Abstract
We have characterized a novel pleiotropic role for CymR, the master regulator of cysteine metabolism. We show here that CymR plays an important role both in stress response and virulence of Staphylococcus aureus. Genes involved in detoxification processes, including oxidative stress response and metal ion homeostasis, were differentially expressed in a ΔcymR mutant. Deletion of cymR resulted in increased sensitivity to hydrogen peroxide-, disulfide-, tellurite- and copper-induced stresses. Estimation of metabolite pools suggests that this heightened sensitivity could be the result of profound metabolic changes in the ΔcymR mutant, with an increase in the intracellular cysteine pool and hydrogen sulfide formation. Since resistance to oxidative stress within the host organism is important for pathogen survival, we investigated the role of CymR during the infectious process. Our results indicate that the deletion of cymR promotes survival of S. aureus inside macrophages, whereas virulence of the ΔcymR mutant is highly impaired in mice. These data indicate that CymR plays a major role in virulence and adaptation of S. aureus for survival within the host.
Author Summary
Staphylococcus aureus is a very harmful human pathogen that is a major cause of nosocomial infections. Humans have developed sophisticated defense strategies against invading bacteria, including the innate immune response, with the generation of an oxidative burst inside phagocytic cells. Staphylococcal infections are extremely difficult to eradicate due to the remarkable capacity of these bacteria to adapt to different environmental conditions both inside and outside the host organism. Sulfur metabolism is essential for all living organisms and is tightly controlled by regulatory proteins. In this paper, we revealed an important role for CymR, a major regulator of sulfur metabolism, in adaptation of S. aureus to the host environment. Inactivation of the gene encoding this regulator in S. aureus leads to a mutant bacterium with increased vulnerability to stress conditions including oxidative stress encountered inside the host. More importantly, the deletion of the cymR gene strongly affected the interaction of S. aureus with its host, leading to impaired virulence in mice. Our results place CymR among the potential targets for attenuation of S. aureus infections.
Introduction
Cysteine, an important sulfur-containing amino acid, plays a major role in cellular physiology. Cysteine residues are required for the biogenesis of [Fe-S] clusters, are found in the catalytic sites of several enzymes and assist in protein folding and assembly through disulfide bond formation [1], [2]. In several pathogenic bacteria, links between bacterial virulence and cysteine metabolism have been described. In toxinogenic clostridia and Bordetella pertussis, toxin synthesis is repressed in the presence of cysteine [3]–[5]. Sulfur metabolism genes are also induced upon interaction of Neisseria meningitidis and Mycobacterium tuberculosis with human cells [6], [7] and decreased virulence of mutants inactivated in various steps of sulfur metabolism has been reported in several microorganisms [6], [8], [9].
Cysteine-containing molecules such as thioredoxin and glutathione play an important role in protecting cells against oxidative stress [10], [11]. In Gram-positive bacteria, mycothiol, coenzyme A and bacillithiol are thought to function as antioxidant thiols [12]–[14]. Several studies have shown that cysteine itself plays a role in bacterial sensitivity to oxidative stress [15]–[21]. More generally, recent data report the existence of links between cysteine metabolism and the response to various stressors such as hydrogen peroxide, superoxide, diamide, nitric oxide, thiol-reactive electrophiles and metal ions [18], [20], [22]–[24].
Due to the reactivity of the SH group of cysteine and to its toxicity, cysteine metabolism is tightly controlled in bacteria. The CymR repressor, belonging to the poorly characterized Rrf2 family of regulators, has recently been identified as the master regulator of cysteine metabolism in Bacillus subtilis and Staphylococcus aureus [15], [25]. CymR forms a regulatory complex with the key cysteine biosynthesis enzyme, CysK (O-acetyl-serine (OAS) thiol-lyase), to repress genes involved in cysteine formation pathways [26]. We have recently compared the expression profiles of the S. aureus ΔcymR mutant and the parental SH1000 strain grown in the presence of cystine to characterize global changes in gene expression. The presence of cystine corresponds to conditions where the CymR repressor is active and binds to its direct targets [25], [26]. This transcriptome analysis identified sulfur metabolism genes including direct CymR targets and cell envelope associated genes as differentially expressed in the ΔcymR mutant. Moreover, we have shown the involvement of the S. aureus CymR regulator in utilization of sulfur sources of human origin and its requirement for efficient biofilm formation [25]. This suggested a potential role for this metabolic regulator in adaptation and survival within the host.
S. aureus is an important human opportunistic pathogen responsible for a broad spectrum of diseases ranging from food poisoning and minor skin lesions to life-threatening postsurgical infections in humans [27]. This bacterium is a major cause of nosocomial infections of increasing importance due to the spread of antibiotic resistance, particularly methicillin-resistant strains [28]. Oxidative stress is one of the challenges S. aureus faces during host infection. Following ingestion by phagocytic cells such as neutrophils and macrophages, bacteria are exposed to an oxidative burst [29]. Reactive oxygen species (ROS), such as superoxide anion (O2 −), hydrogen peroxide (H2O2), and hydroxyl radicals (⋅OH), may also be generated as by-products of endogenous metabolism. Their actions lead to damage of DNA, proteins and lipids [30]. Several other stressors such as diamide, resulting in disulfide stress, and metal ions or metal-containing compounds, including copper and tellurite, can also induce oxidative stress [31]. Staphylococci are highly resistant to potassium tellurite (K2TeO3), a selective agent often used for their isolation [18], [32]. Copper is an essential trace element that is toxic to cells at high concentrations and its homeostasis is maintained by copper uptake and efflux systems [33]. Detoxification enzymes that allow transformation of ROS mediate oxidative stress resistance. In particular, in S. aureus, there are two cytoplasmic manganese superoxide dismutases, SodA and SodM that catalyse superoxide radical dismutation. Hydrogen peroxide resulting from this reaction is then eliminated by the action of catalase (KatA). Oxidative stress response and metal ion homeostasis are tightly controlled via a complex regulatory network involving the PerR, Fur, Zur, and MntR repressors [34]–[37]. The PerR regulator of peroxide response controls the expression of genes encoding antioxidants and iron storage proteins and predominantly protects S. aureus cells against H2O2-induced oxidative stress [34]. Fur, the ferric uptake regulator, represses iron uptake genes and positively controls catalase expression, helping to prevent formation of the toxic hydroxyl radical via the Fenton reaction [35]. In addition, the sodA and sodM superoxide dismutases genes are directly regulated by SarA, a global virulence regulator [38], [39].
In this study, we show that the pleiotropic CymR repressor plays an important role in the response to various stresses and in virulence of S. aureus. Comparative transcriptome analysis showed the significant upregulation of genes involved in detoxification processes, including oxidative stress response and metal ion homeostasis in a ΔcymR mutant. Increased sensitivity of the ΔcymR mutant to H2O2, disulfide, tellurite and copper stresses might be explained by profound metabolic changes in this mutant. We observed increased survival of the ΔcymR mutant inside macrophages but drastically decreased virulence in mice. This indicates for the first time in S. aureus the existence of a direct link between the control of cysteine metabolism and adaptation to the host.
Results
Upregulation of genes involved in oxidative stress response and metal ion homeostasis in the ΔcymR mutant
We have revisited our previously reported expression profiling of ΔcymR mutant and parental SH1000 strains focusing on the role of CymR in the S. aureus stress response. A more detailed analysis of these transcriptome data, carried out by hierarchical clustering (see Materials and Methods), not only showed derepression of directly CymR-dependent sulfur metabolic genes in the ΔcymR mutant [25], but also revealed increased expression of genes involved in detoxification processes such as oxidative stress response and metal ion homeostasis. These include genes belonging to the PerR regulon such as ahpFC, trxB, ftnA, dps, perR and fur [34] as well as the sodA and sodM genes encoding superoxide dismutases [39] (Table 1). Notably, the copAP operon encoding a copper efflux system [33] is also strongly up-regulated in the ΔcymR mutant. Quantitative RT-PCR analysis for selected genes was in accordance with transcriptome data and the ratios obtained were generally greater than those resulting from the expression profiling. In particular, we observed about a ten-fold derepression of the ahpF and copA genes in the ΔcymR mutant (Table 1). In addition, several genes differentially expressed under H2O2, nitrosative, disulfide or paraquat stress conditions also showed altered expression in the ΔcymR mutant as compared to the parental strain (Tables 1 and S1).
