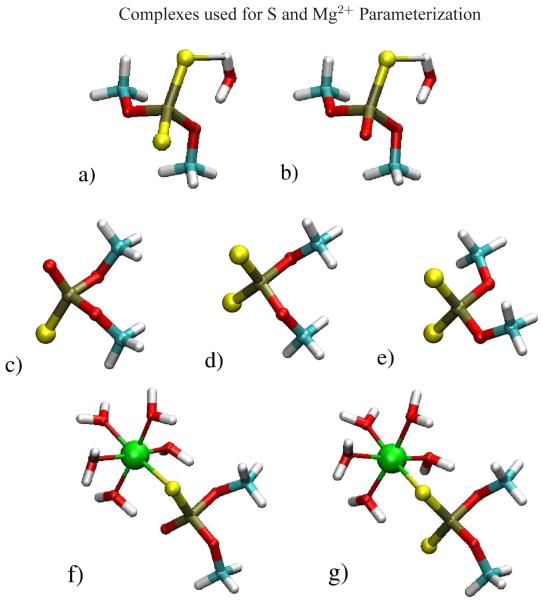
Figure 4.
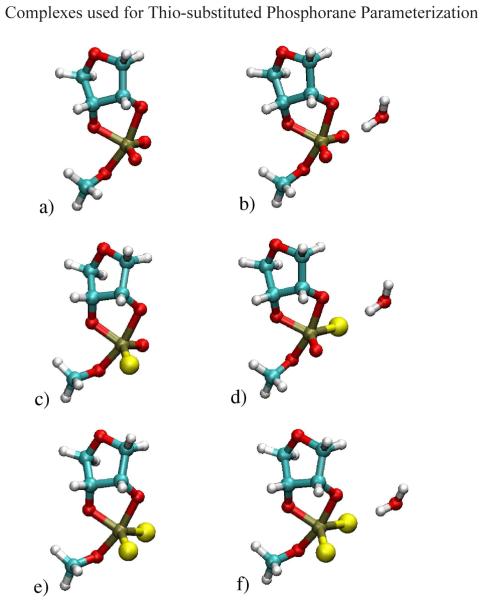
a)DMP-ss−:HOH, b)DMP-so−:HOH, c)DMP-so−, d)DMP-ss−g–g, e)DMP-ss−g–t, f)DMP-so−:[Mg(HOH)5]2+, g)DMP-ss−:[Mg(HOH)5]2+. Color coding of atoms is as follows: magnesium=green, oxygen=red, carbon=turquoise, hydrogen=white, phosphorous=brown, sulfur=yellow.
Figure 5.
a)ribose MePA2−, b)ribose MePA2−:HOH, c)ribose MePA-so2−, d)ribose MePA-so2−:HOH, e)ribose MePA-ss2−, f)ribose MePA-ss−:HOH. Color coding of atoms is as follows: oxygen=red, carbon=turquoise, hydrogen=white, phosphorous=brown, sulfur=yellow.
The CHelpG charges used in the present work to obtain the charge differences with respect to similar standard CHARMM residues are different than the Mulliken charges that were used as a starting point for optimization in the original CHARMM27 force field. In general, the CHelpG charges are larger in magnitude, and due to the constraints, preserve the molecular dipole moments. The Mulliken charges on the other hand are considerably basis set dependent, particularly when diffuse functions are used, and generally smaller in magnitude. It should be emphasized that the Mulliken charges used as a starting point for parameter optimization with CHARMM27 were often significantly altered upon optimization to obtain intermolecular interactions with TIP3P water molecules, the final charges in many cases being closer to the CHelpG charges (e.g., for the non-bridging phosphate oxygens).
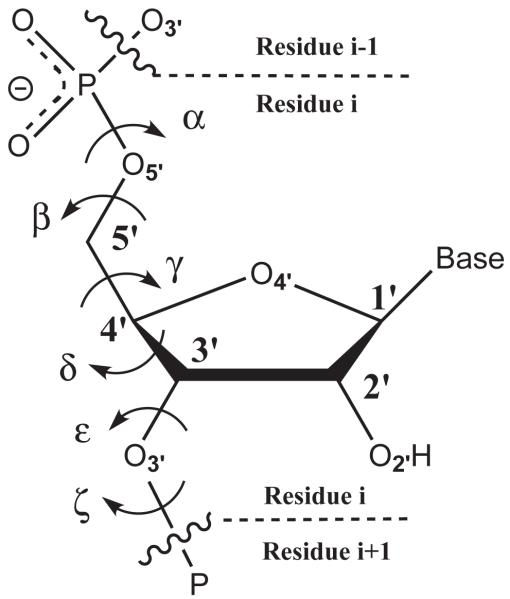
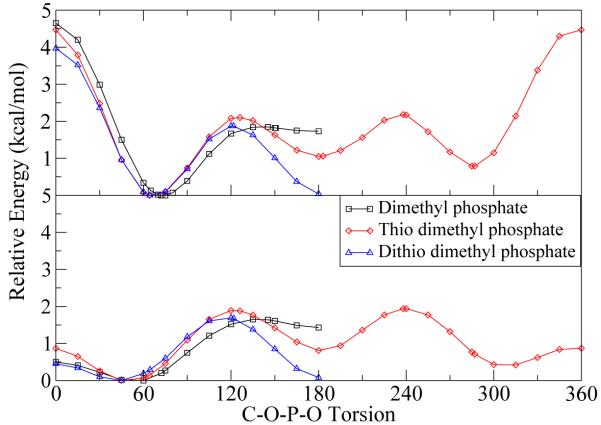
RNA generally has more accessible “folded” conformations than DNA63 and often conformational deformation in the phosphate backbone is requisite for catalysis.64 The CHARMM27 force field contains dihedral parameters for unsubstituted RNA based on ab initio gas-phase calculations of a variety of test compounds and adjusted with respect to A and B form DNA and RNA within the Nucleic Acid Database (NDB).51 These adjustments included the lowering of torsional energy barriers in certain regions after the observation that direct parameterization to the ab initio torsion profiles resulted in simulated dihedral distributions that were inconsistent (e.g., too rigid) compared with those from a large NDB survey of RNA and DNA.42 In the case of nonbridging thio-substitutions, no parameters exist. Therefore, attention was given to the C-O-P-O (i.e., α and ζ in Figure 6) dihedral parameters because of their role along the phosphate backbone and the possibility of significant change upon thio-substitution. As in CHARMM27, dimethyl phosphate was used as the model compound for native RNA as well as singly and doubly thio-substituted dimethyl phosphate to determine the effects of the nonbridging sulfur(s) on this torsion. The dimethyl phosphate and thio substituted torsional potential energy surfaces were generated using B3LYP/6-311++G(3df,2p)//B3LYP/6-31++G(d,p) with all degrees of freedom relaxed except for the dihedral of interest and the symmetric ζ dihedral, which is fixed in the trans conformation to induce symmetry and facilitate parameterization (Fig. 7).
Figure 6.
Phosphate backbone of RNA with torsions labeled.
Figure 7.
Ab initio (top frame) and CHARMM (bottom frame) torsional potential energy surface for the C-O-P-O dihedral of dimethyl phosphate (OO), nonbridging thiosubstituted dimethyl phosphate (SO), and nonbridging dithiosubstituted dimethyl phosphate (SS).
