Abstract
The long-term effects of behaviour and environmental exposures, particularly during childhood, on health outcomes are well documented. Particularly thought provoking is the notion that exposures to different social environments have a long-lasting impact on human physical health. However, the mechanisms mediating the effects of the environment are still unclear. In the last decade, the main focus of attention was the genome, and interindividual genetic polymorphisms were sought after as the principal basis for susceptibility to disease. However, it is becoming clear that recent dramatic increases in the incidence of certain human pathologies, such as asthma and type 2 diabetes, cannot be explained just on the basis of a genetic drift. It is therefore extremely important to unravel the molecular links between the "environmental" exposure, which is believed to be behind this emerging incidence in certain human pathologies, and the disease's molecular mechanisms. Although it is clear that most human pathologies involve long-term changes in gene function, these might be caused by mechanisms other than changes in the deoxyribonucleic acid (DNA) sequence. The genome is programmed by the epigenome, which is composed of chromatin and a covalent modification of DNA by methylation. It is postulated here that "epigenetic" mechanisms mediate the effects of behavioural and environmental exposures early in life, as well as lifelong environmental exposures and the susceptibility to disease later in life. In contrast to genetic sequence differences, epigenetic aberrations are potentially reversible, raising the hope for interventions that will be able to reverse deleterious epigenetic programming.
Keywords: autoimmune disease, demethylation, DNA methylation, epigenetics chromatin, epigenome, histone modification, maternal care, socioeconomic status
Genes, Gene Expression Programs, and Phenotype
The comprehensive sequencing of the human genome has generated great anticipation that by comparing the deoxyribonucleic acid (DNA) sequence between individuals, we will be able to understand the basis of phenotypic diversity between individuals, including the reasons for diseases such as asthma and other autoimmune and atopic states. However, our current understanding suggests that this might not be the complete story. There are clear environmental factors that facilitate the emergence of these pathologies. What are the mechanisms that memorize exposures at different points in life, leading to long-term impact on human health? One of the factors that are known to impact the incidence of asthma is socioeconomic status in early childhood. How can the socioeconomic environment affect physical and physiologic parameters? The genomic theory focuses on differences in gene function as the molecular mechanism of pathologic processes. The principal hypothesis is that differences in gene sequences are behind differences in gene function. However, it is now clear that long-lasting differences in gene function might be brought about by mechanisms other than gene sequence variations, which we define as "epigenetic" processes. These mechanisms are excellent candidates to mediate the long-lasting impact of environmental exposure.
The genome has to be programmed to express its unique patterns of gene expression. Different cell types execute distinctive plans of gene expression, which are highly responsive to developmental, physiologic, pathologic, and environmental cues. The combinations of mechanisms, which confer long-term programming to genes and could bring about a change in gene function without changing the gene sequence, are called epigenetic changes here. The dynamic nature of epigenetic regulation in contrast to the static nature of the gene sequence provides a mechanism for reprogramming gene function in response to changes in lifestyle trajectories. Thus, epigenetics could provide an explanation for welldocumented gene-environment interactions. An important implication of the possible involvement of epigenetics is the potential for therapeutic intervention. Epigenetic mechanisms are dynamic and potentially reversible and are therefore amenable to therapeutic intervention [1]. Drugs that target the epigenetic machinery are currently tested in clinical trials in cancer [2,3] and psychiatric disorders [4]. Moreover, once we understand the rules through which different environmental exposures modify the epigenetic processes, we might be able to design behavioural and therapeutic strategies to prevent and revert deleterious environmentally driven epigenetic alterations.
During the normal processes of development and cellular differentiation, a cell type-specific pattern of epigenetic marks is generated [5]. This normal "pattern" of epigenetic marks defines the normal pattern of gene function in each tissue and cell type [6]. The normal pattern of gene-function is critical for the execution of the normal life necessities, physiologic and behavioural functions. A change in the normal pattern of gene function would result in phenotypic differences. Gene function could change by sequence alterations, which either completely eliminate the function of the gene or alter the function of the protein encoded by the gene, resulting in either an increase or a decrease in its activity. A paradigm of epigenetic silencing is the case of "tumour suppressor" genes in cancer. Tumour suppressor genes are normally active and protect our cells from abnormal growth. The first tumour suppressor gene that was characterized was the retinoblastoma gene, a recessive mutation leading to childhood tumours in either one or two eyes [7,8]. All tumour suppressor genes were originally discovered by looking for a recessive mutation, which led to a specific type of cancer. It was later found that many of these tumour suppressor genes were silenced by epigenetic inactivation in cancer rather than by genetic lesions [9]. Thus, epigenetic silencing and genetic silencing could have similar phenotypic consequences.
The epigenome consists of the chromatin and its modifications, as well as a covalent modification by methylation of cytosine rings found at the dinucleotide sequence CG [10]. The epigenome determines the accessibility of the transcription machinery, which transcribes the genes into messenger ribonucleic acid (RNA), to the DNA. Inaccessible genes are therefore silent, whereas accessible genes are transcribed. We therefore distinguish between open and closed configuration of chromatin [11-15]. Densely packaged chromatin could be visualized microscopically and is called heterochromatin, whereas open accessible chromatin is called euchromatin. Recently, another new level of epigenetic regulation by small noncoding RNAs called microRNAs has been discovered [16]. MicroRNAs regulate gene expression at different levels: silencing of chromatin, degradation of messenger RNA, and blocking translation. MicroRNAs were found to play an important role in cancer [17] and could potentially play an important role in behavioural pathologies as well [18].
Chromatin and Its Modifications
The DNA is wrapped around a protein-based structure called chromatin. The basic building block of chromatin is the nucleosome, which is formed from an octamer of histone proteins. There are five basic forms of histone proteins, H1, H2A, H2B, H3, and H4, [19] as well as other minor variants, which are involved in specific functions, such as DNA repair or gene activation [20]. The octamer structure of the nucleosome is composed of an H3-H4 tetramer flanked on either side with an H2A-H2B dimer [19]. The N-terminal tails of these histones are extensively modified by methylation, [21] phosphorylation, acetylation, [22] and ubiquitination [23]. The state of modification of these tails plays an important role in defining the accessibility of the DNA wrapped around the nucleosome core. It was proposed that the amino terminal tails of H3 and H4 histones that are positively charged form tight interactions with the negatively charged DNA backbone, thus blocking the interaction of transcription factors with the DNA. Modifications of the tails neutralize the charge on the tails, thus relaxing the tight grip of the histone tails. Different histone variants, which replace the standard isoforms, also play a regulatory role and serve to mark active genes in some instances [24]. The specific pattern of histone modifications was proposed to form a "histone code," which delineates the parts of the genome to be expressed at a given point in time in a given cell type [25]. A change in histone modifications around a gene will change its level of expression and could convert an active gene to become silent, resulting in "loss of function," or switch a silent gene to be active, leading to "gain of function."
Histone-Modifying Enzymes
The most investigated histone-modifying enzymes are histone acetyltransferases (HATs), which acetylate H3 histone at the K9 and other residues and H4 tails at a number of residues, and histone deacetylases (HDACs), which deacetylate histone tails [26]. Histone acetylation is believed to be a predominant signal for an active chromatin configuration [27,28]. Deacetylated histones signal inactive chromatin, which is chromatin associated with inactive genes. Many repressors and repressor complexes recruit HDACs to genes, thus causing their inactivation [29]. Histone tail acetylation is believed to enhance the accessibility of a gene to the transcription machinery, whereas deacetylated tails are highly charged and believed to be tightly associated with the DNA backbone, thus limiting accessibility of genes to transcription factors [26].