Table 1. Stress response associated genes differentially expressed in the S. aureus ΔcymR mutant strain compared to SH1000.
| Gene namea (synonym) | Function/similarity | Transcriptome analysisb | qRT-PCRc | Differential expression under stress conditionsd | ||
| ΔcymR/SH1000 expression ratio | P value | ΔcymR/SH1000 expression ratio | ||||
| TSB+ Cys | TSB | |||||
| SA0366 ahpC* | alkyl hydroperoxide reductase, subunit C | 5.88 | <1.0E-16 | Nitrosative, nitrite, H2O2, diamide, paraquat | ||
| SA0365 ahpF* | alkyl hydroperoxide reductase, subunit F | 4.80 | <1.0E-16 | 8.52 | 1.73 | Nitrosative, nitrite |
| SA2481 | rhodanese family protein | 5.55 | <1.0E-16 | |||
| SA2345 copP | copper-ion-binding protein | 3.85 | <1.0E-16 | H2O2, nitrite | ||
| SA2344 copA | cation-transporting ATPase, E1-E2 family | 3.54 | <1.0E-16 | 12.48 | 1.35 | H2O2, nitrite |
| SA1709 ftnA | ferritin | 3.00 | <1.0E-16 | H2O2, nitrosative, nitrite | ||
| SA1941 dps* | general stress protein Dps | 2.83 | <1.0E-16 | 4.51 | 1.44 | H2O2, nitrosative, nitrite |
| SA1382 sodA* | superoxide dismutase | 2.75 | <1.0E-16 | 4.22 | 1.06 | Paraquat, nitrite |
| SA0128 sodM | superoxide dismutase | 1.52 | 1.16E-06 | 2.29 | 1.07 | Paraquat, nitrite |
| SA0719 trxB* | thioredoxin reductase | 1.70 | 3.07E-13 | 3.04 | 1.27 | Nitrite, diamide, paraquat |
| SA0992 trxA | thioredoxin | 1.62 | 3.54E-08 | 1.85 | Nitrite | |
| SA1979* | iron-compound ABC transporter component | 1.58 | 3.92E-05 | Nitrosative | ||
| SA0758 | thioredoxin, putative | 1.48 | 2.52E-06 | |||
| SA0755 | organic hydroperoxide resistance protein | 1.44 | 1.07E-09 | H2O2 | ||
| SA0231 | flavohemoprotein, putative | 1.42 | 5.29E-07 | H2O2, nitrosative, nitrite | ||
| SA1329 fur* | Fur regulator | 1.42 | 0.000036 | 2.37 | 1.42 | Nitrite |
| SA1678 perR* | PerR regulator | 1.42 | 0.0012 | 2.07 | 1.16 | |
| SA0531 proP | osmoprotectant proline transporter | 0.47 | <1.0E-16 | Nitrite | ||
a. The SA numbers (N315 strain) for S. aureus genes correspond to those of AureoList (http://genolist.pasteur.fr/AureoList/).
“*” indicates genes differentially expressed upon internalization of S. aureus in human epithelial cells [58].
b. The results obtained are representative of 8 hybridizations from 4 independent cultures in TSB medium with 2 mM cystine. The generated data sets were loaded into the GenoScript Database (http://genoscript.pasteur.fr)[25].
c. For qRT-PCR analysis, total RNA was extracted from S. aureus strains grown in TSB medium with or without 2 mM cystine. After reverse transcription, specific cDNAs was quantified by qRT-PCR using 16S rRNA gene for normalization.
The CymR regulator functions as a transcriptional repressor. Similar levels of derepression in the ΔcymR mutant were observed for stress-related genes and for previously identified direct targets of CymR [25]. To determine whether CymR also directly controls stress-related genes, we performed electrophoresis mobility shift assays (EMSAs) with promoter regions of the copA, ahpC, sodA, ftnA and dps genes using crude extracts of a S. aureus ΔcymR mutant overexpressing or not cymR. We have previously used this approach successfully to demonstrate specific direct interactions of CymR with several promoter regions [25]. No specific DNA-protein complexes for the promoters of stress-related genes were formed under these conditions, indicating that these genes are likely to be controlled indirectly by CymR (data not shown). This is in agreement with the absence of a CymR binding motif in the promoter regions of stress-related genes [25]. CymR also controlled the synthesis of the two oxidative stress regulators, Fur and PerR, and several genes derepressed in the ΔcymR mutant belong to the PerR regulon (ahpFC, trxB, dps, ftnA). However, CymR does not appear to bind directly to the promoter regions of either fur or perR in EMSAs (data not shown).
To determine the relative roles of CymR, PerR and Fur on the expression of stress-related genes, we carried out quantitative RT-PCR analysis of gene expression in various mutants inactivated for the cymR, perR and/or fur genes. While some genes of the PerR regulon, such as dps and ahpF, are strongly repressed by PerR and negatively affected to a lesser extent by Fur and CymR, we did not observe any synergistic effect of the combined mutations. On the contrary, these experiments revealed an antagonistic effect of the ΔcymR and ΔperR mutations on the expression of the dps and ahpF genes (Table S2). These genes, belonging to the PerR regulon, were strongly derepressed in a perR mutant as compared to the SH1000 strain. However, in a perR cymR mutant, the derepression of these genes was lower, and similar to that observed in the ΔcymR mutant suggesting antagonistic effects of these two mutations. Inactivation of CymR had similar effects in different mutant backgrounds on the expression of other genes including sodA, sodM and copA. Thus, the CymR effect on stress response does not appear to be mainly mediated by the known oxidative stress regulators, PerR and Fur, even though these regulatory systems may interfere with each other.
To compare the changes induced by peroxide stress in the ΔcymR mutant and the parental SH1000 strain, we analyzed the expression of several stress-related genes after an H2O2 challenge (Table S2). As expected, we observed a strong induction of ahpF and dps in strain SH1000. However, the induction of ahpF expression by H2O2 was lost in a ΔcymR mutant while the extent of dps induction was reduced in this mutant as compared to SH1000. This illustrated that the cymR deletion resulted in an altered stress response at the molecular level.
Increased sensitivity of the ΔcymR mutant to disulfide, tellurite, copper and H2O2 stresses
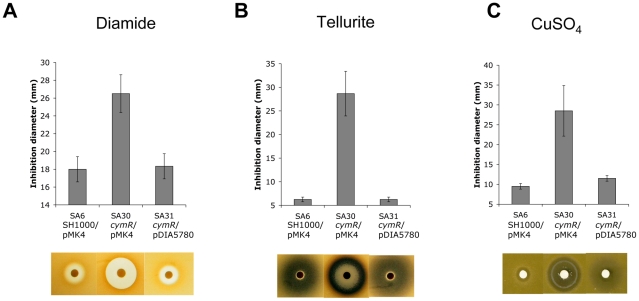
S. aureus can survive a wide range of stresses during its life cycle [29]. As mentioned above, a set of genes involved in stress response was differentially expressed in the ΔcymR mutant as compared with the parental strain. The role of CymR in responses to various stress stimuli was tested using either disk diffusion assays or survival analyses. In disk diffusion assays, the ΔcymR mutant was significantly more sensitive than SH1000 to 1 M diamide, a specific thiol oxidant that causes disulfide stress (Fig. 1A). The SH1000 strain showed high resistance to 200 mM K2TeO3 in disk diffusion assays without a detectable growth inhibition area around the 6 mm-disk. By contrast, the ΔcymR mutant was extremely sensitive to tellurite stress with a growth inhibition area of 28 mm under the same conditions (Fig. 1B). Bacterial detoxification of tellurite leads to formation of insoluble tellurium (Te°), appearing as black deposits in the growth plates. We also observed increased sensitivity of the ΔcymR mutant to copper stress in disk diffusion assays carried out with 200 mM CuSO4 (Fig. 1C). Sensitivity of the ΔcymR mutant to other metal ions (FeCl3, Pb(CH3COO)2, MnSO4, CoCl2, ZnSO4, HgSO4 and NiSO4) was similar to that of the parental strain (data not shown). In all cases, increased stress sensitivity of the ΔcymR mutant could be complemented by the introduction of plasmid pDIA5780 carrying the cymR gene (Fig. 1).
Figure 1. Stress resistance phenotypes of a S. aureus ΔcymR mutant.
Disk diffusion assays were performed with 1 M diamide (A), 200 mM tellurite K2TeO3 (B) or 200 mM CuSO4 (C) in TSB medium. Quantitative analysis of growth inhibition is shown in the upper panels. The lower section shows representative results of stress sensitivity assays. Strains SA6 (SH1000/pMK4), SA30 (ΔcymR/pMK4) and SA31 (ΔcymR/pDIA5780) were used for complementation experiments. Results correspond to the mean values with standard deviations and are representative of at least three independent experiments.
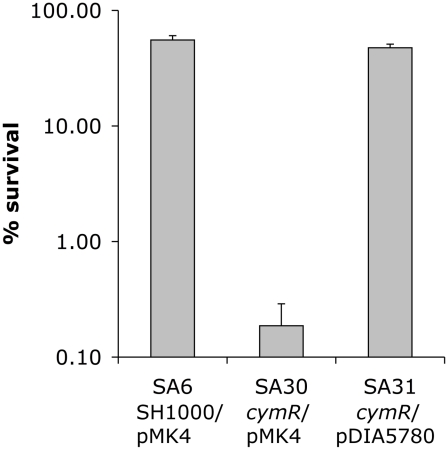
With respect to oxidative stress, no significant differences in sensitivity were observed between the ΔcymR mutant and the parental strain in disk diffusion assays in the presence of paraquat (2 M methyl viologen) (data not shown). Viability of the ΔcymR mutant and the SH1000 strain grown in TSB medium with cystine was also tested 1 h after addition of 20 mM H2O2. A 1000-fold reduction in survival was observed for the ΔcymR mutant as compared to the parental strain and viability was restored in a ΔcymR mutant complemented by pDIA5780 (Fig. 2). We further tested the oxidative stress response in mutants inactivated for CymR, PerR and/or Fur. We observed a 10-fold decreased viability in a ΔperR ΔcymR mutant as compared to the ΔcymR mutant and extremely low survival capacities for the ΔcymR ΔperR Δfur mutant as compared to the ΔperR Δfur mutant (data not shown). Taken together, our results indicate that CymR plays a major role in staphylococcal stress response, independently of other known regulators.