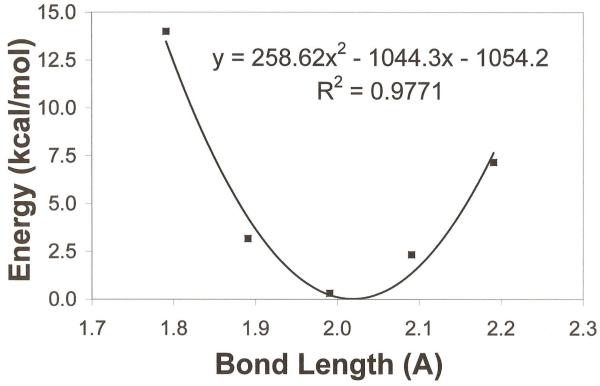
As in the original force field parameterization,42 a self-consistent step-wise optimization approach was taken that involved the iterative adjustments of L-J and internal parameters (not including torsion parameters) until convergence of the fitting function was obtained. Initial force constant values were taken from harmonic fitting to potential energy surface scans of bonds and angles with distortions of 0.2 Å and 2.0o respectively (See example in Figure 7). Starting geometries for the new CHARMM* model compounds were taken directly from the density-functional results and read into CHARMM to perform an Adopted Basis Newton-Raphson (ABNR) minimization65 until a gradient of <10−6 kcal/mol·Å was reached. The results of the CHARMM minimization with the current parameters were used to construct a χ2 fitting function. In the present work, this function took the form:
| (2) |
Where the , , and are the CHARMM energy-minimized bond lengths (Å), bond angles (degrees), torsion angles (degrees) and interaction energies (kcal/mol), respectively, and , , and are the corresponding DFT reference values. The σ parameters, have the same units as their associated geometrical or energetic quantities (indicated by superscript), the inverse squares for which serve as weights in the unitless χ2(λ) merit function. A non-linear optimization procedure is then used to minimize the χ2(λ) merit function with respect to the CHARMM* parameters (this work) denoted by the vector λ. The σ parameters (and hence the weights) were adjusted empirically over the course of a stepwise minimization procedure in order to drive the parameter search in the most realistic and robust region of parameter space (see Supporting Information for for more details). In most cases, the last step-wise minimization procedure used to arrive at the final set of parameters was a quadratically convergent direction set minimization method66 that does not require explicit gradient information. Sets of conjugate directions were generated with the algorithm due to Powell in a series of successive line minimizations achieved by parabolic interpolation. The convergence criterion was 10−4 on the relative change in the function value with respect to its minimum value after a series of line minimizations. While this method works very well for finding the local minima of a multi-variable function, it is limited in its ability to find the absolute minimum of such a function. For this reason, the minimization procedure was carried out at a large number of starting points for each variable until the best overall minima could be found. Once a minimum was reached the torsion angle parameters were adjusted empirically to obtain a reasonable balance between the trends in the relative ab initio curves (native versus sulfur substituted) while maintaining consistency with the standard CHARMM27 torsion profile which does not correspond directly to an ab initio curve. Course grained adjustment of the parameters (0.05 kcal) was seen to be sufficient to provide an acceptable balance. The torsion parameters were adjusted iteratively with the parameters of the non-linear optimization. The complete topology and parameter files of this work are available as Supporting Information.
3 Results and Discussion
3.1 Magnesium Complexes
RNA enzymes show diversity in the number as well as the role of metal ions they require. Metal ions can have both structural and chemical roles in catalysis. Possible (chemical) catalytic roles for metal ions include acting as:11,17
General acid-base catalyst by accepting/donating protons from coordinated water molecules to the nucleophilic (2′) and leaving group (5′) positions
Lewis acid catalyst by direct coordination to either the nucleophilic (2′) or leaving group (5′) positions
Electrophilic catalyst by coordinating to the non-bridging phosphoryl oxygens and assisting in formation of a phosphorane-like transition state or intermediate
One way for a metal ion to act as a general base is to lose a proton from a coordinated water to form a metal hydroxide. This OH− has been postulated to play the role of abstracting a proton from a ribose 2′-OH,67 although the pKa difference between a phosphate coordinated Mg2+ and a ribose 2′OH remains a topic of current interest.68 In one postulated two metal ion mechanism for the hammerhead ribozyme (HR), it has been proposed that a metal hydroxide exists as a μ-bridging OH− between two magnesium ions.39 Other studies suggest a single metal ion mechanism with an outer-sphere coordination of the Mg2+ ion to the 2′OH69 and still others suggests a non-bridging two metal ion mechanism with inner-sphere binding of Mg2+.70 Clearly, MD and QM/MM simulation could help resolve these mechanisms and the catalytic role played by Mg2+ ions.
The force field parameters for Mg2+ and OH− are important to obtain accurate geometries and energies for Mg2+ ligand binding and the formation of di-metal Mg2+ complexes containing a bridging hydroxide. Current non-bonded parameters exist for Mg2+ in the CHARMM27 force field46 and OH− parameters are available from Lee et al.71 However, these parameters are not optimal for the specific problems that involve di-metal bridges or RNA phosphate binding. Hence, the parameters for OH− and Mg2+ have been re-optimized specifically for these types of systems.
DFT calculations have been run on several model systems55 which formed the training set for the Mg2+ and OH− parameterization. Each structure of the training set (Figure 3) contained one or two Mg2+ ions with a first coordination sphere combination of H2O, OH− and/or DMP− ligands. In the structures with two Mg2+ ions present, a OH− ion was placed in a μ-bridging position between the two metal ions. CHelpG charges were calculated for OH− and the L-J parameters for Mg2+ and OH− were iteratively adjusted until the best fit with geometry and water binding energy was obtained (Table 1). Mg2+ coordination distances were significantly improved with the new force field parameters for ligand binding with H2O, OH− and DMP−. This improvement was most pronounced for the di-metal bridging hydroxide structures which lengthen the Mg2+…OH− interaction by almost 0.30 Å and contract the Mg2+…OH…Mg2+ angle by almost 10 degrees. The new parameters also improved the Mg2+ water binding energy by approximately 1.0 kcal/mol. In the current study, only complexes that involve inner-sphere coordination of Mg2+ were used in the parameterization, and transferability to the important case of outer-sphere coordination remains to be tested with simulation.
Table 1.
Mg2+ and OH− parameter fitting results
| Bond (Å) | CHARMM1 | DFT | CHARMM* |
|---|---|---|---|
| HT–OW | 0.97 (0.00) | 0.97 | 0.97 (0.00) |
| MG⋯OH2 | 1.99 (−0.13) | 2.12 | 2.13 (0.01) |
| MG⋯OW | 1.81 (−0.19) | 2.00 | 2.00 (0.00) |
| MG⋯ON3 | 1.86 (−0.17) | 2.03 | 2.01 (−0.02) |
| MG⋯MG | 3.56 (−0.33) | 3.89 | 3.91 (0.02) |
|
| |||
| Angle (°) | CHARMM 1 | DFT | CHARMM* |
|
| |||
| MG⋯OW⋯MG | 155.8 (7.6) | 148.2 | 152.7 (4.5) |
|
| |||
| Energy (kcal/mol) | CHARMM 1 | DFT | CHARMM* |
|
| |||
| (Mg(H20)5 + H2O) → Mg(H20)6 |
−27.4 (0.0) | −27.4 | −28.3 (−0.9) |
Training set (see Figure 3) average errors for geometry and reaction energy fitting. With the exception of the hydroxide OW and HT atom types from Lee et. al.,71 the remaining parameters were orginally from the standard CHARMM27 force field Foloppe et. al.42 OH2 is the CHARMM27 water oxygen and ON3 is the non-bridging oxygen of DMP−. See the Supporting Information for more data on the fitting function weight values used.