Histone modification by methylation is catalyzed by different histone methyltransferases. Some specific methylation events are associated with gene silencing and some with gene activation. For example, methylation of the K9 residue of H3 histone tails is catalyzed by the histone methyltransferase SUV3-9 and is associated with silencing of the associated gene [30]. Particular factors recognize histone modifications and further stabilize an inactive state. For example, the heterochromatin-associated protein HP-1 binds H3 histone tails methylated at the K9 residue and precipitates an inactive chromatin structure [30]. Recently described histone demethylases remove the methylation, causing either activation or repression of gene expression [31,32].
Chromatin Remodeling
Chromatin remodeling complexes, which are adenosine triphosphate dependent, alter the position of nucleosomes around the transcription initiation site and define its accessibility to the transcription machinery [15]. It is becoming clear that there is an interrelationship between chromatin modification and chromatin remodeling. For example, BRG1, the catalytic subunit of SWI/SNF-related chromatin remodeling complexes, is required for histone acetylation and regulation of β-globin expression during development [33].
Targeting of Chromatin-Modifying Enzymes to Specific Genes
A basic principle in epigenetic regulation is targeting. Histone-modifying enzymes are generally not gene specific. Specific transcription factors and transcription repressors recruit histone-modifying enzymes to specific genes and thus define the gene-specific profile of histone modification [25]. Transcription factors and repressors recognize specific cisacting sequences in genes, bind to these sequences, and attract the specific chromatin-modifying enzymes to these genes through protein-protein interactions. The cis-acting sequences act as area codes, whereas the transcription factors that read these codes deliver a load of chromatin-modifying and - remodeling enzymes. Specific transacting factors are responsive to cellular signaling pathways such as those signaling through increased cyclic adenosine monophosphate (cAMP). One of the transcription factors that respond to increased cAMP is CREB (cAMP response element binding protein). CREB binds cAMP response elements in certain genes. CREB also recruits CREB binding protein (CBP). CBP is a HAT that acetylates histones [34]. Thus, elevation of cAMP levels in response to an extracellular signal would result in a change in the state of histone acetylation in specific genes.
DNA Methylation
In addition to chromatin, which is associated with DNA, DNA itself is chemically modified by methyl residues at the 5' position of the cytosine rings in the dinucleotide sequence CG in vertebrates (Figure 1) [10]. What distinguishes DNA methylation in vertebrate genomes is the fact that not all CGs are methylated in any given cell type [10]. Distinct CGs are methylated in different cell types, generating cell type-specific patterns of methylation (Figure 2). Thus, the DNA methylation pattern confers on the genome its cell type identity [10]. Active regions of the chromatin, which enable gene expression, are associated with hypomethylated DNA, whereas hypermethylated DNA is packaged in inactive chromatin (Figure 3) [10,35]. It is generally accepted that DNA methylation plays an important role in regulating gene expression (Figure 4). DNA methylation in distinct regulatory regions is believed to mark silent genes. There are now overwhelming data indicating that aberrant silencing of tumour suppressor genes by DNA methylation is a common mechanism in cancer [36].
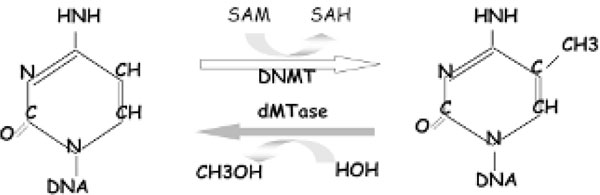
Figure 1.
Methylation and demethylation reactions. DAM = S-adenosylmethionine; dMTase = demethylase; DNMT = DNA methyltransferase.
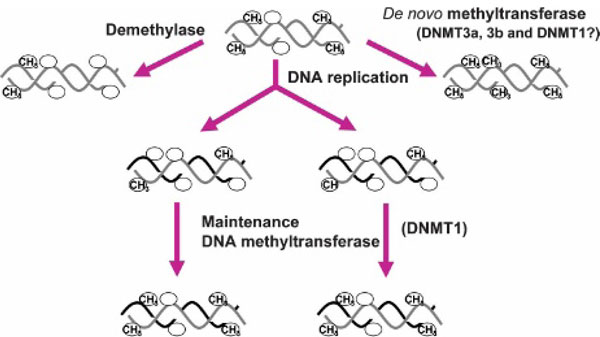
Figure 2.
The DNA methylation pattern is sculpted during development by methylation and demethylation reactions to generate a cell type-specific pattern of methylation. Circle = CG site; CH3 methylated CG site; dark line = nascent DNA strand; grey line = parental DNA strand.
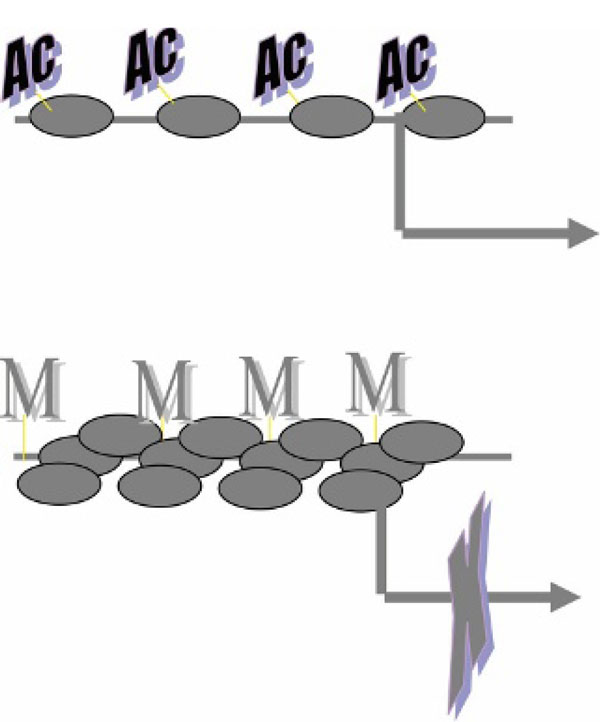
Figure 3.
Chromatin structure, gene expression, and DNA methylation are tightly correlated; DNA methylation and chromatin program and control gene expression. Ac = acetylated histone tails; horizontal arrow = transcription; M = methylated DNA.
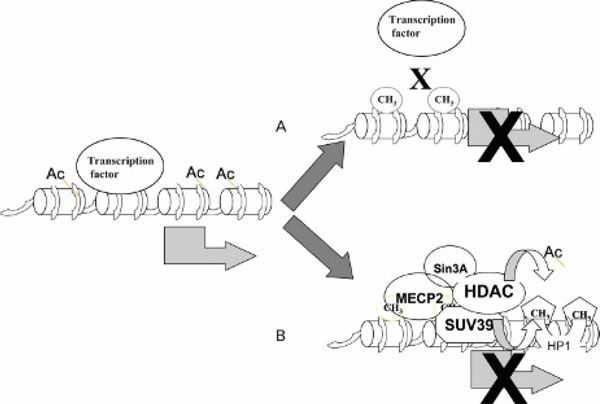
Figure 4.