Figure 2. Oxidative stress sensitivity of a S. aureus ΔcymR mutant.
Viability of SA6 (SH1000/pMK4), SA30 (ΔcymR/pMK4) and SA31 (ΔcymR/pDIA5780) was tested. Exponential-phase cells grown in TSB medium with 2 mM cystine were treated for 1 h with 20 mM H2O2 and plated on BHI. Results represent the mean values for survival with standard deviations and are representative of at least two independent experiments.
Increased oxidative stress sensitivity of the ΔcymR mutant could be due to elevated intracellular cysteine pools, driving production of hydroxyl radicals via the Fenton reaction and leading to cellular damage [20]. We tested the effect of cystine on stress sensitivity in a ΔcymR background. We observed decreased sensitivity to tellurite and copper stress of the ΔcymR mutant in the presence of cystine in several genetic backgrounds including perR and/or fur mutants (Fig. S1 and S2). By contrast, the ΔcymR mutant showed increased sensitivity to both diamide and H2O2 stress in the presence of cystine (data not shown). Similarly, the decrease in H2O2 and diamide stress resistance in the ΔcymR mutant due to perR inactivation was more pronounced in the presence of cystine (data not shown). In agreement with altered oxidative stress response, the presence of cystine affected the expression of genes associated with stress response in the ΔcymR mutant as shown by quantitative RT-PCR analysis (Table 1). These genes were differentially expressed in a ΔcymR mutant only in the presence of cystine.
Metabolic changes of the ΔcymR mutant in the presence of cystine
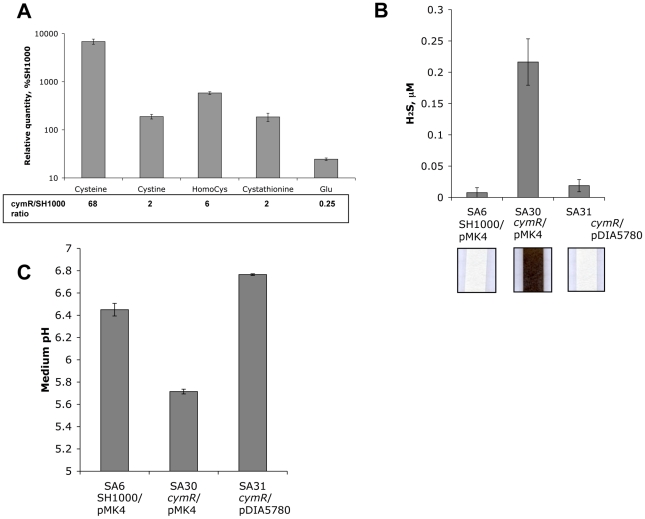
The altered stress response linked to cymR inactivation can be explained by an imbalance in thiol redox status. The derepression of genes involved in cystine uptake and cysteine biosynthesis from sulfide and homocysteine may result in cysteine accumulation in the ΔcymR mutant. Analysis of the intracellular pools of several metabolites using HPLC revealed a strong up to 68-fold increase of the intracellular cysteine concentration in the ΔcymR mutant in comparison with the parental strain during growth in TSB medium with cystine (Fig. 3A and Table S3). We also observed a 2-fold increase in cystine and cystathionine content and a 6-fold increase in homocysteine content. This analysis also revealed a 36-fold increase in the cysteine to cystine ratio in the ΔcymR mutant as compared to the SH1000 strain, reflecting the imbalance in thiol redox status of the cell in the absence of CymR. The estimated glutamate concentration decreased 4-fold while the concentration of other amino acids was unchanged in the ΔcymR mutant as compared to the SH1000 strain. As cysteine is probably toxic for the cell at high concentrations, it may then be rapidly transformed into hydrogen sulfide, pyruvate and ammonia by cysteine desulfhydrases. The MccB, MetC and CysK enzymes have cysteine desulfhydrase activities in B. subtilis [40] and orthologous proteins are present in S. aureus. We then compared production of hydrogen sulfide, the main product of cysteine catabolism, in the ΔcymR mutant and SH1000 strains grown in the presence of cystine. An important increase in H2S production was observed in the ΔcymR mutant in a qualitative lead-acetate-paper assay and a 40-fold increase was further confirmed by an H2S quantification assay (Fig. 3B). The introduction of a plasmid carrying the intact cymR gene into the ΔcymR mutant led to a level of H2S production similar to that observed in the parental SH1000 strain (Fig. 3B). In the absence of cystine, H2S production was undetectable in the ΔcymR mutant and SH1000 strains (data not shown). We also measured the pH of the TSB medium after 16 h culture in the presence of cystine. A significant acidification of the medium was observed with the ΔcymR mutant as compared with the parental strain (Fig. 3C). This may be associated with pyruvate production from cysteine and/or with a decreased capacity to catabolize organic acids. These changes in the pH of the growth medium of the ΔcymR mutant could be reversed by the introduction of a plasmid carrying the cymR gene.
Figure 3. Metabolic changes in the ΔcymR mutant after growth in the presence of cystine.
The strains were grown in TSB medium with 2 mM cystine. A. Intracellular metabolite concentrations were estimated by HPLC for the ΔcymR mutant (SA17) and the parental SH1000 strain. The ΔcymR/SH1000 ratio is indicated. The complete data on metabolite concentrations are given in Table S3. B. H2S production measurement was performed using the quantitative methylene blue method and a Na2S standard curve. Representative results of lead-acetate paper assays are shown in the lower section. Strains SA6 (SH1000/pMK4), SA30 (ΔcymR/pMK4) and SA31 (ΔcymR/pDIA5780) were used. C. The pH of the medium was measured after an overnight culture (16 h) at 37°C. Strains SA6 (SH1000/pMK4), SA30 (ΔcymR/pMK4) and SA31 (ΔcymR/pDIA5780) were used. Results correspond to the mean values with standard deviations and are representative of at least two independent experiments.
The ΔcymR mutation favors S. aureus survival within macrophages
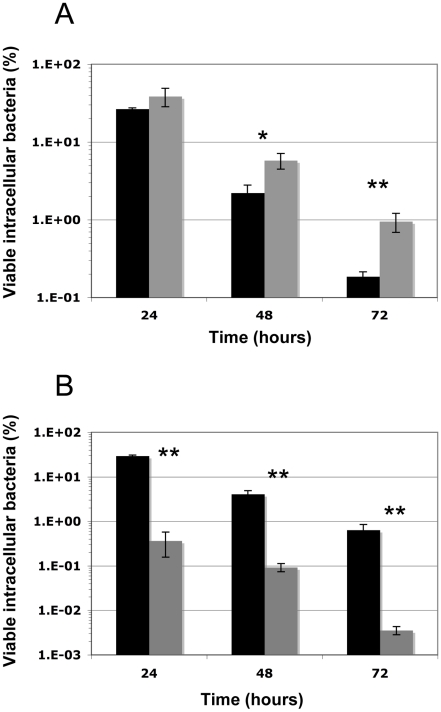
Since the CymR regulator was shown to be involved in stress adaptation, we tested its role in the S. aureus survival inside macrophages. Professional phagocytes are the first line of defense encountered by pathogens during the infection process. Since it has been shown that S. aureus is particularly efficient in persisting within professional phagocytes [41], [42], the survival of the parental SH1000 strain and the ΔcymR derivative inside RAW 264.7 murine macrophages was investigated over a 3-day period (Fig. 4A). We also examined the survival of a sodAsodM mutant as a positive control of macrophage stress generation. As shown in Fig. 4B, clearance of the sodAsodM mutant was much faster than that of the wild type strain, directly correlating its increased stress sensitivity with lowered survival within RAW 264.7 murine macrophages. We measured the internalization rates of the parental and ΔcymR mutant strains, which were identical (about 90% of entry for a multiplicity of infection (m.o.i) = 5). Viable bacterial counts inside macrophages over time allowed us to demonstrate that the ΔcymR mutant is more resistant to macrophage stress than the parental strain (Fig. 4A).
Figure 4. Intracellular survival of S. aureus SH1000, the ΔcymR and sodAsodM mutants in RAW 264.7 macrophages.
Macrophages were infected as described (See Materials and Methods) with the SH1000 parental strain (black) or the ΔcymR mutant (panel A) or the sodAsodM mutant (panel B) (grey). Viable intracellular bacteria at 24, 48, and 72 h were counted and expressed as a percentage of internalized bacteria. One representative experiment (out of 3) is shown, performed in triplicate (means±SD). *: P<0.05; **: P<0.01.