3.2 2′O Deprotonated Sugar Ring
The first step of transesterification for a RNA phosphate is deprotonation of the sugar 2′OH. This creates a nucleophile of sufficient strength to attack the phosphorous of an adjacent or nearby phosphate group.16 A “patch”42 has been prepared for the standard CHARMM27 RNA nucleotide which removes the 2′OH hydrogen and modifies the 2′O− charge, L-J interactions and geometry of nearby atoms. Parameterization of this patch is discussed below.
To derive new charges for a deprotonated nucleotide residue, a ribose methyl phosphate in its protonated and deprotonated states was optimized using DFT. It was found that an “in-line” geometry minimum did not exist in the gas phase without the solvated ribozyme environment. In order to parameterize a deprotonated ribose methyl phosphate (ribose MeP−) residue to a structure that models the in-line phosphate attack conformation, a minimization was performed with the corresponding α, ∊ and ζ dihedrals of the phosphate-sugar backbone fixed to the minimum energy geometry found for the phosphorane structure described in Section 3.3 (Figure 2). In this way, the backbone of the ribose MeP− was constrained to be “in-line”. CHelpG charges, constrained to reproduce molecular dipole moment for neutral molecules, were then calculated for the atoms of the optimized structures (Table 2). Since the original CHARMM27 charges (qo) were obtained via a slightly different procedure than those of the present DFT/CHelpG method (q), adjustments to the CHelpG charges (q’) were made to create charges more consistent with the constrained CHARMM27 force field parameterization. For instance, in the CHARMM27 force field, all non-polar hydrogens are constrained to have a charge of 0.09 e while polar hydroxyl hydrogens are assigned a charge of 0.43 e. The calculated CHelpG charges were adjusted for this by changing the value of the hydrogens to be consistent with the CHARMM27 force field while adding the difference created from this adjustment to the adjoining heavy atoms (Table 2). The differences between the adjusted CHelpG charges for the protonated and deprotonated ribose methyl phosphate were then calculated and used to modify the original CHARMM27 charges of a neutral nucleotide as follows:
| (3) |
| (4) |
where q’P and q’D are the adjusted DFT/CHelpG charges for the protonated and deprotonated structures, respectively, qo are the CHARMM27 charges, and are the new charges for the deprotonated structure. Final partial atomic charge parameters are shown in Table 2. It should be noted that constraints on the charge fitting to create unit charge groups within residues was not used in this method.
Table 2.
CHelpG charge fitting for deprotonated ribose phosphate
| Atom | qo | qP | q’P | qD | q’D | δq | q*D |
|---|---|---|---|---|---|---|---|
| C1′ | 0.07 | 0.19 | 0.00 | 0.31 | −0.19 | −0.19 | −0.12 |
| H1′ | 0.09 | 0.03 | 0.09 | −0.12 | 0.09 | 0.00 | 0.09 |
| H1” | 0.09 | −0.04 | 0.09 | −0.20 | 0.09 | 0.00 | 0.09 |
| C2′ | 0.14 | 0.35 | 0.21 | 0.70 | 0.30 | 0.09 | 0.23 |
| H2” | 0.09 | −0.05 | 0.09 | −0.31 | 0.09 | 0.00 | 0.09 |
| O2′ | −0.66 | −0.71 | −0.72 | −0.97 | −0.97 | −0.25 | −0.91 |
| H2 ′ | 0.43 | 0.42 | 0.43 | - | - | −0.43 | - |
| C3′ | 0.01 | 0.30 | 0.20 | 0.65 | 0.37 | 0.17 | 0.18 |
| H3′ | 0.09 | −0.01 | 0.09 | −0.19 | 0.09 | 0.00 | 0.09 |
| O3′ | −0.57 | −0.57 | −0.57 | −0.63 | −0.63 | −0.06 | −0.63 |
| P | 1.50 | 1.23 | 1.23 | 1.37 | 1.37 | 0.14 | 1.64 |
| O1P | −0.78 | −0.77 | −0.77 | −0.84 | −0.83 | −0.06 | −0.84 |
| O2P | −0.78 | −0.76 | −0.76 | −0.81 | −0.83 | −0.06 | −0.84 |
| O3P | −0.57 | −0.50 | −0.50 | −0.58 | −0.58 | −0.08 | −0.65 |
| C3T | −0.17 | 0.31 | −0.10 | 0.36 | −0.16 | −0.06 | −0.23 |
| H3T1 | 0.09 | −0.06 | 0.09 | −0.08 | 0.09 | 0.00 | 0.09 |
| H3T2 | 0.09 | −0.05 | 0.09 | −0.08 | 0.09 | 0.00 | 0.09 |
| H3T3 | 0.09 | −0.03 | 0.09 | −0.09 | 0.09 | 0.00 | 0.09 |
| C4′ | 0.07 | 0.27 | 0.06 | 0.14 | −0.12 | −0.18 | −0.11 |
| H4′ | 0.09 | −0.03 | 0.09 | −0.07 | 0.09 | 0.00 | 0.09 |
| H4” | 0.09 | 0.00 | 0.09 | −0.01 | 0.09 | 0.00 | 0.09 |
| O4′ | −0.50 | −0.52 | −0.52 | −0.55 | −0.55 | −0.03 | −0.53 |
Adjusted charges (q’P/q’D) were determined by changing all CHelpG hydrogen charges to be consistent with CHARMM27 by the procedure outlined in Eqn. 3 and 4 of the text. This difference between the CHARMM27 (qo) and CHelpG charges (qP/qD) was then added into the nearest heavy atom charge. Once the adjusted atomic charge differences between the protonated and deprotonated structures were determined (δq), these differences were then added to the standard CHARMM27 charges to correct for deprotonation (q*D).
Once charges were calculated for the deprotonated ribose 2′O− oxygen, L-J parameters were determined by fitting to the 2′O− water coordination distance (1.713 Å) and interaction energy (-18.4 kcal/mol) obtained from DFT optimization of a ribose MeP− (Table 3). Bond and angle parameters were fit to the differences between the gas-phase DFT geometry of protonated and ribose MeP− (Table 4). Force constants were predicted by fitting to relaxed potential energy surface scans. All dihedrals except for H3′-C3′-C2′-O2′ were set to zero in the original protonated CHARMM27 structure and remained so in the new parameterization. The H3′-C3′-C2′-O2′, which has a Kχ of 0.195 kcal/mol/radian2 and 0.0° phase for the 3-fold term, was retained. Final fitting was performed for the residue patch within the RNA sequence UCA taken from the “early intermediate” active site geometry in the x-ray crystal structure of a hammerhead ribozyme.10 ABNR minimization was performed for this sequence with the U and A residues fixed and, where necessary to improve fitting, small adjustments to the internal parameters were made manually. The final results (Table 4) were able to reproduce the DFT structure and water binding energy very closely.