DNA methylation silences gene expression by two mechanisms. A, Methylation interferes with binding of a transcription factor to its recognition element. B, Methylated DNA attracts methylated DNA binding proteins such as MeCP2, which recruits histone deacetylase (HDAC), corepressor Sin3A, histone methyltransferases such as SuV39, and methyl K9 H3-histone binding protein (HP1).
DNA Methylation Enzymes
The DNA methylation pattern is not copied by the DNA replication machinery but by independent enzymatic machinery, the DNA methyltransferase(s) (DNMT) [35]. The methylation of DNA occurs immediately after replication by a transfer of a methyl moiety from the donor S-adenosyl-L-methionine (AdoMet; SAM) in a reaction catalyzed by DNMTs (see Figure 1). Three distinct phylogenetic DNMTs were identified in mammals. DNMT1 shows preference for hemimethylated DNA in vitro, which is consistent with its role as a maintenance DNMT, whereas DNMT3a and DNMT3b methylate unmethylated and methylated DNA at an equal rate, which is consistent with a de novo DNMT role [37]. Two additional DNMT homologues were found: DNMT2, whose substrate and methylation activity is unclear, [38] and DNMT3L, which belongs to the DNMT3 family of DNMTs by virtue of its sequence. It is essential for the establishment of maternal genomic imprints but lacks key methyltransferase motifs and is possibly a regulator of methylation rather than an enzyme that methylates DNA [39]. Knockout mouse data indicate that DNMT1 is responsible for a majority of DNA methylation in the mouse genome, [40] whereas DNMT3a and DNMT3b are responsible for some but not all de novo methylation during development [41].
DNA Demethylation Enzymes
It was a long-held belief that the DNA methylation pattern is solely dependent on DNMTs and that the reverse reaction cannot occur. Thus, according to the classic model, DNA methylation patterns were generated during development but were then copied faithfully by the maintenance DNMT. The only reaction that takes place according to this model in differentiated cells is maintenance DNA methylation during cell division. The answer to the question of whether the DNA methylation is reversible has important implications for the possibility that DNA methylation is dynamic and responsive to physiologic and environmental signals throughout life. This issue of the reversibility of the DNA methylation reaction has important implications for our understanding of the role of DNA methylation in nondividing tissues such as neurons. If DNA methylation happens only when DNMT is copying DNA methylation patterns during cell division, as suggested by the classic model, there is no need for DNMTs in neurons. Nevertheless, DNMTs are present in neurons, [42] and there are data suggesting that DNMT levels in neurons change in certain pathologic conditions, such as schizophrenia [43]. The presence of DNMT in neurons would make sense only if the DNA methylation is dynamic in postmitotic tissues and is a balance of methylation and demethylation reactions (see Figure 1) [1]. Without active demethylation, there is no need for DNA methylation in neurons.
We proposed awhile ago that the DNA methylation pattern is a balance of methylation and demethylation reactions that are responsive to physiologic and environmental signals and thus forms a platform for gene-environment interactions (see Figures 1 and 5) [44]. There is a long list of data from both cell culture and early mouse development supporting the hypothesis that active methylation occurs in embryonal and somatic cells. There are now convincing examples of active, replication-independent DNA demethylation during development, as well as in somatic tissues. Active demethylation was reported for the myosin gene in differentiating myoblast cells, [45] the interleukin-2 gene on T-cell activation, [46] the interferon-c gene on antigen exposure of memory CD8 T cells, [47] and the glucocorticoid receptor (GR) gene promoter in adult rat brains on treatment with the HDAC inhibitor trichostatin A (TSA) [48].
Figure 5.
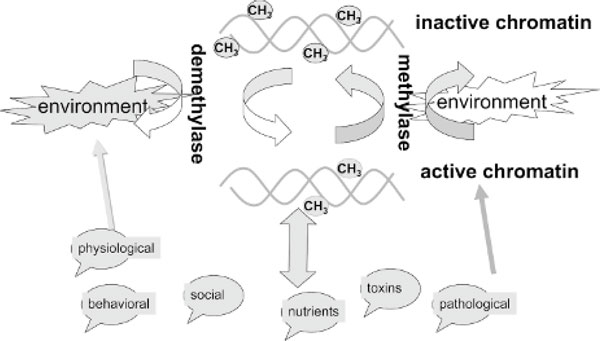
The steady-state methylation pattern is a dynamic equilibrium between methylase and demethylase activities. Different environmental exposures trigger signaling pathways, which affect chromatin structure and, in turn, affect DNA methylation.
The main challenge of the field is identifying the enzymes responsible for demethylation.
The characteristics of the enzymes responsible for active demethylation are controversial. One proposal has been that a G/T mismatch repair glycosylase also functions as a 5-methylcytosine DNA glycosylase, recognizes methyl cytosines, and cleaves the bond between the sugar and the base. The abasic site is then repaired and replaced with a nonmethylated cytosine, resulting in demethylation [49]. An additional protein with a similar activity was recently identified, methylated DNA binding protein 4 (MBD4) [50]. Although such a mechanism can explain site-specific demethylation, global demethylation by a glycosylase would involve extensive damage to DNA that would compromise genomic integrity. Another report has proposed that methylated binding protein 2 (MBD2) has demethylase activity. MBD2b (a shorter isoform of MBD2) was shown to directly remove the methyl group from methylated cytosine in methylated CpGs [51]. This enzyme was therefore proposed to reverse the DNA methylation reaction. However, other groups disputed this finding [52]. Our recent data further support the role of MBD2 in active demethylation [53-55]. Very recent data suggest that active demethylation early in embryogenesis and in somatic cells is catalyzed by a nucleotide excision repair mechanism, whereby methylated cytosines are replaced by unmethylated cytosines, which involves the growth arrest and damage response protein Gadd45a and the DNA repair endonuclease XPG [56]. The main problem with this mechanism is that it involves the risk of extensive damage to the DNA. Although a number of biochemical processes were implicated in demethylation, it is unclear how and when these different enzymes participate in shaping and maintaining the overall pattern of methylation and how these activities respond to different environmental exposures.
Targeting DNA Methylation and Demethylation: Chromatin and DNA Methylation
Methylation and demethylation enzymes do not have exquisite sequence specificity; how could these enzymes maintain highly specific DNA methylation patterns? Methylation and demethylation enzymes have to be targeted to specific genes to either preserve or change in a regulated manner their pattern of methylation. The picture that is currently emerging is that the DNA methylation pattern is tightly coordinated with the chromatin structure; that is, "opening" of chromatin leads to demethylation, and a "closed configuration" of chromatin leads to methylation. Thus, we propose that the direction of the DNA methylation reaction is defined by the state of chromatin and as discussed above (see Figures 5 and 6). The gene specificity of the state of chromatin is defined by sequence-specific trans-acting factors that recruit chromatin-modifying enzymes to specific genes. Chromatin configuration then gates the accessibility of genes to either DNA methylation or demethylation machineries [57,58]. In support of this hypothesis, we have previously shown that the HDAC inhibitor TSA, which causes histone hyperacetylation, also causes active DNA demethylation [57]. A change in histone acetylation is normally caused by transcription factors, which recruit HATs (see Figure 6). Thus, binding of transcription factors to a specific sequence in a gene could recruit HATs, which would cause histone acetylation, facilitating, in turn, demethylation. We propose that a similar mechanism mediates the effects of cellular signaling pathways fired by environmental exposures on the state of DNA methylation.