The ΔcymR mutation has a drastic effect on S. aureus virulence
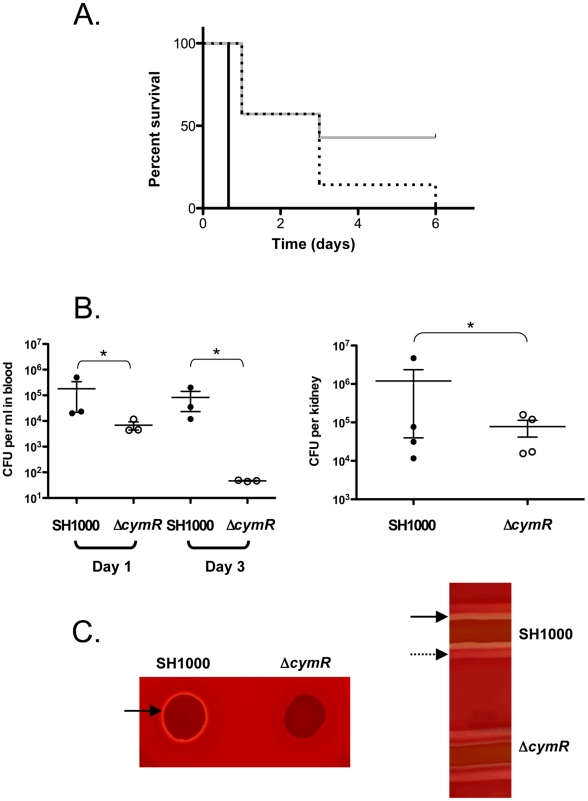
Professional phagocytes are part of the host anti-microbial defense and the ΔcymR mutation seems to favor intracellular survival of S. aureus. We therefore tested whether this selective advantage could have an effect on global S. aureus virulence, using a murine intraperitoneal infection model with BALB/c mice since this lineage has been shown to be susceptible to S. aureus infection [43]. We infected mice intraperitoneally with 3.108 colony-forming units (CFU) of either the SH1000 strain or the ΔcymR mutant. As shown in Fig. 5A, while mice infected by the parental strain were all dead (7 mice/7) 18 h after inoculation (black curve), those infected with the ΔcymR mutant displayed a significant extension of time-to-death and 3 mice (out of 7) were still alive 6 days post-infection (grey curve). As a control, we tested a sodA sodM mutant previously described as impaired in its capacity to develop abscesses in a mouse subcutaneous infection model [39]. We observed that decreased mouse mortality linked to the bacterial sodA sodM inactivation was equivalent to that caused by cymR inactivation (Fig. 5A, dotted lines). In order to follow bacterial dissemination within the animal, we also infected mice with a sub-lethal dose (5.107 CFU) of either the SH1000 strain or the ΔcymR mutant. We followed bacteraemia at 1 and 3 days post-infection and quantified the renal load 7 days post-infection. The bacterial load drastically decreased in the cymR mutant (Fig. 5B). In the blood, one -day post-infection, there was at least 1-log-unit decrease for the ΔcymR mutant compared to the wild type strain (Fig. 5B, left panel). The difference between the two strains increased 3 days post-infection with more than 3-log-unit less bacteria in the blood in a ΔcymR background. The bacterial load in the kidneys also showed a colonization defect of the ΔcymR mutant since 7 days post-infection there was more than 1-log-unit less CFU with the ΔcymR strain than with SH1000 (Fig. 5B, right panel). Thus, although the ΔcymR mutation appears to be beneficial with regard to survival within professional phagocytes, it largely decreases global S. aureus virulence.
Figure 5. Role of CymR during S. aureus infection.
A. Survival of BALB/c mice following intraperitoneal challenge with 3.108 CFU of the SH1000 parental strain (black), the ΔcymR derivative (grey), and the sodAsodM mutant (dotted lines). Comparison of survival curves was performed using the log-rank test (P<0.005). Results are representative of at least two independent experiments. B. Bacterial counts of the SH1000 strain and the ΔcymR mutant in blood (left panel) and kidneys (right panel). BALB/c mice were infected by the i.p. route with 5.107 CFU of each strain. Bacteraemia was measured 1 and 3 days post-infection, and the bacterial load in kidneys was determined 7 days post-infection. Bars represent mean CFU. *, P>0.001 (Student's t test). C. Hemolytic activity assays. Overnight cultures of the SH1000 wild type strain and the ΔcymR derivatives were spotted (20 µl) on horse blood agar plates or streaked on sheep agar plates. Full and dotted arrows indicate δ- and β-hemolysins, respectively.
In order to determine whether the production of major virulence factors is affected in the ΔcymR mutant, we performed hemolytic activity assays on blood agar plates. These experiments clearly show that the ΔcymR mutant is impaired in its capacity to produce δ-hemolysin as compared to the parental SH1000 strain (Fig. 5C, left panel). On sheep blood agar plates, we can distinguish between δ- and β-hemolysin [44]. As shown on Fig. 5C (right panel) we have confirmed that the δ-hemolysin production was strongly reduced in the cymR mutant strain with respect to the parental strain, whereas β-hemolytic activity was not affected by the mutation. α-hemolytic activity was not tested since it is inhibited by β-hemolysin [45]. This significantly altered δ-hemolysin production likely contributes to the virulence defect observed in the absence of CymR.
Discussion
In S. aureus, diamide and H2O2-induced stresses result in induction of several direct CymR target genes including mccAB, cysM, tcyABC and metNPQ, indicating an increased requirement for cysteine under these conditions [46], [47]. Conversely, here we show an upregulation of part of the peroxide stress PerR regulon, of superoxide stress (sodA and sodM) and copper efflux system (copAP) genes together with other stress-related genes in a S. aureus mutant lacking the master regulator of cysteine metabolism, CymR. However, the effect of CymR on these genes appears to be indirect. We investigated possible connections between CymR and the PerR and Fur regulators of oxidative stress response. CymR appears to affect stress response independently of these regulators, since the effect of cymR inactivation on stress sensitivity and gene expression is still observed in perR and/or fur mutant backgrounds.
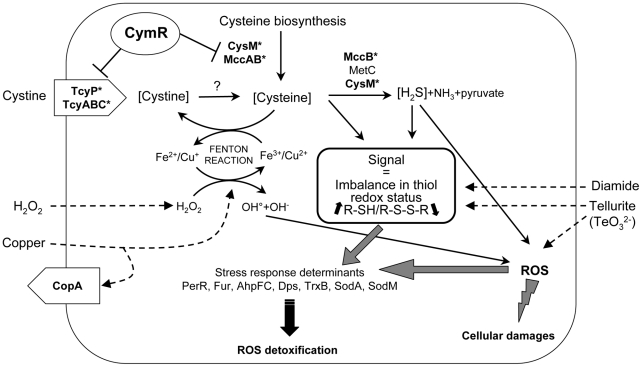
We propose that these different stress response systems may recognize a common stress signal that is present in the ΔcymR mutant. This signal could be related to thiol-redox homeostasis imbalance and to increases in intracellular cysteine pools or changes in other cysteine-related compound content including H2S (Fig. 6). Cysteine is one of the major cellular thiols in S. aureus. Metabolite content estimation revealed a 36-fold increase in the cysteine to cystine ratio in the ΔcymR mutant reflecting the imbalance in thiol redox status in the absence of CymR. It is worth noting that the simultaneous induction of the PerR and CymR regulons as well as metal-ion efflux systems, including CopA, by thiol-reactive electrophiles leading to imbalance of thiol-redox homeostasis has been reported in B. subtilis [24]. In agreement with this metabolic hypothesis, the addition of cystine to the culture medium affected stress-related phenotypes of the ΔcymR mutant. Recent studies suggested the existence of links between cysteine and/or cysteine-containing molecules and oxidative stress defense in several bacterial systems with positive or negative effects of this amino acid. Cysteine protects Lactobacillus reuteri from H2O2 stress while cysteine or thiol-derived compounds such as glutathione are important for defense against damages [10], [19]. By contrast, in E. coli, a 8-fold increase in intracellular cysteine concentrations promotes oxidative DNA damages by driving the Fenton reaction due to the efficient reduction of Fe3+ by cysteine [20]. We observed a strong 68-fold increase in the intracellular cysteine pool in the ΔcymR mutant grown in the presence of cystine, leading to a 1000-fold increase in sensitivity to H2O2 stress. However, the addition of extracellular or cell-penetrating iron and copper chelators (dipyridyl, desferal, neocuproine and ferrozine) had no positive effect on viability of the ΔcymR mutant after an H2O2 challenge (data not shown). This suggests more complex mechanisms of altered stress response in addition to the Fenton reaction-mediated process, as recently proposed for other microorganisms [48], [49]. High cysteine levels are correlated with the production of H2S by cysteine desulfhydrases (MccB, MetC, and CysM) (Fig. 6). H2S increases the formation of H2O2 and other ROS in several organisms and inhibits human superoxide dismutase activity and S. aureus catalase activity in acid medium [50]–[52]. This could also contribute to the oxidative stress sensitivity of the ΔcymR mutant.
Figure 6. Proposed model for the role of CymR in the stress response in S. aureus.
In the absence of CymR, the derepression of genes involved in pathways leading to cystine/cysteine uptake (e.g. tcyP, tcyABC encoding cystine transporters) and biosynthesis (e.g. cysM encoding OAS thiol-lyase and mccAB encoding homocysteine to cysteine conversion enzymes) leads to increased intracellular cysteine levels. High amounts of cysteine promote oxidative DNA damage by driving the Fenton reaction. Iron and copper are both capable of catalyzing the formation of hydroxyl radicals from H2O2. Cysteine may be catabolized into hydrogen sulfide (H2S), pyruvate and ammonia (NH3) by cysteine desulfhydrases (MccB, MetC, CysM). High H2S levels may also induce oxidative stress by the formation of reactive oxygen species (ROS). The altered stress response may be explained by an imbalance in thiol redox status induced by cymR inactivation. Different stress response systems including PerR regulon members (AhpFC, Dps, TrxB, PerR and Fur), superoxide dismutases (SodA, SodM) and a copper efflux system (CopA) may recognize a common stress signal in the ΔcymR mutant. This signal may either be an increase in intracellular cysteine pools or changes in levels of other cysteine-related compounds. Tellurite, copper, H2O2 and diamide can cause imbalance in the thiol status of the cytoplasm and oxidative stress. A question mark indicates a step, which remains to be characterized. Asterisks indicate directly controlled CymR target genes.