Table 3.
RNA L-J parameters
| Parameter | Protonated Methyl Ribose |
Deprotonated Methyl Ribose |
Phosphorane Methyl Ribose |
2′,3′-cyclic Methyl Ribose |
|---|---|---|---|---|
| Rmin/2O2′ | 1.77 | 1.75 | 1.76 | 1.77 |
| ∊ O2′ | −0.1521 | −0.3236 | −0.2378 | −0.1521 |
L-J parameters for the deprotonated ribose phosphate were fit to the water binding coordination distance (1.713 Å) and the water binding coordination energy (18.4 kcal/mol) with TIP3P water (See Table 4 as well). L-J parameters for axial phosphorane oxygens (bond order ≈ 0.5) were determined by averaging between the protonated (bond order ≈ 0.0) and 2′,3′-cyclic (bond order ≈ 1.0) structures.
Table 4.
Geometry fitting results of deprotonated ribose phosphate
| Bond (Å) | Protonated DFT |
Deprotonated DFT |
Relative Difference |
Protonated CHARMM |
Adjusted CHARMM* |
|---|---|---|---|---|---|
| O2′-C2′ | 1.43 | 1.33 | −0.10 | 1.43 | 1.33 (0.00) |
|
| |||||
| O2′-C2′-H2′ | 109.6 | 115.8 | 6.2 | 109.9 | 116.1 (0.0) |
| O2′-C2′-C1′ | 110.2 | 112.7 | 2.5 | 114.0 | 116.5 (0.0) |
| O2′-C2′-C3′ | 109.8 | 116.4 | 6.6 | 112.2 | 118.8 (0.0) |
| Coordination Distance | Coordination Energy | |||
|---|---|---|---|---|
| Reaction | DFT | CHARMM* | DFT | CHARMM* |
| ribose MeP− + H2O → ribose MeP−:H2O (kcal/mol) |
1.71 | 1.70 (0.00) | −18.4 | −18.4 (0.0) |
Where ribose MeP− stands for the deprotonated ribose methyl phosphate. Final fitting for the Adjusted CHARMM* residue was performed within the RNA sequence active site taken from the “early intermediate” hammerhead ribozyme x-ray crystal structure.10
3.3 Phosphate Transition State Analog
The deprotonated ribose 2′O− attack at the phosphate produces a phosphorane that is a transition state in the gas phase (Figure 2). Partial charges for the phosphorane atoms were determined similarly to those for the 2′O− ribose MeP− described above. However, due to the instability of dianionic phosphorane in the gas phase, DFT optimization was carried out with the axial P-O2′ bond length fixed to the P-O2′ bond length of a 2′O− ribose methyl phosphorane (ribose MePA2−) optimized in the aqueous phase (1.986 Å) using the Polarizable Continuum Model (PCM) solvation model.72,73 Comparison of similar neutral and anionic phosphorane structures optimized in both the gas and aqueous phases, suggests that the error for this constraint should be less than 0.02 Å for all but the axial P-O bonds which may have errors as high as 0.07 Å. Calculated CHelpG charges for hydrogen atoms were adjusted to 0.09 e with heavy atoms taking up the difference as before (Table 5). Final charges (q*TS) for the phosphorane structure were determined by adding the adjusted CHelpG charge difference (δq) between the ribose methyl phosphorane and the 2′O− ribose MeP− to the previously determined CHARMM* charges (q*D) for the the 2′O− ribose MeP− (see Table 2).
Table 5.
CHelpG charge fitting for ribose phosphorane
| Atom | q*D | qD | q’D | qTS | q’TS | δq | q*TS |
|---|---|---|---|---|---|---|---|
| C1′ | −0.12 | 0.31 | −0.19 | 0.21 | −0.09 | 0.10 | −0.02 |
| H1′ | 0.09 | −0.12 | 0.09 | −0.10 | 0.09 | 0.00 | 0.09 |
| H1″ | 0.09 | −0.20 | 0.09 | −0.02 | 0.09 | 0.00 | 0.09 |
| C2′ | 0.23 | 0.70 | 0.30 | 0.42 | 0.17 | −0.13 | 0.10 |
| H2″ | 0.09 | −0.31 | 0.09 | −0.16 | 0.09 | 0.00 | 0.09 |
| O2′ | −0.91 | −0.97 | −0.97 | −0.74 | −0.74 | 0.23 | −0.68 |
| C3′ | 0.18 | 0.65 | 0.37 | 0.54 | 0.29 | −0.08 | 0.10 |
| H3′ | 0.09 | −0.19 | 0.09 | −0.16 | 0.09 | 0.00 | 0.09 |
| O3′ | −0.63 | −0.63 | −0.63 | −0.62 | −0.62 | 0.01 | −0.62 |
| P | 1.64 | 1.37 | 1.37 | 1.54 | 1.54 | 0.17 | 1.81 |
| O1P | −0.84 | −0.84 | −0.83 | −0.88 | −0.90 | −0.08 | −0.92 |
| O2P | −0.84 | −0.81 | −0.83 | −0.92 | −0.90 | −0.08 | −0.92 |
| O3P | −0.65 | −0.58 | −0.58 | −0.68 | −0.68 | −0.10 | −0.75 |
| C3T | −0.23 | 0.36 | −0.16 | 0.54 | −0.21 | −0.05 | −0.28 |
| H3T1 | 0.09 | −0.08 | 0.09 | −0.14 | 0.09 | 0.00 | 0.09 |
| H3T2 | 0.09 | −0.08 | 0.09 | −0.16 | 0.09 | 0.00 | 0.09 |
| H3T3 | 0.09 | −0.09 | 0.09 | −0.18 | 0.09 | 0.00 | 0.09 |
| C4′ | −0.11 | 0.14 | −0.12 | 0.02 | −0.14 | −0.02 | −0.13 |
| H4′ | 0.09 | −0.07 | 0.09 | −0.04 | 0.09 | 0.00 | 0.09 |
| H4″ | 0.09 | −0.01 | 0.09 | 0.02 | 0.09 | 0.00 | 0.09 |
| O4′ | −0.53 | −0.55 | −0.55 | −0.53 | −0.53 | 0.02 | −0.51 |
CHelpG adjusted charges for determining ribose phosphorane partial atomic charges. New charges were created by taking the adjusted CHelpG charge difference δq between the ribose MeP− charges calculated in Table 2 and the ribose phosphorane. qTS are the ribose phosphorane CHelpG charges, q’TS are the adjusted CHelpG charges and q*TS are the final CHARMM* charges. See Eqn. 3 and 4 of text for further details.
Bond length and angle equilibrium parameters for the ribose MePA2− were determined from fitting to the gas phase DFT optimization of the partially frozen dianionic ribose MePA2− structure described previously. Force constants for bonds and angles were calculated by fitting to relaxed potential energy surface (PES) scans for each P-O bond and O-P-O angle of a monoanionic phosphorane (which is stable in the gas phase). The final ribose MePA2− residue was fit within the RNA sequence UCA taken from the near in-line geometry in the x-ray crystal structure of a hammerhead ribozyme “late-intermediate”.74 ABNR minimization was performed for this sequence with the U and A residues fixed and where necessary, small adjustments to the internal parameters were made to improve fitting. Because the penta-coordinated geometry of the phosphorane is quite rigid, the dihedrals had negligible effect, and hence were set to zero. The final fitting results matched with the DFT results almost exactly (Tables 6).