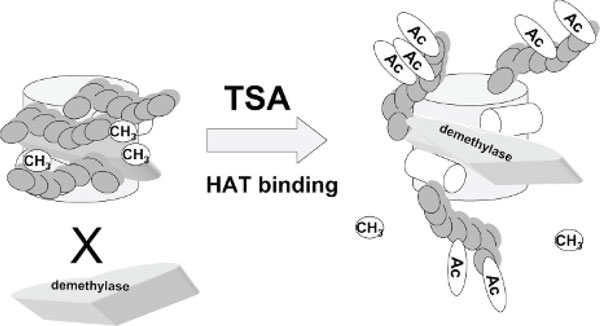
Figure 6.
Activation of chromatin by increasing acetylation facilitates demethylation. Acetylation of histones could be increased by either recruitment of histone acetyltransferases (HAT) or pharmacologic inhibition of histone deacetylases with trichostatin A (TSA). Histone acetylation facilitates interaction of demethylases with the DNA and DNA demethylation.
There is evidence to support this model. Histone modification enzymes interact with DNA-methylating enzymes and participate in recruiting them to specific targets. A growing list of histone-modifying enzymes has been shown to interact with DNMT1, such as HDAC1 and HDAC2, the histone methyltransferases SUV3-9 and EZH2, and a member of the multiprotein polycomb complex PRC2, which methylates H3 histone at the K27 residue [59-62]. DNMT3a was recently also shown to interact with EZH2, which targets the DNA methylation-histone modification multiprotein complexes to specific sequences in DNA [62]. Trans-acting repressors target both histonemodifying enzymes and DNMTs to specific cis-acting signals in regulatory regions of particular genes, causing gene-specific DNA methylation and chromatin modification. For example, the promyelocytic leukemia PML-RAR fusion protein engages histone deacetylases and DNMTs to its target binding sequences and produces de novo DNA methylation of adjacent genes [63].
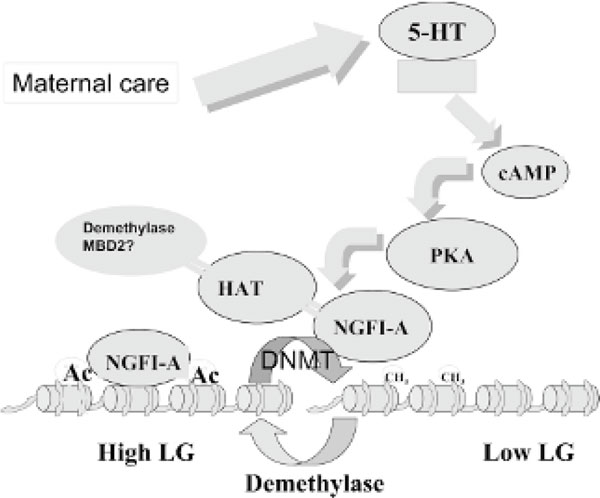
Evidence is emerging that supports the hypothesis that sequence-specific transcription factors target demethylation to specific genes. Transcription factors recruit HATs to specific genes, causing gene-specific acetylation, and thus facilitate their demethylation. For example, the intronic kappa chain enhancer and the transcription factor nuclear factor κB are required for B cell-specific demethylation of the kappa immunoglobulin gene [64]. We discuss below how maternal care is employing this mechanism to program gene expression through recruitment of the transcription factor NGFI-A to one of the GR gene promoters in the hippocampus [65].
How Does DNA Methylation Silence Gene Expression?
There are two main mechanisms by which cytosine methylation suppresses gene expression (see Figure 4). The first mechanism involves direct interference of the methyl residue with the binding of a transcription factor to its recognition element in the gene. The interaction of transcription factors with genes is required for activation of the gene; lack of binding of a transcription factor would result in silencing of gene expression [66,67]. This form of inhibition of transcription by methylation requires that the methylation events occur within the recognition sequence for a transcription factor. A second mechanism is indirect. A certain density of DNA methylation moieties in the region of the gene attracts the binding of methylated-DNA binding proteins such as MeCP2 [68]. MeCP2 recruits other proteins, such as SIN3A, and histone-modifying enzymes, which lead to formation of a "closed" chromatin configuration and silencing of gene expression [68]. Several methylated-DNA binding proteins, such as MBD1, MBD2, and MBD3, suppress gene expression by a similar mechanism [52,69,70].
Maternal Care Model and Its Implications for Epigenetics as a Mediator and Effector of Social Environment on Gene Function
Our hypothesis is that the social environment would trigger long-term changes in gene expression that could lead to pathology by eliciting signaling pathways in the brain, which will, in turn, cause epigenetic reprogramming. The best-documented case to date of epigenetic programming triggered by the social environment is the long-term impact that maternal care has on expression of the GR gene in the hippocampus of the offspring in the rat. In the rat, the adult offspring of mothers that exhibit increased levels of pup licking/grooming (ie, high licking/grooming [LG] mothers) over the first week of life show increased hippocampal GR expression, enhanced glucocorticoid feedback sensitivity, decreased hypothalamic corticotropin-releasing factor expression, and more modest hypothalamic-pituitary-adrenal stress responses compared with animals reared by low LG mothers [71,72]. Crossfostering studies suggest an epigenetic mechanism rather than a genetic mechanism since the fostering mother and not the biologic mother defined the stress response of its adult offspring [70,71]. The critical question was obviously the mechanism. How could the behaviour of the caregiver cause a stable change in gene expression in the offspring long after the caregiver was gone? We postulated an epigenetic mechanism; that is, we hypothesized that the maternal behaviour of the caregiver triggered an epigenetic change in the brain of the offspring [73].
This model has two nodal implications for our understanding of the relationship between behaviour and epigenetics. First, the social behaviour of one subject can affect epigenetic programming in another subject. Thus, our model provides a molecular mechanism mediating the effects of nurture on nature. Second, epigenetic programming can have a long-term impact on behaviour, stress response, and health status.
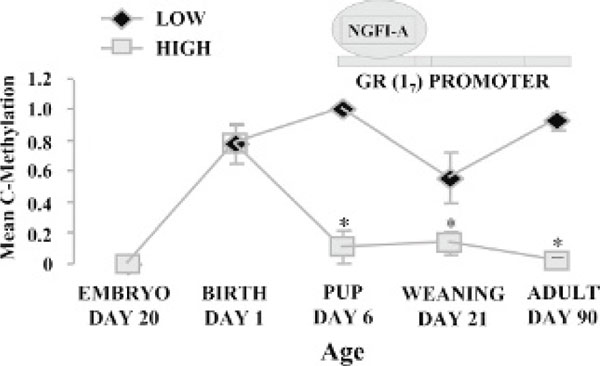
Increased maternal LG is associated with histone acetylation increased transcription factor NGFI-A occupancy and demethylation of the exon 17 GR promoter [48]. The difference in the methylation status of this CpG site between the offspring of high and low LG mothers emerges over the first week of life, is reversed with cross-fostering, persists into adulthood, and is associated with altered histone acetylation and NGFI-A binding to the GR promoter (Figure 7) [48]. We have also shown that maternal care early in life affected the expression of hundreds of genes in the adult hippocampus, [74] thus illustrating the profound effect of the social environment early in life on gene expression programming throughout life. These results have quite tantalizing implications. They imply that differences in maternal care early in life can result in gene expression changes, which remain persistent into adulthood in numerous genes. This range of change in gene expression would have required simultaneously mutating hundreds of genes had it been accomplished by genetic means. This illustrates the potential power of epigenetic processes in modulating our genomic inheritance.