The S. aureus ΔcymR mutant exhibited increased susceptibility to disulfide, copper, tellurite, and H2O2-induced oxidative stresses. Diamide, tellurite and copper can each cause both oxidative stress as well as an imbalance in the thiol redox status of the cytoplasm (Fig. 6). A recent proteomic study that analyzed the diverse S. aureus responses to H2O2, diamide and paraquat [47] indicates a close relationship between disulfide and H2O2 stress responses, in agreement with the similar behavior of the ΔcymR mutant toward these compounds. Tellurite (TeO3 2−) is toxic for most forms of life, even at very low concentrations. The genetic and biochemical basis underlying bacterial tellurite toxicity is still poorly understood [32]. However, several tellurite resistance determinants have been identified, mainly in E. coli, suggesting mechanisms involving cysteine metabolism and cellular oxidative stress due to its strong oxidizing ability. Cysteine synthases from various bacteria and molecules containing cysteine including glutathione are involved in tellurite resistance via reductive detoxification of this compound [32]. In S. aureus, the cysM mutant defective in cysteine synthase is more sensitive to tellurite, probably due to cysteine depletion [18]. Inactivation of cymR also leads to extreme sensitivity to tellurite, even greater than that of the cysM mutant. However, the addition of cystine to the culture medium resulted in a drastic decrease in tellurite toxicity in both the cymR and cysM mutants (Fig. S1 and data not shown). The accumulation of cysteine and/or H2S (Fig. 3) under these conditions could promote tellurite detoxification leading to the formation of nontoxic tellurium. As observed with tellurite, a ΔcymR mutant is more sensitive to copper stress than the parental SH1000 strain, and this effect is more pronounced in the absence of cystine (Fig. S1). The copA and copP genes encoding a copper efflux system involved in maintaining copper homeostasis in S. aureus [33] are strongly upregulated in the ΔcymR mutant in the presence of cystine (Table 1). Further studies will be required to characterize the molecular mechanisms linking CymR to tellurite and copper sensitivity.
The intracellular cysteine level is kept within a narrow range to address both the cysteine supply for protein synthesis and the production of other essential molecules and the necessity of maintaining cysteine levels below the toxicity threshold. Elevated cysteine or H2S levels must also be avoided as they may lead to cysteine autooxidation, the production of ROS and protein thiol oxidation [53], [54]. The CymR regulator in S. aureus plays an essential role in maintaining intracellular cysteine levels. However, H2S together with cysteine may be a signal recognized by several oxidative stress defense systems in S. aureus. During infection, this pathogen must cope with host phagocytic attack, accompanied by the release of a number of ROS including superoxide anion, hydrogen peroxide, hydroxyl radical, peroxynitrite and hypochlorous acid [29], [55]. In this study, we showed that the ΔcymR mutant has an increased long-term survival rate within macrophages. This result could be related to increased transcription in the ΔcymR mutant of a number of genes known to be differentially expressed under several host-related stress conditions, including H2O2, nitrite and nitrosative stresses (Tables 1 and S1). The differences observed in vitro after a H2O2 challenge and in vivo in macrophages may be explained by variations in the level of H2O2 formed as well as a multitude of reactive species produced inside macrophages. Despite the fact that cymR inactivation promotes survival of S. aureus inside the macrophages, virulence of the ΔcymR mutant in mice is drastically impaired as previously observed for an S. aureus strain lacking catalase and beta-toxin [56]. During the infectious process, the CymR regulator influences different virulence pathways. Indeed, we have shown that mice infected with a lethal dose of the SH1000 strain died very rapidly (less than 18 hours post-inoculation), suggesting that toxemia is responsible. Accordingly, we observed that the ΔcymR mutant has impaired hemolytic activity. Reduced hemolysin production may be responsible at least in part for the virulence defect observed in the absence of CymR. In addition, bacteraemia and the bacterial load in kidneys following infection with a sub-lethal dose were significantly decreased in the absence of CymR.
Bacterial metabolism has been linked to virulence of Staphylococci by several studies [57]. Some CymR regulon cysteine metabolic genes (mccA, cysM and tcyAB) were shown to be differentially expressed upon internalization of S. aureus in human epithelial cells [58]. In the ΔcymR mutant, we also observed differential expression of genes known to be affected upon internalization in human cells (Table S4).
Our results suggest that the link between cysteine metabolism control by CymR, stress response and virulence is likely indirect and may be integrated into the general concept that alterations of the bacterial metabolic status create metabolic signals that may be “sensed” by the regulatory network controlling virulence determinants, as proposed by Somerville and Proctor [57]. One hypothesis may be that the alteration in redox cell status and metal ion homeostasis modulates the activity of virulence and stress-response regulators, including SarA, SarZ and PerR. Indeed, recent results have shown that the central virulence regulator, SarA, is responsive to redox and pH [59] and that SarZ is a redox active regulator [60], [61]. Thus, cymR inactivation may affect redox-mediated virulence control in S. aureus at several levels of the regulatory network. Indeed, a number of genes differentially expressed in strains deficient for virulence regulators (such as SarA, AgrA, ArlSR, SaeSR, Rot and MgrA) showed altered expression in the ΔcymR mutant in comparison with the parental SH1000 strain (Table S4).
The role of CymR in virulence is most likely multifactorial since, as we show here, it controls several steps in the infectious process, including dissemination within the host and colonization of different organs. The ΔcymR mutant is also affected in biofilm formation and in synthesis of exotoxins (hemolysins) and cell envelope components, functions that could be important for host colonization [25]. Our data bring important insights into understanding the interactions between sulfur metabolism and virulence of this major pathogen and suggest interesting possibilities for metabolic strategies to attenuate S. aureus infection. Proteins involved in controlling cysteine metabolism may therefore represent potential targets for antibacterial compounds aimed at treating staphylococcal infections.
Materials and Methods
Ethics statement
All the animal experiments described in the present study were conducted at the Institut Pasteur according to the European Union guidelines for the handling of laboratory animals (http://ec.europa.eu/environment/chemicals/lab_animals/home_en.htm) and were approved by the Institut Pasteur animal care and use committee.
Bacterial strain construction and growth conditions
Bacterial strains used in this study are listed in Table 2. S. aureus was grown in brain heart infusion (BHI) (Oxoid) or tryptic soy broth/agar (TSB/TSA) (Difco) [25]. Antibiotics were added at the following concentrations: chloramphenicol, 5 µg ml−1; erythromycin, 1 or 5 µg ml−1, tetracycline, 5 µg ml−1 and kanamycin, 50 µg ml−1. S. aureus was transformed by electroporation [62]. The chromosomal perR, fur, sodA and sodM mutations [34], [35], [39](Table 2) were introduced into the SH1000 strain or ΔcymR mutant by Φ11 phage transduction [63].
Table 2. Strains and plasmids used in this study.
| Strain or plasmid | Genotype or description | Reference or source |
| Strain | ||
| SH1000 | Functional rsbU+ derivative of 8325-4 wild-type strain | [70] |
| S897 | 8325-4 perR::kan a | [34] |
| S906 | 8325-4 fur::tet | [35] |
| S1799 | SH1000 sodA::Tn917(ery) | [39] |
| S739 | SH1000 sodM::tet | [39] |
| SA17 | SH1000 ΔcymR | [25] |
| SA6 | SH1000/pMK4 | [25] |
| SA30 | SH1000 ΔcymR/pMK4 | [25] |
| SA31 | SH1000 ΔcymR/pDIA5780 | [25] |
| SA37 | SH1000 perR::kan | This study |
| SA39 | SH1000 ΔcymR perR::kan | This study |
| SA41 | SH1000 fur::tet | This study |
| SA42 | SH1000 ΔcymR fur::tet | This study |
| SA53 | SH1000 perR::kan fur::tet | This study |
| SA61 | SH1000 ΔcymR perR::kan fur::tet | This study |
| SA63 | SH1000 sodA::ery sodM::tet | This study |
| Plasmids | ||
| pMK4 | E. coli – S. aureus catr shuttle vector | [71] |
| pDIA5780 | pMK4 derivative carrying the cymR gene for complementation | [25] |
akan, ery, tet and cat genes encode proteins leading to kanamycin, erythromycin, tetracycline and chloramphenicol resistance.
Stress response analysis
Disk diffusion assays were performed as follows: 5 ml of TSB or BHI top agar (0.7%, wt/vol) was seeded with 100 µl of an exponential-phase S. aureus culture in TSB or BHI medium (OD600 = 0.2) and used as an overlay on a TSA or BHI agar plates. When indicated 2 mM cystine was added to the culture medium and to the agar plates. Sterile 6 mm disks were placed on top of the overlay, and 10 µl of either 1 M diamide, 200 mM K2TeO3, 200 mM CuSO4, 10 M H2O2 or 2 M paraquat (methyl viologen) (Sigma) was added to the disk. Diameters of growth inhibition zones were measured after 24 h of incubation at 37°C. Hydrogen peroxide resistance assays were carried out as previously described with some modifications [18]. Cells were grown in TSB medium with or without 2 mM cystine. At exponential phase (OD600 = 0.2), H2O2 was added to a final concentration of 20 mM in TSB medium. After 1 h of incubation, cells were serially diluted in BHI medium and viability was assessed by overnight growth on BHI agar.