Table 6.
Geometry fitting results for ribose phosphorane parameterization
| Bond (Å) | DFT | CHARMM* |
|---|---|---|
| C2-O2 | 1.361 | 1.361 (0.00) |
| P-O2 | 1.99 | 1.99 (0.00) |
| P-O3 | 1.79 | 1.79 (0.00) |
| P-O5 | 1.84 | 1.84 (0.00) |
| P-OR/S | 1.53 | 1.53 (0.00) |
| O2-O5 | 3.78 | 3.79 (0.01) |
| Angle (°) | DFT | CHARMM* |
|---|---|---|
| C1-C2-O2 | 113.5 | 113.7 (0.2) |
| C3-C2-O2 | 105.9 | 105.9 (0.0) |
| C2-O2-P | 109.3 | 109.3 (0.0) |
| C3-O3-P | 117.5 | 117.5 (0.0) |
| O2-P-O5 | 163.5 | 164.0 (0.5) |
| OR-P-OS | 129.1 | 129.1 (0.0) |
| O3-P-OR/S | 115.3 | 115.3 (0.0) |
| O2-P-OR/S | 91.4 | 91.4 (0.0) |
| O5-P-OR/S | 95.4 | 95.3 (0.1) |
| O2-P-O3 | 81.0 | 81.0 (0.0) |
| O3-P-O5 | 83.3 | 83.3 (0.0) |
Final fitting for the Adjusted CHARMM* residue was performed within the RNA sequence active site taken from the “late intermediate” hammerhead ribozyme x-ray crystal structure.74
3.4 2′,3′-Cyclic Phosphate
Transesterification terminates in the creation of a 2′,3′-cyclic phosphate after exocyclic P-O5′ bond cleavage. The 2′,3′-cyclic phosphate is similar to a standard RNA ribose methyl phosphate residue except for the cyclization of the O2′-P-O3′-C3′-C2′ atoms (see Figure 2 and the parameter file in the Supporting Information). In the cyclic residue, the O2′ is bonded to the phosphorus in the same manner as the O3′. Due to the near symmetric equivalence between the O2′ and O3′ cyclic ribose phosphate sequences, the bond length, angle, and dihedral parameters are assumed to be identical. Therefore, O2′ was assigned the same atom type as O3′ (type ON2). Adjusted CHelpG charges for the 2′,3′-cyclic phosphate ring were calculated and force constants were determined through fitting to relaxed PES scans. Parameters were fit to the DFT data using ABNR minimization of the CHARMM* residue. New dihedrals were set to zero analogous those of the CHARMM27 ribose ring. Final parameters fit with very small errors with respect to the DFT results (Table 6).
3.5 Thio Substitutions
To determine the role of divalent metal ions in a reaction, it is often useful to perform thio effect experiments.75,76 Changing the oxygen atom to a sulfur effects how strongly a divalent metal can bind to this position due to the differences between their sizes, polarizabilities, and bond lengths with phosphorous. While a “hard” ion like Mg2+ will bind very tightly to a correspondingly “hard” oxygen, it will bind much more weakly to a “softer”, more diffuse sulfur.77 If the divalent metal ion binding at a certain position is required for catalysis, a large decrease in the reaction rate should be seen upon thio substitution at this position. However, the interpretation of thio affect results are sometimes inconclusive21,23,78,79 especially when possible conformational changes induced by the sulfur are considered. Molecular simulation of thio-substituted phosphates and phosphoranes and their role on structure would aid in the interpretation of experimental thio effects.
A training set of phosphate compounds, bound and unbound to hexa-coordinated Mg2+, with both single and double sulfur substitutions was constructed (Figure 4). Adjusted CHelpG partial atomic charge calculations were calculated for both mono- and di-thio DMP− substitutions. Iterative optimization of the L-J parameters for sulfur was made until the best possible fit to the DFT calculated sulfur water-binding distance and relative water-binding energy between DMP− and di-thio substituted DMP− was obtained. Force constants and equilibrium geometry parameters were then optimized iteratively along with the L-J parameters until minimization of the χ2 function was achieved.
In order to improve the differential binding of Mg2+ to the non-bridging phosphoryl O and S atoms, a NBFIX term42 was used to parameterize specifically for Mg2+-S interactions; i.e., a specific value for the 2-body non-bonded van der Waals parameters was included explicitly for Mg2+-S rather than using the Lorentz-Berthelot combining rules to derive the 2-body parameters from 1-body Mg2+ and S parameters. The resulting fit was further improved by the introduction of three new atom types. Two for the non-bridging oxygen (ONS) and sulfur (SO) of a mono-substituted phosphate and one for the non-bridging sulfur (SS) of a di-thiosubstituted phosphate. The DFT training set average binding distance for a Mg2+-S was 2.55 Å as compared to the CHARMM* average value of 2.06 Å (Table 6). The shortened distances arose out of the need to strengthen the binding energy results that, in the absence of explicit polarization on the soft sulfur atoms, is considerably underestimated with non-polarizable force fields. In the case of water binding to the substituted sulfurs, the distances are in better agreement and the relative binding energies are all within 1.0 kcal/mol of the DFT values.
The ζ ab initio torsional potential energy surfaces for the native and nonbridging thio-substituted DMP− are shown in Figure 7 (top frame). The minima identified agree well with fully relaxed structures in previous work by Florian et al.80 A significant difference between the thio-substituted and unsubstituted surfaces is the minima at the staggered position, 180°, which is negligible for the native DMP−, but of almost equal energy to the gauche minima in the case of dithio-substituted DMP−. Also noteworthy is the shift of the gauche minima (70°) and gauche-staggered barrier with thio substitution. New torsional parameters, summarized in Table 6, are defined along the S-P-O-C and O-P-O-C dihedrals to qualitatively reproduce the changes indicated by the ab initio calculations, while maintaining consistency with the CHARMM27 force field.
A scan of the DMP− and thio-substituted torsions with the same constraints as the ab initio calculations using the CHARMM27 parameters and those developed in this work are shown in Figure 7 (bottom frame). As indicated in Foloppe et al.,42 the barrier between gauche-gauche states in the unsubstituted DMP− has been significantly lowered to reproduce results from experiment, while the gauche-staggered barrier is left relatively unchanged. The thio-substituted structures retain the lowered gauche-gauche barrier and the gauche minima is shifted as indicated by the ab initio calculations. The staggered conformation has been lowered in energy for both thio-substituted molecules. For the singly substituted case, the minima at 300° was also lowered in addition to the minima at 60°, but the relative energy of the two gauche and eclipsed conformations were maintained. The lowering staggered conformation may be of particular importance to catalytic differences in native and thio-substituted structures since conformational deformation is a prerequisite to reaction.32,63,81,82
L-J parameters for the non-bridging atoms of the penta-coordinated di-anionic phosphorane structures were parameterized using a training set which contained methyl ribose phosphorane (ribose MePA2+), mono-thio substituted ribose MePA-so2+ and di-thio substituted ribose MePA-ss2+, with and without water bound (see Figure 5). In the case of the unsubstituted phosphorane, it was required to freeze the P-O2′ bond as discussed in Section 3.3. Stable gas-phase sulfur substituted phosphoranes were used for the other structures. Both the non-bridging S and O atoms were reparameterized in this case due to the large differences in geometry and charge distribution of a phosphate and phosphorane. As before, equilibrium geometry values were fit to DFT structures with force constants for all modified bonds and angles determined through fitting to relaxed PES scans. Geometry fitting results are shown in Table 6.