Figure 7.
Timeline of demethylation of hippocampal glucocorticoid receptor (17) in response to maternal care.
Epigenetic Programming that Occurred Early in Life in Response to Social Exposure Is Reversible in the Adult Animal
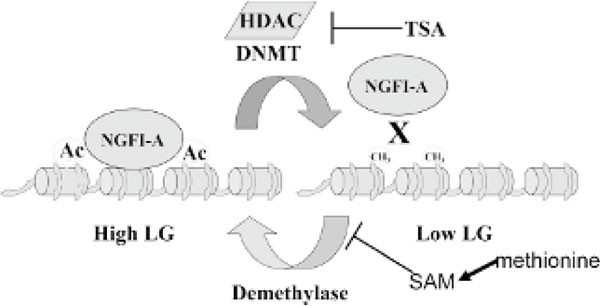
Although epigenetic programming by maternal care is highly stable and results in long-term changes in gene expression, it is nevertheless reversible (Figure 8). The combination of reversibility and stability is one of the appealing aspects of epigenetics and might have immense implications for therapeutic approaches to many late-onset diseases, such as asthma, diabetes, and others. We previously proposed as discussed above that chromatin states and DNA methylation states were linked, so opening up of chromatin by increasing histone acetylation would tilt the balance of the DNA methylation equilibrium toward demethylation (see Figures 5 and 7) [57,75]. Treating adult offspring of low licking/grooming and arched-back nursing (LG-ABN) maternal care with an HDAC inhibitor, TSA, reversed the epigenetic marks on the GR exon 17 promoter; histone acetylation increased, the gene was demethylated, and there was increased occupancy of the promoter with the transcription factor NGFI-A, resulting in increased GR exon 17 promoter expression (see Figure 8). The epigenetic reversal was accompanied by a behavioural change, so the stress response of the TSAtreated adult offspring of low LG-ABN mothers was indistinguishable from the offspring of high LG-ABN mothers [76]. These data illustrate the tight association between the DNA methylation and histone acetylation equilibriums in the adult brain and the potential reversibility of the DNA methylation pattern in the nondividing adult neuron.
Figure 8.
In the adult (day 90) rat, hippocampal glucocorticoid receptor methylation of low licking/grooming and arched-back nursing (LG-ABN) offspring is reversed by trichostatin A and hypomethylation of the high LG-ABN offspring is reversed by methionine.
If the DNA methylation state remains in equilibrium of methylation-demethylation in adult neurons throughout life, it should be possible also to reverse the DNA methylation in the opposite direction by increasing DNA methylation (see Figure 8). We previously demonstrated that the methyl donor SAM inhibits the demethylation reaction [55]. Thus, changing SAM levels would alter the DNA methylation equilibrium by either increasing the rate of the DNA methylation reaction, by inhibiting the demethylation reaction, or both. Since SAM is an unstable compound, we injected the precursor of SAM, the amino acid L-methionine, into the brain of adult offspring of either high or low LG-ABN mothers. Systemic injection of methionine was previously shown to increase SAM concentrations in the brain [77]. Injection of methionine to the brain led to hypermethylation and reduced expression of the GR exon 17 expression in the adult hippocampus of the offspring of high LG-ABN mothers and reversal of its stress response to a pattern, which was indistinguishable from that of the offspring of low LG-ABN mothers [78]. Thus, maternal epigenetic programming could be reversed later in life in both directions. Methionine is especially interesting since the levels of methionine in cells are influenced by diet. Thus, this might provide an example of a potential link between dietary intake and alteration in epigenetic programming in the brain.
Mechanisms Leading from Maternal Care to Epigenetic Programming
How would LG-ABN result in distinct epigenetic changes in certain genes? In vivo and in vitro studies suggest that maternal LG or postnatal handling, which increase maternal LG, increase GR gene expression in the offspring through a thyroid hormone-dependent increase in serotonin (5-hydroxytryptamine [5-HT]) activity at 5-HT7 receptors and the subsequent activation of cAMP and cAMP-dependent protein kinase A [79-81]. Both the in vitro effects of 5-HT and the in vivo effects of maternal behaviour on GR messenger RNA expression are accompanied by increased hippocampal expression of NGFI-A transcription factor. The GR exon 17 promoter region contains a binding site for NGFI-A [82]. Our findings are consistent with the hypothesis that maternal LG-ABN results in increased targeting of NGFI-A to the GR exon 17 promoter and that this targeting leads to increased binding of CBP (a histone acetyltransferase), increased acetylation, and DNA demethylation [65]. Thus, our data depict a conduit leading from exposure to maternal behaviour down to targeting of gene-specific epigenetic reprogramming (Figure 9).
Figure 9.
Behavioural gene programming. Maternal care elicits a signaling pathway in hippocampal neurons, leading to epigenetic reprogramming of the glucocorticoid receptor exon 17 promoter.
To test a causal link between NGFI-A binding and epigenetic reprogramming of the GR exon 17 promoter, we resorted to cell culture experiments. The GR exon 17 promoter was introduced into a reporter vector that contained the complementary DNA encoding the firefly luciferase enzyme under its direction to report for the transcriptional activity of this promoter. The promoter was methylated with a CG-specific bacterial DNA methyltransferase in vitro to completion; thus, all of the CG dinucleotides in the plasmids were methylated. The methylated reporter plasmid was then introduced into HEK 293 cells.
Our results show that in cell culture, DNA methylation causes a significant inhibition of GR exon 17 promoterluciferase transcription activity, reduced NGFI-A binding, reduced CBP binding, and reduced histone acetylation when transfected into HEK 293 cells, thus confirming that DNA methylation plays a causal role in the silencing of GR exon 17 promoter. However, if an expression vector expressing high levels of NGFI-A is cotransfected with the methylated GR exon 17 promoter-luciferase, the transcription activity of the promoter is induced, there is an increased recruitment of NGFI-A to the promoter as expected, increased recruitment of CBP, increased histone acetylation, and methylation mapping indicating that the GR exon 17 promoter was demethylated. We suggest that the role that NGFI-A plays in regulation of the GR exon 17 promoter is bimodal. Under low concentrations of NGFIA, binding to the target sequence is inhibited by DNA methylation. However, under conditions of high NGFI-A activity, some NGFI-A interacts with the methylated GR exon 17 promoter, launching a cascade of events leading to demethylation of the promoter. Thus, increased activation of NGFI-A triggered by a repetitive and frequent behaviour such as maternal LG leads to binding of NGFI-A to the methylated promoter and recruitment of CBP. We proposed that the recruitment of CBP led to increased histone acetylation that resulted in demethylation [65]. This sequence of events is consistent with our working hypothesis on the relationship between histone acetylation and DNA demethylation [57,75]. Thus, we show that, similar to acetylation in response to pharmacologic administration of TSA, targeted acetylation by recruitment of a transcription factor leads to demethylation of DNA [65].