Hierarchical clustering analysis
Previously obtained transcriptome data [25] were analyzed using hierarchical clustering as the less a priori-based method for transcriptome data exploitation. Uncentered Pearson correlation was used for distance calculation, and the average-linkage clustering was performed on logarithmically transformed data for gene expression ratio in SH1000 versus ΔcymR mutant. We used the Michael Eisen Cluster software program, followed by tree diagram visualization with TreeView [64]. This analysis revealed several specific clusters including the group of genes upregulated in the ΔcymR mutant and involved in detoxification processes.
Estimation of metabolite content
Strains were grown in TSB medium with 2 mM cystine to an OD600 of 1 (with 1/10 medium-to-flask volume ratio at 160 rpm shaking). H2S production was revealed using lead-acetate paper (Macherey-Nagel) which turned black following incubation for up to 3 h at 37°C. H2S production was quantified by the modified methylene blue reaction as previously described [65]. Intracellular concentrations of amino acids and other ninhydrin-reactive compounds were estimated using high-pressure liquid chromatography (HPLC) [26], [66]. Briefly, cells were suspended in a sulfosalicylic acid buffer (3% final concentration) and disrupted using a FastPrep apparatus (Bio101). Supernatant samples were analyzed by cation-exchange chromatography, followed by ninhydrin postcolumn derivatization as previously described [66].
RNA extraction and quantitative real-time PCR
Total RNA was isolated from S. aureus strains grown in TSB with or without 2 mM cystine as previously described [15]. For H2O2 stress induction bacteria were incubated with 20 mM H2O2 for 10 minutes followed by RNA extraction. Quantitative real-time PCR analysis was performed as previously described [25]. Oligonucleotides used in this study are listed in Table S5.
Electrophoresis mobility shift assays
DNA fragments containing various promoter regions were amplified by PCR using specific primers and chromosomal DNA of S. aureus strain SH1000. PCR products were labeled using [γ32P]ATP 5′-end labeled specific primers. Protein-DNA complexes were formed in 10 µl reaction volumes, by incubating labeled DNA fragments with various amounts of crude extracts of the S. aureus ΔcymR mutant carrying either pDIA5780 (pMK4-cymR) or pMK4 as previously described [66].
Macrophage survival assays
Murine macrophage RAW 264.7 cells were used for bacterial survival assays as previously described [67] with some modifications. Briefly, bacteria were grown in TSB until OD600 ∼2. Cultures were washed in PBS and adjusted to the desired inoculum in RPMI 1640 medium (Gibco), and CFU counts were verified by plating serial dilutions on TSA plates. Macrophages grown to confluence were counted and incubated with bacteria (m. o. i. ∼5) in RPMI 1640 at 37°C with 5% CO2 for 1 h to allow bacterial phagocytosis. They were then washed once with RPMI and incubated in RPMI-10%Fetal Calf Serum-streptomycin (100 µg ml−1)/penicillin (100 U ml−1). At the indicated times, infected macrophages were washed once with RPMI and then lysed by incubation in ice-cold water for 15 min. CFU counts were determined by plating serial dilutions on TSA plates.
Mouse virulence assays
Female inbred BALB/c mice (4 to 5 weeks old) were obtained from Janvier Laboratories (Le Genest-St-Isle, France). S. aureus strains (SH1000 and the ΔcymR or the sodA sodM derivatives) were grown in TSB until OD600 ∼2, cells were pelleted and resuspended to the appropriate concentration in sterile PBS. Mice were injected by the intraperitoneal route with ∼3.108 CFU (mortality assays), or 5.107 CFU (sub-lethal dose) in 0.2 ml PBS.
For mortality rate assays, mice were monitored daily for signs of illness and death. At the end of the experiment, surviving mice were humanely sacrificed (CO2 asphyxiation). Results were statistically analyzed by the log-rank test using Prism 5.0b software (GraphPad Software, San Diego, CA).
For measuring the bacterial load in blood and kidneys, animals were followed during 7 days post-infection. Blood samples were collected from the retro orbital sinus 1 and 3 days post-infection, immediately mixed with heparin and plated on TSA plates. Seven days post-inoculation, mice were sacrificed (CO2 asphyxiation), and the kidneys were removed and homogenized for determination of CFU counts.
Hemolytic activity assays
Hemolysis was detected on Columbia blood agar plates (BioMérieux). Strains were grown overnight in TSB medium and then either spotted (20 µl) on horse blood agar plates or streaked on sheep blood agar plates. The plates were incubated for 24 hours at 37°C, and specific hemolytic activities (β- and δ-hemolysins) were identified as previously described [44].
Supporting Information
Cystine effect on stress resistance phenotypes of an S. aureus ΔcymR mutant. Disk diffusion assays were performed with 200 mM tellurite K2TeO3 (A) or 200 mM CuSO4 (B) in TSB medium with (white) and without (grey) 2 mM cystine. Strains SA6 (SH1000/pMK4), SA30 (cymR/pMK4) and SA31 (cymR/pDIA5780) were used for complementation experiments. Results represent the mean values with standard deviations and are representative of at least three independent experiments.
(0.62 MB TIF)
Cystine effect on stress resistance phenotypes of S. aureus mutant strains. Disk diffusion assays were performed with 200 mM tellurite K2TeO3 (A) or 200 mM CuSO4 (B) in TSB medium with (white) and without (grey) 2 mM cystine. Results represent the mean values with standard deviations and are representative of at least three independent experiments.
(0.56 MB TIF)
Additional stress-regulated genes differentially expressed in the S. aureus ΔcymR mutant strain compared with strain SH1000.
(0.11 MB PDF)
Relative expression levels of stress response-associated genes in S. aureus mutant strains compared to SH1000.
(0.09 MB PDF)
Intracellular metabolite estimation in S. aureus cymR mutant strain compared to SH1000.
(0.08 MB PDF)
Virulence or host-interaction associated genes differentially expressed in the S. aureus ΔcymR mutant strain compared to SH1000.
(0.11 MB PDF)
Oligonucleotides used in this study.
(0.07 MB PDF)
Acknowledgments
We are grateful to S. Foster for providing us with S. aureus strains and valuable discussions. We thank O. Sismeiro, C. Lacroix and J.-Y. Coppee for macroarray preparation, S. Rousseau and P. Vantadour for loading transcriptome data onto the Genoscript database and P. Courtin for metabolite analysis.
Footnotes
The authors have declared that no competing interests exist.