4 Conclusions
Molecular simulation force field parameters are presented for a series of non-standard residues important in RNA catalysis. Parameters are based on density-functional calculations and developed to be consistent with the CHARMM27 all-atom empirical force field for nucleic acids. Parameters have been developed for an activated (2′O deprotonated) ribose phosphate representing an early reactive state, a penta-coordinate phosphorane intermediate/transition state model, and a 2′,3′-cyclic phosphate transesterification product. In addition, parameters for thio-substituted analogs important in the study of experimental thio effects, and specific single and di-metal Mg2+ complexes and μ-bridging OH− ion parameters have been developed. These parameters, which are optimized specifically for the present systems, will allow the simulation of reactive intermediates and experimentally modified residues important in the study of ribozyme mechanisms. Further validation and testing of these parameters in simulations is important, although this will likely be a lengthy and tedious endeavor due to the lack of available experimental structural data for the reactive intermediates and thio-substituted nucleic acids. Ultimately, it is the hope that simulations of these systems, together with experiment, will help paint a more detailed picture of the catalytic mechanisms of RNA catalysis.
Supplementary Material
Figure 8.
Harmonic fitting to ab initio potential energy surface scan of thio-substituted dimethyl phosphate P-S bond.
Table 7.
Geometry fitting results for 2′,3′-cyclic phosphate parameterization
| Bond (Å) | DFT | CHARMM* |
|---|---|---|
| O2′-P | 1.71 | 1.71 (0.00) |
| O3′-P | 1.71 | 1.71 (0.00) |
| P-O1P | 1.50 | 1.50 (0.00) |
| P-O2P | 1.50 | 1.50 (0.00) |
| Angle (°) | DFT | CHARMM* |
|---|---|---|
| C2′-O2′-PC | 109.4 | 109.4 (0.0) |
| C3′-O3′-PC | 109.4 | 109.4 (0.0) |
| O2′-PC-O3′ | 91.0 | 91.0 (0.0) |
| O2′-PC-O1/2C | 109.0 | 109.0 (0.0) |
| O3′-PC-O1/2C | 109.0 | 109.0 (0.0) |
| O1C-PC-O2C | 124.5 | 124.5 (0.0) |
Training set average geometries for geometry fitting of 2′,3′-cyclic phosphate.
Table 8.
Geometry and binding energy results for phosphate thio-substitution parameterization
| Bond (Å) | Relative DFT | CHARMM* |
|---|---|---|
| SO⋯OT | 2.65 | 2.41 (−0.24) |
| SS⋯OT | 2.65 | 2.34 (−0.31) |
| SO⋯MG | 2.56 | 2.06 (−0.50) |
| SS⋯MG | 2.54 | 2.05 (−0.49) |
|
| ||
| SO-P | 2.01 | 2.00 (−0.01) |
| SS-P | 2.00 | 2.00 ( 0.00) |
| Angle (°) | DFT | CHARMM* |
|---|---|---|
| ON2-P-SO | 107.9 | 107.9 ( 0.0) |
| ON2-P-SS | 108.4 | 108.0 (−0.4) |
| ONS-P-SO | 121.0 | 120.6 (−0.4) |
| SS-P-SS | 121.4 | 121.1 (−0.3) |
| Binding Energy (kcal/mol) | Relative DFT | CHARMM* |
|---|---|---|
| DMP(oo)/(so)⋯H2O | −2.7 | −2.8 (−0.1) |
| DMP(oo)/(ss)⋯H2O | −3.4 | −3.8 (−0.4) |
| DMP(oo)/(ss)-(g-t)/(g-g) | −1.8 | −1.5 (−0.2) |
| Mg(HOH)5(DMP)-(oo)/(so)⋯H2O | −4.5 | −5.2 (−0.7) |
| Mg(HOH)5(DMP)-(oo)/(ss)⋯H2O | −6.8 | −7.5 (−0.7) |
Training set average geometries for geometry and reaction energy for L-J fitting. DMP-oo/ss⋯H2O above is the relative energy difference between water binding to DMP− with and without thio substitution at the non-bridging oxygens. The binding distance for SO⋯MG and SS⋯MG were fit using a nonbond fix (NBFIX). Geometries were fit to the shift predicted by DFT relative to the unsubstituted DMP−. (g-t) vs. (g-g) indicates the gauche-trans vs. gauche-gauche conformations. See the Supporting Information for more data on the fitting function weight values used.
Table 9.
Geometry fitting results for thio substituted ribose phosphorane parameterization
| Bond (Å) | DFT | CHARMM* |
|---|---|---|
| P-SR/S | 2.10 | 2.10 (0.00) |
| Angle (°) | DFT | CHARMM* |
|---|---|---|
| OR-P-SS | 125.4 | 125.3 (0.1) |
| SR-P-SS | 125.3 | 125.3 (0.0) |
| O3-P-SR/S | 113.4 | 113.5 (0.1) |
| O2-P-SR/S | 91.4 | 91.4 (0.0) |
| O5-P-SR/S | 97.3 | 97.3 (0.0) |
|
| ||
| ribose MePA-(oo/os)2− + H2O → ribose MePA-(oo/os)2−:H2O (kcal/mol) |
6.6 | 7.9 (1.3) |
| ribose MePA-(oo/ss)2− + H2O → ribose MePA-(oo/ss)2−:H2O (kcal/mol) |
6.4 | 7.7 (1.3) |
Training set average geometries and energies for geometry fitting and L-J fitting of dianionic ribose methyl phosphorane.
Table 10.
Dihedral parameters of nonbridging oxygen and sulfur for thio-substituted phosphate
| Atom Types | Force Constant | Fold | Phase | ||||
|---|---|---|---|---|---|---|---|
| Thio substitution | ONS | P | ON2 | CN9 | 0.20 | 2 | 0.00 |
| ONS | P | ON2 | CN9 | 0.20 | 3 | 0.00 | |
| SO | P | ON2 | CN9 | 0.40 | 1 | 180.00 | |
| SO | P | ON2 | CN9 | 0.10 | 3 | 0.00 | |
|
| |||||||
| Dithio substitution | SS | P | ON2 | CN9 | 0.45 | 1 | 180.00 |
| SS | P | ON2 | CN9 | 0.20 | 2 | 0.00 | |
| SS | P | ON2 | CN9 | 0.10 | 3 | 0.00 | |
5 Acknowledgment
DY is grateful for financial support provided by the National Institutes of Health (Grant GM62248), and the Army High Performance Computing Research Center (AHPCRC) under the auspices of the Department of the Army, Army Research Laboratory (ARL) under Cooperative Agreement number DAAD19-01-2-0014. Computational resources were provided by the Minnesota Supercomputing Institute. ADM is grateful for financial support provided by the National Institutes of Health (Grant GM51501).