We then tested the hypothesis that MBD2, which we previously characterized to be a demethylase, [51] mediated the demethylation of GR exon 17 promoter. We first tested whether MBD2 interacted with the GR exon 17 promoter in the hippocampi of day 6 pups at the point in life when the pups are licked and groomed by their mother. Our results indicate that MBD2 binds the GR exon 17 promoter in the hippocampi of day 6 pups and that this binding is increased with high maternal LG-ABN. Using a transient transfection assay, we showed that ectopically expressed MBD2 transcriptionally activates in vitro the methylated GR exon 17 promoter-luciferase reporter construct, increases the interaction of CBP, and increases histone acetylation to the promoter. A combination of chromatin immunoprecipitation (ChIP) and bisulfite mapping of DNA methylation indicated that MBD2-bound GR exon 17 promoter molecules were demethylated at a CG site found in the NGFI-A recognition element. Using a double-ChIP approach, which involves immunoprecipitation sequentially with both NGFI-A and MBD2 antibodies, we show that both proteins simultaneously bind the same GR exon 17 promoter molecule (Weaver IC, 2008) (see Figure 9).
In summary, our studies establish a first working of the hypothesis on how maternal behaviour can result in epigenetic reprogramming in the offspring. Neurotransmitter release results in activation of a signaling pathway that leads to recruitment of particular transcription factors such as NGFI-A to their recognition elements in front of specific genes. Our hypothesis is that NGFI-A facilitates MBD2 interaction through recruitment of CBP and that the ensuing increased acetylation of the GR exon 17 promoter opens up the chromatin configuration, thus increasing the accessibility of the sequence to MBD2.
Epigenetic Programming and Human Behavioural Exposure
A fundamental question that remains to be answered is whether a mechanism similar to the mechanism described in the rat operates in generating interindividual differences in humans and that exposure to different social behaviour results in differences in epigenetic programming of gene expression, leading to altered gene function with consequences on health status. The hypothesis is obviously attractive; social adversity in early childhood similar to low LG-ABN might result in aberrant epigenetic programming, causing changes in gene expression, which will stably impact on behaviour and physiologic functions later in life. Similarly strong environmental exposures later in life might reverse or alter epigenetic programming of the genes regulating human behaviour. The main impediment in studying epigenetic programming in living humans is obviously the inaccessibility of the brain and other tissues to epigenetic analysis. Although candidate genes could be a reasonable approach to identify differentially methylated targets, a nonbiased approach might identify other unanticipated candidates. Thus, whole-epigenome analyses should enable the identification of hitherto unknown epigenetic markers of human behaviour exposures.
Summary
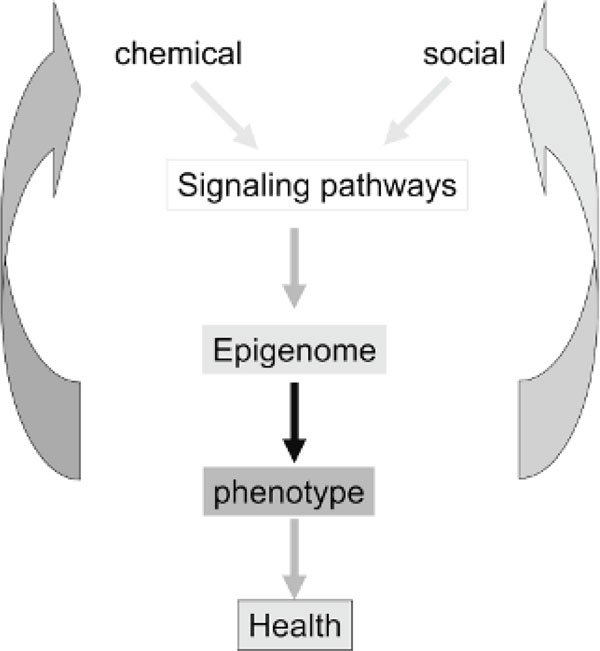
Recent data from the rat maternal care model chart a pathway leading from the behaviour of the mother to long-term programming of gene expression in the offspring (see Figure 9). This pathway involves the firing of neurotransmitter receptors in response to the behaviour and signaling pathways, which activate sequence-specific transcription factors such as NGFI-A. NGFI-A interacts with its recognition element in the GR exon 17 promoter and recruits the HAT CBP to the gene. This results in acetylation of chromatin and recruitment of DNA demethylases such as MBD2, leading to demethylation and stable activation of this gene. These data point to a thought-provoking notion that epigenetic processes play a role in shaping human behaviour in response to different levels of social adversity early in life and later during adulthood (Figure 10). Preliminary data examining a few genes, the ribosomal RNA genes, and the GR exon 1f in the hippocampi of suicide victims and their matched controls suggest that differences in epigenetic programming do exist between individuals and that these variations might associate with exposure to social adversity early in life (MacGowen PO, unpublished data). The rapid development of high-throughput sequencing techniques will enable in the future the unbiased mapping of epigenomes and identification of candidate genes, which exhibit epigenetic differences among individuals. The possibility that epigenetic mechanisms might be playing a role in generating interindividual differences in behaviour has tremendous potential to provide a mechanism for the ageold question of the relationship of nurture and nature (see Figure 10 for a model).
Figure 10.
The implications of a lifelong dynamic epigenome. Environmental exposures impact our epigenome, which, in turn, impacts our response to these exposures. The final outcome is a change in phenotype, including effects on health status.
Similar mechanisms might be at play in autoimmune disease and in asthma. It is well established that alteration in DNA methylation plays a role in the autoimmune disease systemic lupus erythematosus (SLE) [83]. Drugs known to induce lupus, such as procainamide and hydralazine, also induce demethylation, [84-86] and the demethylating agent 5-azacytidine induces lupus [87]. The genome of T cells from SLE patients is demethylated in comparison with T cells from control subjects [88]. It is proposed that demethylation activates genes whose expression confers on T cells' self-reactivity. Interestingly, MBD2, which was implicated to be responsible for epigenetic reprogramming and demethylation in response to maternal care in our studies, detailed above, was also found to be upregulated in T cells from lupus patients [88]. Lupus serves as a nice illustration of the notion that exposure to different xenobiotics, such as procainamide, could precipitate a pathologic state, which is mediated by epigenetic reprogramming. Our studies on maternal care suggest that similar signaling pathways that lead to lupus, such as MBD2 induction, could also be triggered by behavioural exposures. Unraveling the mechanisms linking environmental exposures, including the social environment, to health outcomes could open new horizons in our understanding of the late-onset diseases, including autoimmune disease and asthma. More importantly, since epigenetic processes are reversible, this might lead to a paradigm shift in prophylactic and therapeutic approaches to late-onset pathologies.
Note
These studies were supported by a grant from the Canadian Institutes for Health Research (CIHR) to M.J.M. and M.S. and from the National Cancer Institute of Canada to M.S. M.J.M. is supported by a CIHR Senior Scientist Award, and the project was supported by a Distinguished Investigator Award to M.J.M. from the National Alliance for Research on Schizophrenia and Affective Disorders.