I. M-V. and O. S. are full and assistant professors at the Université Paris 7, respectively. Research was supported by grants from the Centre National de la Recherche Scientifique (CNRS URA 2171 and 2172), the Institut Pasteur (Grand Programme Horizontal N°9 and PTR N°256) and the European Commission for the group of T. M. (Grants StaphDynamics, LHSM-CT-2006-019064, and BaSysBio, LSHG-CT-2006-037469). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ayala-Castro C, Saini A, Outten FW. Fe-S cluster assembly pathways in bacteria. Microbiol Mol Biol Rev. 2008;72:110–125. doi: 10.1128/MMBR.00034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ito K, Inaba K. The disulfide bond formation (Dsb) system. Curr Opin Struct Biol. 2008;18:450–458. doi: 10.1016/j.sbi.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Bogdan JA, Nazario-Larrieu J, Sarwar J, Alexander P, Blake MS. Bordetella pertussis autoregulates pertussis toxin production through the metabolism of cysteine. Infect Immun. 2001;69:6823–6830. doi: 10.1128/IAI.69.11.6823-6830.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gooder H, Gehring LB. Inhibition by cystine of lecithinase (alpha-toxin) production in Clostridium welchii (perfringens) BP6K. Nature. 1954;174:1054–1055. doi: 10.1038/1741054b0. [DOI] [PubMed] [Google Scholar]
- 5.Karlsson S, Lindberg A, Norin E, Burman LG, Akerlund T. Toxins, butyric acid, and other short-chain fatty acids are coordinately expressed and down-regulated by cysteine in Clostridium difficile. Infect Immun. 2000;68:5881–5888. doi: 10.1128/iai.68.10.5881-5888.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhave DP, Muse WB, 3rd, Carroll KS. Drug targets in mycobacterial sulfur metabolism. Infect Disord Drug Targets. 2007;7:140–158. doi: 10.2174/187152607781001772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Grifantini R, Bartolini E, Muzzi A, Draghi M, Frigimelica E, et al. Previously unrecognized vaccine candidates against group B meningococcus identified by DNA microarrays. Nat Biotechnol. 2002;20:914–921. doi: 10.1038/nbt728. [DOI] [PubMed] [Google Scholar]
- 8.Ejim LJ, D'Costa VM, Elowe NH, Loredo-Osti JC, Malo D, et al. Cystathionine beta-lyase is important for virulence of Salmonella enterica serovar Typhimurium. Infect Immun. 2004;72:3310–3314. doi: 10.1128/IAI.72.6.3310-3314.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lestrate P, Delrue RM, Danese I, Didembourg C, Taminiau B, et al. Identification and characterization of in vivo attenuated mutants of Brucella melitensis. Mol Microbiol. 2000;38:543–551. doi: 10.1046/j.1365-2958.2000.02150.x. [DOI] [PubMed] [Google Scholar]
- 10.Masip L, Veeravalli K, Georgiou G. The many faces of glutathione in bacteria. Antioxid Redox Signal. 2006;8:753–762. doi: 10.1089/ars.2006.8.753. [DOI] [PubMed] [Google Scholar]
- 11.Zeller T, Klug G. Thioredoxins in bacteria: functions in oxidative stress response and regulation of thioredoxin genes. Naturwissenschaften. 2006;93:259–266. doi: 10.1007/s00114-006-0106-1. [DOI] [PubMed] [Google Scholar]
- 12.delCardayre SB, Stock KP, Newton GL, Fahey RC, Davies JE. Coenzyme A disulfide reductase, the primary low molecular weight disulfide reductase from Staphylococcus aureus. Purification and characterization of the native enzyme. J Biol Chem. 1998;273:5744–5751. doi: 10.1074/jbc.273.10.5744. [DOI] [PubMed] [Google Scholar]
- 13.Newton GL, Arnold K, Price MS, Sherrill C, Delcardayre SB, et al. Distribution of thiols in microorganisms: mycothiol is a major thiol in most actinomycetes. J Bacteriol. 1996;178:1990–1995. doi: 10.1128/jb.178.7.1990-1995.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Newton GL, Rawat M, La Clair JJ, Jothivasan VK, Budiarto T, et al. Bacillithiol is an antioxidant thiol produced in Bacilli. Nat Chem Biol. 2009;5:625–627. doi: 10.1038/nchembio.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Even S, Burguière P, Auger S, Soutourina O, Danchin A, et al. Global control of cysteine metabolism by CymR in Bacillus subtilis. J Bacteriol. 2006;188:2184–2197. doi: 10.1128/JB.188.6.2184-2197.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hochgrafe F, Mostertz J, Pother DC, Becher D, Helmann JD, et al. S-cysteinylation is a general mechanism for thiol protection of Bacillus subtilis proteins after oxidative stress. J Biol Chem. 2007;282:25981–25985. doi: 10.1074/jbc.C700105200. [DOI] [PubMed] [Google Scholar]
- 17.Hung J, Cooper D, Turner MS, Walsh T, Giffard PM. Cystine uptake prevents production of hydrogen peroxide by Lactobacillus fermentum BR11. FEMS Microbiol Lett. 2003;227:93–99. doi: 10.1016/S0378-1097(03)00653-0. [DOI] [PubMed] [Google Scholar]
- 18.Lithgow JK, Hayhurst EJ, Cohen G, Aharonowitz Y, Foster SJ. Role of a cysteine synthase in Staphylococcus aureus. J Bacteriol. 2004;186:1579–1590. doi: 10.1128/JB.186.6.1579-1590.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lo R, Turner MS, Barry DG, Sreekumar R, Walsh TP, et al. Cystathionine gamma-lyase is a component of cystine-mediated oxidative defense in Lactobacillus reuteri BR11. J Bacteriol. 2009;191:1827–1837. doi: 10.1128/JB.01553-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park S, Imlay JA. High levels of intracellular cysteine promote oxidative DNA damage by driving the fenton reaction. J Bacteriol. 2003;185:1942–1950. doi: 10.1128/JB.185.6.1942-1950.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Weber H, Engelmann S, Becher D, Hecker M. Oxidative stress triggers thiol oxidation in the glyceraldehyde-3-phosphate dehydrogenase of Staphylococcus aureus. Mol Microbiol. 2004;52:133–140. doi: 10.1111/j.1365-2958.2004.03971.x. [DOI] [PubMed] [Google Scholar]
- 22.Gusarov I, Nudler E. NO-mediated cytoprotection: instant adaptation to oxidative stress in bacteria. Proc Natl Acad Sci U S A. 2005;102:13855–13860. doi: 10.1073/pnas.0504307102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liebeke M, Pother DC, van Duy N, Albrecht D, Becher D, et al. Depletion of thiol-containing proteins in response to quinones in Bacillus subtilis. Mol Microbiol. 2008;69:1513–1529. doi: 10.1111/j.1365-2958.2008.06382.x. [DOI] [PubMed] [Google Scholar]
- 24.Nguyen TT, Eiamphungporn W, Mader U, Liebeke M, Lalk M, et al. Genome-wide responses to carbonyl electrophiles in Bacillus subtilis: control of the thiol-dependent formaldehyde dehydrogenase AdhA and cysteine proteinase YraA by the MerR-family regulator YraB (AdhR). Mol Microbiol. 2009;71:876–894. doi: 10.1111/j.1365-2958.2008.06568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soutourina O, Poupel O, Coppee JY, Danchin A, Msadek T, et al. CymR, the master regulator of cysteine metabolism in Staphylococcus aureus, controls host sulfur source utilization and plays a role in biofilm formation. Mol Microbiol. 2009;73:194–211. doi: 10.1111/j.1365-2958.2009.06760.x. [DOI] [PubMed] [Google Scholar]
- 26.Tanous C, Soutourina O, Raynal B, Hullo MF, Mervelet P, et al. The CymR Regulator in Complex with the Enzyme CysK Controls Cysteine Metabolism in Bacillus subtilis. J Biol Chem. 2008;283:35551–35560. doi: 10.1074/jbc.M805951200. [DOI] [PubMed] [Google Scholar]
- 27.Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 28.Gordon RJ, Lowy FD. Pathogenesis of methicillin-resistant Staphylococcus aureus infection. Clin Infect Dis. 2008;46(Suppl 5):S350–359. doi: 10.1086/533591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clements MO, Foster SJ. Stress resistance in Staphylococcus aureus. Trends Microbiol. 1999;7:458–462. doi: 10.1016/s0966-842x(99)01607-8. [DOI] [PubMed] [Google Scholar]
- 30.Imlay JA. Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem. 2008;77:755–776. doi: 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leichert LI, Scharf C, Hecker M. Global characterization of disulfide stress in Bacillus subtilis. J Bacteriol. 2003;185:1967–1975. doi: 10.1128/JB.185.6.1967-1975.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chasteen TG, Fuentes DE, Tantalean JC, Vasquez CC. Tellurite: history, oxidative stress, and molecular mechanisms of resistance. FEMS Microbiol Rev. 2009;33:820–832. doi: 10.1111/j.1574-6976.2009.00177.x. [DOI] [PubMed] [Google Scholar]
- 33.Sitthisak S, Knutsson L, Webb JW, Jayaswal RK. Molecular characterization of the copper transport system in Staphylococcus aureus. Microbiology. 2007;153:4274–4283. doi: 10.1099/mic.0.2007/009860-0. [DOI] [PubMed] [Google Scholar]
- 34.Horsburgh MJ, Clements MO, Crossley H, Ingham E, Foster SJ. PerR controls oxidative stress resistance and iron storage proteins and is required for virulence in Staphylococcus aureus. Infect Immun. 2001;69:3744–3754. doi: 10.1128/IAI.69.6.3744-3754.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horsburgh MJ, Ingham E, Foster SJ. In Staphylococcus aureus, fur is an interactive regulator with PerR, contributes to virulence, and is necessary for oxidative stress resistance through positive regulation of catalase and iron homeostasis. J Bacteriol. 2001;183:468–475. doi: 10.1128/JB.183.2.468-475.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Horsburgh MJ, Wharton SJ, Cox AG, Ingham E, Peacock S, et al. MntR modulates expression of the PerR regulon and superoxide resistance in Staphylococcus aureus through control of manganese uptake. Mol Microbiol. 2002;44:1269–1286. doi: 10.1046/j.1365-2958.2002.02944.x. [DOI] [PubMed] [Google Scholar]
- 37.Lindsay JA, Foster SJ. zur: a Zn(2+)-responsive regulatory element of Staphylococcus aureus. Microbiology. 2001;147:1259–1266. doi: 10.1099/00221287-147-5-1259. [DOI] [PubMed] [Google Scholar]
- 38.Ballal A, Manna AC. Regulation of superoxide dismutase (sod) genes by SarA in Staphylococcus aureus. J Bacteriol. 2009;191:3301–3310. doi: 10.1128/JB.01496-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karavolos MH, Horsburgh MJ, Ingham E, Foster SJ. Role and regulation of the superoxide dismutases of Staphylococcus aureus. Microbiology. 2003;149:2749–2758. doi: 10.1099/mic.0.26353-0. [DOI] [PubMed] [Google Scholar]