References
- [1].Stark BC, Kole R, Bowman EJ, Altman S. Proc Natl Acad Sci USA. 1978;75:3717–3721. doi: 10.1073/pnas.75.8.3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Guerrier-Takada C, Gardiner K, Maresh T. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- [3].Cech T, Zaug A, Grabowski P. Proc Natl Acad Sci USA. 1979;76:5051–5055. doi: 10.1073/pnas.76.10.5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Zaug AJ, Cech TR. Science. 1986;231:470–475. doi: 10.1126/science.3941911. [DOI] [PubMed] [Google Scholar]
- [5].Novina CD, Murray MF, Dykxhoorn DM, Beresford PJ, Riess J, Lee S-K, Collman RG, Lieberman J, Shankar P, Sharp PA. Nat Med July. 2002;8:681–686. doi: 10.1038/nm725. [DOI] [PubMed] [Google Scholar]
- [6].Buryyanovskii LN, Shved AD. Biopolim Kletka. 1996;12:20–23. [Google Scholar]
- [7].Holmlund JT. Curr Opin Mol Ther. 1999;1:372–385. [PubMed] [Google Scholar]
- [8].Yeakley JM, Fan J-B, Doucet D, Luo L, Wickham E, Ye Z, Chee MS, Fu X-D. Nat Biotechnol. 2002 April;20:353–358. doi: 10.1038/nbt0402-353. [DOI] [PubMed] [Google Scholar]
- [9].Soukup GA, Breaker RR. Trends Biotechnol. 1999;17:469–476. doi: 10.1016/s0167-7799(99)01383-9. [DOI] [PubMed] [Google Scholar]
- [10].Scott WG, Murray JB, Arnold JRP, Stoddard BL, Klug A. Science. 1996;274:2065–2069. doi: 10.1126/science.274.5295.2065. [DOI] [PubMed] [Google Scholar]
- [11].Scott WG. Q Rev Biophys. 1999;32:241–294. doi: 10.1017/s003358350000353x. [DOI] [PubMed] [Google Scholar]
- [12].Burke JM. Nature Struct Biol. 2001;8:382–384. doi: 10.1038/87527. [DOI] [PubMed] [Google Scholar]
- [13].Rupert PB, Massey AP, Sigurdsson ST, Ferré-D’Amaré AR. Science. 2002;298:1421–1424. doi: 10.1126/science.1076093. [DOI] [PubMed] [Google Scholar]
- [14].Ferré-D’Amaré Adrian R., Zhou K, Doudna JA. Nature. 1998;395:567–574. doi: 10.1038/26912. [DOI] [PubMed] [Google Scholar]
- [15].Shih I.-h., Been MD. Annu Rev Biochem. 2002;71:887–917. doi: 10.1146/annurev.biochem.71.110601.135349. [DOI] [PubMed] [Google Scholar]
- [16].Doherty EA, Doudna JA. Annu Rev Biophys Biomol Struct. 2001;30:457–475. doi: 10.1146/annurev.biophys.30.1.457. [DOI] [PubMed] [Google Scholar]
- [17].Takagi Y, Ikeda Y, Taira K. Top Curr Chem. 2004;232:213–251. [Google Scholar]
- [18].Takagi Y, Taira K. J Am Chem Soc. 2002;124:3850–3852. doi: 10.1021/ja011044q. [DOI] [PubMed] [Google Scholar]
- [19].He Q-C, Zhou J-M, Zhou D-M, Nakamatsu Y, Baba T, Taira K. Biomacromol. 2002;3:69–83. doi: 10.1021/bm010095c. [DOI] [PubMed] [Google Scholar]
- [20].Sawata S, Komiyama M, Taira K. J Am Chem Soc. 1995;117:2357–2358. [Google Scholar]
- [21].Suzumura K, Takagi Y, Orita M, Taira K. J Am Chem Soc. 2004;126:15504–15511. doi: 10.1021/ja0472937. [DOI] [PubMed] [Google Scholar]
- [22].Laura M, Hunsicker VJD. J Inorg Biochem. 2000;80:271–281. doi: 10.1016/s0162-0134(00)00079-9. [DOI] [PubMed] [Google Scholar]
- [23].Scott EC, Uhlenbeck OC. Nucleic Acids Res. 1999;27:479–484. doi: 10.1093/nar/27.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Pley HW, Lindes DS, DeLuca-Flaherty C, McKay DB. J Biol Chem. 1993;268:19656–19658. [PubMed] [Google Scholar]
- [25].Murray JB, Szöke H, Szöke A, Scott WG. Mol Cell. 2000;5:279–287. doi: 10.1016/s1097-2765(00)80423-2. [DOI] [PubMed] [Google Scholar]
- [26].Beveridge DL, McConnell KJ. Curr Opin Struct Biol. 2000;10:182–196. doi: 10.1016/s0959-440x(00)00076-2. [DOI] [PubMed] [Google Scholar]
- [27].Auffinger P, Westhof E. Curr Opin Struct Biol. 1998;8:227–236. doi: 10.1016/s0959-440x(98)80044-4. [DOI] [PubMed] [Google Scholar]
- [28].Cornell WD, Cieplak P, Bayly CI, Gould IR, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. J Am Chem Soc. 1995;117:5179–5197. [Google Scholar]
- [29].Giudice E, Lavery R. Acc Chem Res. 2002;35:350–357. doi: 10.1021/ar010023y. [DOI] [PubMed] [Google Scholar]
- [30].Cheatham TE, III, A.Young M. Biopolymers. 2001;56:232–256. doi: 10.1002/1097-0282(2000)56:4<232::AID-BIP10037>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- [31].MacKerell AD., Jr. J Comput Chem. 2004;25:1584–1604. doi: 10.1002/jcc.20082. [DOI] [PubMed] [Google Scholar]
- [32].Torres RA, Bruice TC. J Am Chem Soc. 2000;122:781–791. [Google Scholar]
- [33].Sarzynska J, Nilsson L, Kulinski T. Biophys J. 2003;85:1522–1542. doi: 10.1016/S0006-3495(03)74766-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Dvorsky Radovan, Sevcik Josef, Caves Leo S. D., Hubbard Roderick E., Verma Chandra S. J Phys Chem B. 2000;104:10387–10397. [Google Scholar]
- [35].Réblová K, Špačková N, Štefl R, Csaszar K, Koča J, Leontis NB, Šponer J. Biophys J. 2003;84:3564–3582. doi: 10.1016/S0006-3495(03)75089-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Boero M, Terakura K, Tateno M. J Am Chem Soc. 2002;124:8949–8967. doi: 10.1021/ja017843q. [DOI] [PubMed] [Google Scholar]
- [37].Le S-Y, Chen J-H, V.Maizel NPJ., Jr J Biomol Struct Dyn. 1998;6:1–11. doi: 10.1080/07391102.1998.10508221. [DOI] [PubMed] [Google Scholar]
- [38].Auffinger P, Westhof E. J Mol Biol. 1997;269:326–341. doi: 10.1006/jmbi.1997.1022. [DOI] [PubMed] [Google Scholar]
- [39].Hermann T, Auffinger P, Scott WG, Westhof E. Nucleic Acids Res. 1997;25:3421–3427. doi: 10.1093/nar/25.17.3421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Hermann T, Auffinger P, Westhof E. Eur Biophys J. 1998;27:153–165. doi: 10.1007/s002490050121. [DOI] [PubMed] [Google Scholar]