References
- Szyf M. Towards a pharmacology of DNA methylation. Trends Pharmacol Sci. 2001;22:350–4. doi: 10.1016/S0165-6147(00)01713-2. [DOI] [PubMed] [Google Scholar]
- Weidle UH, Grossmann A. Inhibition of histone deacetylases: a new strategy to target epigenetic modifications for anticancer treatment. Anticancer Res. 2000;20:1471–85. [PubMed] [Google Scholar]
- Kramer OH, Gottlicher M, Heinzel T. Histone deacetylase as a therapeutic target. Trends Endocrinol Metab. 2001;12:294–300. doi: 10.1016/S1043-2760(01)00438-6. [DOI] [PubMed] [Google Scholar]
- Simonini MV, Camargo LM, Dong E. The benzamide MS-275 is a potent, long-lasting brain region-selective inhibitor of histone deacetylases. Proc Natl Acad Sci USA. 2006;103:1587–92. doi: 10.1073/pnas.0510341103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A, Riggs AD. DNA methylation and gene function. Science. 1980;210:604–10. doi: 10.1126/science.6254144. [DOI] [PubMed] [Google Scholar]
- Razin A, Szyf M. DNA methylation patterns. Formation and function. Biochim Biophys Acta. 1984;782:331–42. doi: 10.1016/0167-4781(84)90043-5. [DOI] [PubMed] [Google Scholar]
- Neel JV, Falls HF. The rate of mutation of the gene responsible for retinoblastoma in man. Science. 1951;114:419–22. doi: 10.1126/science.114.2964.419. [DOI] [PubMed] [Google Scholar]
- Sparkes RS, Murphree AL, Lingua RW. Gene for hereditary retinoblastoma assigned to human chromosome 13 by linkage to esterase D. Science. 1983;219:971–3. doi: 10.1126/science.6823558. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Zulueta M, Bender CM, Yang AS. Methylation of the 59 CpG island of the p16/CDKN2 tumor suppressor gene in normal and transformed human tissues correlates with gene silencing. Cancer Res. 1995;55:4531–5. [PubMed] [Google Scholar]
- Razin A. CpG methylation, chromatin structure and gene silencing-a three-way connection. Embo J. 1998;17:4905–8. doi: 10.1093/emboj/17.17.4905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groudine M, Eisenman R, Gelinas R, Weintraub H. Developmental aspects of chromatin structure and gene expression. Prog Clin Biol Res. 1983;134:159–82. [PubMed] [Google Scholar]
- Marks PA, Sheffery M, Rifkind RA. Modulation of gene expression during terminal cell differentiation. Prog Clin Biol Res. 1985;191:185–203. [PubMed] [Google Scholar]
- Ramain P, Bourouis M, Dretzen G. Changes in the chromatin structure of Drosophila glue genes accompany developmental cessation of transcription in wild type and transformed strains. Cell. 1986;45:545–53. doi: 10.1016/0092-8674(86)90286-2. [DOI] [PubMed] [Google Scholar]
- Grunstein M. Histone acetylation in chromatin structure and transcription. Nature. 1997;389:349–52. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- Varga-Weisz PD, Becker PB. Regulation of higher-order chromatin structures by nucleosome-remodelling factors. Curr Opin Genet Dev. 2006;16:151–6. doi: 10.1016/j.gde.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Bergmann A, Lane ME. HIDden targets of microRNAs for growth control. Trends Biochem Sci. 2003;28:461–3. doi: 10.1016/S0968-0004(03)00175-0. [DOI] [PubMed] [Google Scholar]
- Zhang B, Pan X, Cobb GP, Anderson TA. MicroRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- Vo N, Klein ME, Varlamova O. A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc Natl Acad Sci USA. 2005;102:16426–31. doi: 10.1073/pnas.0508448102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch JT, Lutter LC, Rhodes D. Structure of nucleosome core particles of chromatin. Nature. 1977;269:29–36. doi: 10.1038/269029a0. [DOI] [PubMed] [Google Scholar]
- Sarma K, Reinberg D. Histone variants meet their match. Nat Rev Mol Cell Biol. 2005;6:139–49. doi: 10.1038/nrm1567. [DOI] [PubMed] [Google Scholar]
- Jenuwein T. Re-SET-ting heterochromatin by histone methyltransferases. Trends Cell Biol. 2001;11:266–73. doi: 10.1016/S0962-8924(01)02001-3. [DOI] [PubMed] [Google Scholar]
- Wade PA, Pruss D, Wolffe AP. Histone acetylation: chromatin in action. Trends Biochem Sci. 1997;22:128–32. doi: 10.1016/S0968-0004(97)01016-5. [DOI] [PubMed] [Google Scholar]
- Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem. 2006;75:243–69. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- Henikoff S, McKittrick E, Ahmad K. Epigenetics, histone H3 variants, and the inheritance of chromatin states. Cold Spring Harb Symp Quant Biol. 2004;69:235–43. doi: 10.1101/sqb.2004.69.235. [DOI] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–80. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- Kuo MH, Allis CD. Roles of histone acetyltransferases and deacetylases in gene regulation. Bioessays. 1998;20:615–26. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Perry M, Chalkley R. Histone acetylation increases the solubility of chromatin and occurs sequentially over most of the chromatin. A novel model for the biological role of histone acetylation. J Biol Chem. 1982;257:7336–47. [PubMed] [Google Scholar]
- Lee DY, Hayes JJ, Pruss D, Wolffe AP. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell. 1993;72:73–84. doi: 10.1016/0092-8674(93)90051-Q. [DOI] [PubMed] [Google Scholar]
- Wolffe AP. Histone deacetylase: a regulator of transcription. Science. 1996;272:371–2. doi: 10.1126/science.272.5260.371. [DOI] [PubMed] [Google Scholar]
- Lachner M, O'Carroll D, Rea S. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–20. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–53. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- Tsukada Y, Fang J, Erdjument-Bromage H. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–6. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- Bultman SJ, Gebuhr TC, Magnuson T. A Brg1 mutation that uncouples ATPase activity from chromatin remodeling reveals an essential role for SWI/SNF-related complexes in beta-globin expression and erythroid development. Genes Dev. 2005;19:2849–61. doi: 10.1101/gad.1364105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogryzko VV, Schiltz RL, Russanova V. The transcriptional coactivators p300 and CBP are histone acetyltransferases. Cell. 1996;87:953–9. doi: 10.1016/S0092-8674(00)82001-2. [DOI] [PubMed] [Google Scholar]
- Razin A, Cedar H. Distribution of 5-methylcytosine in chromatin. Proc Natl Acad Sci USA. 1977;74:2725–8. doi: 10.1073/pnas.74.7.2725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baylin SB, Esteller M, Rountree MR. Aberrant patterns of DNA methylation, chromatin formation and gene expression in cancer. Hum Mol Genet. 2001;10:687–92. doi: 10.1093/hmg/10.7.687. [DOI] [PubMed] [Google Scholar]
- Okano M, Xie S, Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat Genet. 1998;19:219–20. doi: 10.1038/890. [DOI] [PubMed] [Google Scholar]
- Vilain A, Apiou F, Dutrillaux B, Malfoy B. Assignment of candidate DNA methyltransferase gene (DNMT2) to human chromosome band 10p15.1 by in situ hybridization. Cytogenet Cell Genet. 1998;82:120. doi: 10.1159/000015083. [DOI] [PubMed] [Google Scholar]