- 40.Auger S, Gomez MP, Danchin A, Martin-Verstraete I. The PatB protein of Bacillus subtilis is a C-S-lyase. Biochimie. 2005;87:231–238. doi: 10.1016/j.biochi.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 41.Kubica M, Guzik K, Koziel J, Zarebski M, Richter W, et al. A potential new pathway for Staphylococcus aureus dissemination: the silent survival of S. aureus phagocytosed by human monocyte-derived macrophages. PLoS One. 2008;3:e1409. doi: 10.1371/journal.pone.0001409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nizet V. Understanding how leading bacterial pathogens subvert innate immunity to reveal novel therapeutic targets. J Allergy Clin Immunol. 2007;120:13–22. doi: 10.1016/j.jaci.2007.06.005. [DOI] [PubMed] [Google Scholar]
- 43.von Kockritz-Blickwede M, Rohde M, Oehmcke S, Miller LS, Cheung AL, et al. Immunological mechanisms underlying the genetic predisposition to severe Staphylococcus aureus infection in the mouse model. Am J Pathol. 2008;173:1657–1668. doi: 10.2353/ajpath.2008.080337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakoulas G, Eliopoulos GM, Moellering RC, Jr, Wennersten C, Venkataraman L, et al. Accessory gene regulator (agr) locus in geographically diverse Staphylococcus aureus isolates with reduced susceptibility to vancomycin. Antimicrob Agents Chemother. 2002;46:1492–1502. doi: 10.1128/AAC.46.5.1492-1502.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Traber KE, Lee E, Benson S, Corrigan R, Cantera M, et al. agr function in clinical Staphylococcus aureus isolates. Microbiology. 2008;154:2265–2274. doi: 10.1099/mic.0.2007/011874-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang W, Small DA, Toghrol F, Bentley WE. Global transcriptome analysis of Staphylococcus aureus response to hydrogen peroxide. J Bacteriol. 2006;188:1648–1659. doi: 10.1128/JB.188.4.1648-1659.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wolf C, Hochgrafe F, Kusch H, Albrecht D, Hecker M, et al. Proteomic analysis of antioxidant strategies of Staphylococcus aureus: diverse responses to different oxidants. Proteomics. 2008;8:3139–3153. doi: 10.1002/pmic.200701062. [DOI] [PubMed] [Google Scholar]
- 48.Almeida CE, Felicio DL, Galhardo RS, Cabral-Neto JB, Leitao AC. Synergistic lethal effect between hydrogen peroxide and neocuproine (2,9-dimethyl 1,10-phenanthroline) in Escherichia coli. Mutat Res. 1999;433:59–66. doi: 10.1016/s0921-8777(98)00064-0. [DOI] [PubMed] [Google Scholar]
- 49.Macomber L, Rensing C, Imlay JA. Intracellular copper does not catalyze the formation of oxidative DNA damage in Escherichia coli. J Bacteriol. 2007;189:1616–1626. doi: 10.1128/JB.01357-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Phan TN, Kirsch AM, Marquis RE. Selective sensitization of bacteria to peroxide damage associated with fluoride inhibition of catalase and pseudocatalase. Oral Microbiol Immunol. 2001;16:28–33. doi: 10.1034/j.1399-302x.2001.160105.x. [DOI] [PubMed] [Google Scholar]
- 51.Truong DH, Eghbal MA, Hindmarsh W, Roth SH, O'Brien PJ. Molecular mechanisms of hydrogen sulfide toxicity. Drug Metab Rev. 2006;38:733–744. doi: 10.1080/03602530600959607. [DOI] [PubMed] [Google Scholar]
- 52.Yaegaki K, Qian W, Murata T, Imai T, Sato T, et al. Oral malodorous compound causes apoptosis and genomic DNA damage in human gingival fibroblasts. J Periodontal Res. 2008;43:391–399. doi: 10.1111/j.1600-0765.2007.01052.x. [DOI] [PubMed] [Google Scholar]
- 53.Kari C, Nagy Z, Kovacs P, Hernadi F. Mechanism of the growth inhibitory effect of cysteine on Escherichia coli. J Gen Microbiol. 1971;68:349–356. doi: 10.1099/00221287-68-3-349. [DOI] [PubMed] [Google Scholar]
- 54.Stipanuk MH, Dominy JE, Jr, Lee JI, Coloso RM. Mammalian cysteine metabolism: new insights into regulation of cysteine metabolism. J Nutr. 2006;136:1652S–1659S. doi: 10.1093/jn/136.6.1652S. [DOI] [PubMed] [Google Scholar]
- 55.Palazzolo-Ballance AM, Suquet C, Hurst JK. Pathways for intracellular generation of oxidants and tyrosine nitration by a macrophage cell line. Biochemistry. 2007;46:7536–7548. doi: 10.1021/bi700123s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martinez-Pulgarin S, Dominguez-Bernal G, Orden JA, de la Fuente R. Simultaneous lack of catalase and beta-toxin in Staphylococcus aureus leads to increased intracellular survival in macrophages and epithelial cells and to attenuated virulence in murine and ovine models. Microbiology. 2009;155:1505–1515. doi: 10.1099/mic.0.025544-0. [DOI] [PubMed] [Google Scholar]
- 57.Somerville GA, Proctor RA. At the crossroads of bacterial metabolism and virulence factor synthesis in Staphylococci. Microbiol Mol Biol Rev. 2009;73:233–248. doi: 10.1128/MMBR.00005-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Garzoni C, Francois P, Huyghe A, Couzinet S, Tapparel C, et al. A global view of Staphylococcus aureus whole genome expression upon internalization in human epithelial cells. BMC Genomics. 2007;8:171. doi: 10.1186/1471-2164-8-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fujimoto DF, Higginbotham RH, Sterba KM, Maleki SJ, Segall AM, et al. Staphylococcus aureus SarA is a regulatory protein responsive to redox and pH that can support bacteriophage lambda integrase-mediated excision/recombination. Mol Microbiol. 2009;74:1445–1458. doi: 10.1111/j.1365-2958.2009.06942.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen PR, Nishida S, Poor CB, Cheng A, Bae T, et al. A new oxidative sensing and regulation pathway mediated by the MgrA homologue SarZ in Staphylococcus aureus. Mol Microbiol. 2009;71:198–211. doi: 10.1111/j.1365-2958.2008.06518.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Poor CB, Chen PR, Duguid E, Rice PA, He C. Crystal structures of the reduced, sulfenic acid, and mixed disulfide forms of SarZ, a redox active global regulator in Staphylococcus aureus. J Biol Chem. 2009;284:23517–23524. doi: 10.1074/jbc.M109.015826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cucarella C, Solano C, Valle J, Amorena B, Lasa I, et al. Bap, a Staphylococcus aureus surface protein involved in biofilm formation. J Bacteriol. 2001;183:2888–2896. doi: 10.1128/JB.183.9.2888-2896.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Novick RP. Genetic systems in staphylococci. Methods Enzymol. 1991;204:587–636. doi: 10.1016/0076-6879(91)04029-n. [DOI] [PubMed] [Google Scholar]
- 64.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lopez del Castillo Lozano M, Tache R, Bonnarme P, Landaud S. Evaluation of a quantitative screening method for hydrogen sulfide production by cheese-ripening microorganisms: the first step towards L-cysteine catabolism. J Microbiol Methods. 2007;69:70–77. doi: 10.1016/j.mimet.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 66.Hullo MF, Auger S, Soutourina O, Barzu O, Yvon M, et al. Conversion of methionine to cysteine in Bacillus subtilis and its regulation. J Bacteriol. 2007;189:187–197. doi: 10.1128/JB.01273-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Palmqvist N, Patti JM, Tarkowski A, Josefsson E. Expression of staphylococcal clumping factor A impedes macrophage phagocytosis. Microbes Infect. 2004;6:188–195. doi: 10.1016/j.micinf.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 68.Schlag S, Nerz C, Birkenstock TA, Altenberend F, Gotz F. Inhibition of staphylococcal biofilm formation by nitrite. J Bacteriol. 2007;189:7911–7919. doi: 10.1128/JB.00598-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Richardson AR, Dunman PM, Fang FC. The nitrosative stress response of Staphylococcus aureus is required for resistance to innate immunity. Mol Microbiol. 2006;61:927–939. doi: 10.1111/j.1365-2958.2006.05290.x. [DOI] [PubMed] [Google Scholar]
- 70.Horsburgh MJ, Aish JL, White IJ, Shaw L, Lithgow JK, et al. sigmaB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J Bacteriol. 2002;184:5457–5467. doi: 10.1128/JB.184.19.5457-5467.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sullivan MA, Yasbin RE, Young FE. New shuttle vectors for Bacillus subtilis and Escherichia coli which allow rapid detection of inserted fragments. Gene. 1984;29:21–26. doi: 10.1016/0378-1119(84)90161-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Cystine effect on stress resistance phenotypes of an S. aureus ΔcymR mutant. Disk diffusion assays were performed with 200 mM tellurite K2TeO3 (A) or 200 mM CuSO4 (B) in TSB medium with (white) and without (grey) 2 mM cystine. Strains SA6 (SH1000/pMK4), SA30 (cymR/pMK4) and SA31 (cymR/pDIA5780) were used for complementation experiments. Results represent the mean values with standard deviations and are representative of at least three independent experiments.
(0.62 MB TIF)
Cystine effect on stress resistance phenotypes of S. aureus mutant strains. Disk diffusion assays were performed with 200 mM tellurite K2TeO3 (A) or 200 mM CuSO4 (B) in TSB medium with (white) and without (grey) 2 mM cystine. Results represent the mean values with standard deviations and are representative of at least three independent experiments.
(0.56 MB TIF)
Additional stress-regulated genes differentially expressed in the S. aureus ΔcymR mutant strain compared with strain SH1000.
(0.11 MB PDF)
Relative expression levels of stress response-associated genes in S. aureus mutant strains compared to SH1000.
(0.09 MB PDF)
Intracellular metabolite estimation in S. aureus cymR mutant strain compared to SH1000.
(0.08 MB PDF)
Virulence or host-interaction associated genes differentially expressed in the S. aureus ΔcymR mutant strain compared to SH1000.
(0.11 MB PDF)
Oligonucleotides used in this study.
(0.07 MB PDF)