- [41].MacKerell AD, Jr., Brooks B, Brooks CL, III, Nilsson L, Roux B, Won Y, Karplus M. CHARMM: the energy function and its parameterization with an overview of the program. In: v. R. Schleyer P, Allinger NL, Clark T, Gasteiger J, Kollman PA, Schaefer HF III, Schreiner PR, editors. Encyclopedia of Computational Chemistry. Vol. 1. John Wiley & Sons; Chichester: 1998. pp. 271–277. [Google Scholar]
- [42].Foloppe N, MacKerell AD., Jr. J Comput Chem. 2000;21:86–104. [Google Scholar]
- [43].MacKerell AD, Jr., Bashford D, Bellott M, Dunbrack RL, Jr., Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, III, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiórkiewicz-Kuczera J, Yin D, Karplus M. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- [44].MacKerell AD, Jr, Banavali N, Foloppe N. Biopolymers. 2001;56:257–265. doi: 10.1002/1097-0282(2000)56:4<257::AID-BIP10029>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- [45].Stone A. The Theory of Intermolecular Forces, volume 32 of International Series of Monographs in Chemistry. Clarendon Press; Oxford: 1996. [Google Scholar]
- [46].MacKerell AD, Jr., Banavali NK. J Comput Chem. 2000;21:105–120. [Google Scholar]
- [47].MacKerell AD, Jr., Wiórkiewicz-Kuczera J, Karplus M. J Am Chem Soc. 1995;117:11946–11975. [Google Scholar]
- [48].Yang L, Pettitt BM. J Phys Chem A. 1996;100:100–102. [Google Scholar]
- [49].Feig M, Pettitt BM. J Phys Chem B. 1997;101:7361–7363. [Google Scholar]
- [50].Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. J Chem Phys. 1983;79:926–935. [Google Scholar]
- [51].Nucliec acids database. http://ndbserver.rutgers.edu/
- [52].Becke AD. J Chem Phys. 1993;98:5648–5652. [Google Scholar]
- [53].Lee C, Yang W, Parr RG. Phys Rev B. 1988;37:785–789. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- [54].Range K, McGrath MJ, Lopez X, York DM. J Am Chem Soc. 2004;126:1654–1665. doi: 10.1021/ja0356277. [DOI] [PubMed] [Google Scholar]
- [55].Mayaan E, Range K, York DM. J Biol Inorg Chem. 2004;9:807–817. doi: 10.1007/s00775-004-0583-7. [DOI] [PubMed] [Google Scholar]
- [56].López CS, Faza ON, Gregersen BA, Lopez X, de Lera AR, York DM. Chem Phys Chem. 2004;5:1045–1049. doi: 10.1002/cphc.200400091. [DOI] [PubMed] [Google Scholar]
- [57].López CS, Faza ON, R. de Lera A, York DM. Chem Eur J. 2005;11:2081–2093. doi: 10.1002/chem.200400790. [DOI] [PubMed] [Google Scholar]
- [58].Liu Y, Lopez X, York DM. Chem Commun. 2005;31:3909–3911. doi: 10.1039/b502568k. [DOI] [PubMed] [Google Scholar]
- [59].Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JA, Jr., Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA. Gaussian 03, Revision B.05. Gaussian, Inc.; Wallingford, CT: 2003. [Google Scholar]
- [60].Chirlian LE, Francl MM. J Comput Chem. 1987;8:894–905. [Google Scholar]
- [61].Breneman CM, Wiberg KB. J Comput Chem. 1990;11:361–373. [Google Scholar]
- [62].Bayly CI, Cieplak P, Cornell WD, Kollman PA. J Phys Chem. 1993;97:10269–10280. [Google Scholar]
- [63].Atereshko V, Wallace ST, Usman N, Wincott FE, Egli M. RNA. 2001;7:405–420. doi: 10.1017/s1355838201001935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Soukup GA, Breaker RR. RNA. 1999;5:1308–1325. doi: 10.1017/s1355838299990891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. J Comput Chem. 1983;4:187–217. [Google Scholar]
- [66].Press WH, Teukolsky SA, Vetterling WT, Flannery WP. Numerical Recipes in Fortran. 2nd edition Cambridge University Press; Cambridge: 1992. [Google Scholar]
- [67].Hertel KJ, Uhlenbeck OC. Biochemistry. 1995;34:1744–1749. doi: 10.1021/bi00005a031. [DOI] [PubMed] [Google Scholar]
- [68].Lyne PD, Karplus M. J Am Chem Soc. 2000;122:166–167. [Google Scholar]
- [69].Koizumi M, Ohtsuka E. Biochemistry. 1991;30:5145–5150. doi: 10.1021/bi00235a005. [DOI] [PubMed] [Google Scholar]
- [70].Cunningham LA, Li J, Lu Y. J Am Chem Soc. 1998;120:4518–4519. [Google Scholar]
- [71].Lee H, Darden TA, Pedersen LG. J Chem Phys. 1995;102:3830–3834. [Google Scholar]
- [72].Mineva T, Russo N, Sicilia E. J Comput Chem. 1998;19:290–299. [Google Scholar]
- [73].Cossi M, Scalmani G, Rega N, Barone V. J Chem Phys. 2002;117:43–54. [Google Scholar]
- [74].Murray JB, Terwey DP, Maloney L, Karpeisky A, Usman N, Beigelman L, Scott WG. Cell. 1998;92:665–673. doi: 10.1016/s0092-8674(00)81134-4. [DOI] [PubMed] [Google Scholar]
- [75].Herschlag D, Piccirilli JA, Cech TR. Biochemistry. 1991;30:4844–4854. doi: 10.1021/bi00234a003. [DOI] [PubMed] [Google Scholar]
- [76].Oivanen M, Kuusela S, Lönnberg H. Chem Rev. 1998;98:961–990. doi: 10.1021/cr960425x. [DOI] [PubMed] [Google Scholar]
- [77].Pearson RG. J Chem Educ. 1987;64:562–581. [Google Scholar]
- [78].Zhou D-M, He Q-C, Zhou J-M, Taira K. FEBS Lett. 1998;431:154–160. doi: 10.1016/s0014-5793(98)00734-0. [DOI] [PubMed] [Google Scholar]
- [79].Yoshinari K, Taira K. Nucleic Acids Res. 2000;28:1730–1742. doi: 10.1093/nar/28.8.1730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Florián J, Štrajbl M, Warshel A. J Am Chem Soc. 1998;120:7959–7966. [Google Scholar]
- [81].Murray JB, Dunham CM, Scott WG. J Mol Biol. 2002;315:121–130. doi: 10.1006/jmbi.2001.5145. [DOI] [PubMed] [Google Scholar]
- [82].Scott WG. Curr Opin Struct Biol. 1998;8:720–726. doi: 10.1016/s0959-440x(98)80091-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.