- Bourc'his D, Xu GL, Lin CS. Dnmt3L and the establishment of maternal genomic imprints. Science. 2001;294:2536–9. doi: 10.1126/science.1065848. [DOI] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–26. doi: 10.1016/0092-8674(92)90611-F. [DOI] [PubMed] [Google Scholar]
- Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–57. doi: 10.1016/S0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- Goto K, Numata M, Komura JI. Expression of DNA methyltransferase gene in mature and immature neurons as well as proliferating cells in mice. Differentiation. 1994;56:39–44. doi: 10.1046/j.1432-0436.1994.56120039.x. [DOI] [PubMed] [Google Scholar]
- Veldic M, Guidotti A, Maloku E. In psychosis, cortical interneurons overexpress DNA-methyltransferase 1. Proc Natl Acad Sci USA. 2005;102:2152–7. doi: 10.1073/pnas.0409665102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramchandani S, Bhattacharya SK, Cervoni N, Szyf M. DNA methylation is a reversible biological signal. Proc Natl Acad Sci USA. 1999;96:6107–12. doi: 10.1073/pnas.96.11.6107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucarelli M, Fuso A, Strom R, Scarpa S. The dynamics of myogenin site-specific demethylation is strongly correlated with its expression and with muscle differentiation. J Biol Chem. 2001;276:7500–6. doi: 10.1074/jbc.M008234200. [DOI] [PubMed] [Google Scholar]
- Bruniquel D, Schwartz RH. Selective, stable demethylation of the interleukin-2 gene enhances transcription by an active process. Nat Immunol. 2003;4:235–40. doi: 10.1038/ni887. [DOI] [PubMed] [Google Scholar]
- Kersh EN, Fitzpatrick DR, Murali-Krishna K. Rapid demethylation of the IFN-gamma gene occurs in memory but not naive CD8 T cells. J Immunol. 2006;176:4083–93. doi: 10.4049/jimmunol.176.7.4083. [DOI] [PubMed] [Google Scholar]
- Weaver IC, Cervoni N, Champagne FA. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7:847–54. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- Jost JP. Nuclear extracts of chicken embryos promote an active demethylation of DNA by excision repair of 5-methyldeoxycytidine. Proc Natl Acad Sci USA. 1993;90:4684–8. doi: 10.1073/pnas.90.10.4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Zheng Y, Hess D. 5-Methylcytosine-DNA glycosylase activity is present in a cloned G/T mismatch DNA glycosylase associated with the chicken embryo DNA demethylation complex. Proc Natl Acad Sci USA. 2000;97:5135–9. doi: 10.1073/pnas.100107597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya SK, Ramchandani S, Cervoni N, Szyf M. A mammalian protein with specific demethylase activity for mCpG DNA. Nature. 1999;397:579–83. doi: 10.1038/17533. [DOI] [PubMed] [Google Scholar]
- Ng HH, Zhang Y, Hendrich B. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat Genet. 1999;23:58–61. doi: 10.1038/12659. [DOI] [PubMed] [Google Scholar]
- Detich N, Theberge J, Szyf M. Promoter-specific activation and demethylation by MBD2/demethylase. J Biol Chem. 2002;277:35791–4. doi: 10.1074/jbc.C200408200. [DOI] [PubMed] [Google Scholar]
- Detich N, Bovenzi V, Szyf M. Valproate induces replicationindependent active DNA demethylation. J Biol Chem. 2003;278:27586–92. doi: 10.1074/jbc.M303740200. [DOI] [PubMed] [Google Scholar]
- Detich N, Hamm S, Just G. The methyl donor Sadenosylmethionine inhibits active demethylation of DNA: a candidate novel mechanism for the pharmacological effects of Sadenosylmethionine. J Biol Chem. 2003;278:20812–20. doi: 10.1074/jbc.M211813200. [DOI] [PubMed] [Google Scholar]
- Barreto G, Schafer A, Marhold J. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–5. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- Cervoni N, Szyf M. Demethylase activity is directed by histone acetylation. J Biol Chem. 2001;276:40778–87. doi: 10.1074/jbc.M103921200. [DOI] [PubMed] [Google Scholar]
- D'Alessio AC, Szyf M. Epigenetic tete-a-tete: the bilateral relationship between chromatin modifications and DNA methylation. Biochem Cell Biol. 2006;84:463–76. doi: 10.1139/O06-090. [DOI] [PubMed] [Google Scholar]
- Fuks F, Burgers WA, Brehm A. DNA methyltransferase Dnmt1 associates with histone deacetylase activity. Nat Genet. 2000;24:88–91. doi: 10.1038/71750. [DOI] [PubMed] [Google Scholar]
- Fuks F, Hurd PJ, Wolf D. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J Biol Chem. 2003;278:4035–40. doi: 10.1074/jbc.M210256200. [DOI] [PubMed] [Google Scholar]
- Rountree MR, Bachman KE, Baylin SB. DNMT1 binds HDAC2 and a new co-repressor, DMAP1, to form a complex at replication foci. Nat Genet. 2000;25:269–77. doi: 10.1038/77023. [DOI] [PubMed] [Google Scholar]
- Vire E, Brenner C, Deplus R. The Polycomb group protein EZH2 directly controls DNA methylation. Nature. 2006;439:871–4. doi: 10.1038/nature04431. [DOI] [PubMed] [Google Scholar]
- Di Croce L, Raker VA, Corsaro M. Methyltransferase recruitment and DNA hypermethylation of target promoters by an oncogenic transcription factor. Science. 2002;295:1079–82. doi: 10.1126/science.1065173. [DOI] [PubMed] [Google Scholar]
- Lichtenstein M, Keini G, Cedar H, Bergman Y. B cell-specific demethylation: a novel role for the intronic kappa chain enhancer sequence. Cell. 1994;76:913–23. doi: 10.1016/0092-8674(94)90365-4. [DOI] [PubMed] [Google Scholar]
- Szyf M, Weaver I, Meaney M. Maternal care, the epigenome and phenotypic differences in behavior. Reprod Toxicol. 2007;24:9–19. doi: 10.1016/j.reprotox.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Comb M, Goodman HM. CpG methylation inhibits proenkephalin gene expression and binding of the transcription factor AP-2. Nucleic Acids Res. 1990;18:3975–82. doi: 10.1093/nar/18.13.3975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inamdar NM, Ehrlich KC, Ehrlich M. CpG methylation inhibits binding of several sequence-specific DNA-binding proteins from pea, wheat, soybean and cauliflower. Plant Mol Biol. 1991;17:111–23. doi: 10.1007/BF00036811. [DOI] [PubMed] [Google Scholar]
- Nan X, Campoy FJ, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471–81. doi: 10.1016/S0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- Fujita N, Takebayashi S, Okumura K. Methylation-mediated transcriptional silencing in euchromatin by methyl-CpG binding protein MBD1 isoforms. Mol Cell Biol. 1999;19:6415–26. doi: 10.1128/mcb.19.9.6415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol. 1998;18:6538–47. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Diorio J, Tannenbaum B. Maternal care, hippocampal glucocorticoid receptors, and hypothalamic-pituitary-adrenal responses to stress. Science. 1997;277:1659–62. doi: 10.1126/science.277.5332.1659. [DOI] [PubMed] [Google Scholar]
- Francis D, Diorio J, Liu D, Meaney MJ. Nongenomic transmission across generations of maternal behavior and stress responses in the rat. Science. 1999;286:1155–8. doi: 10.1126/science.286.5442.1155. [DOI] [PubMed] [Google Scholar]
- Meaney MJ, Szyf M. Maternal care as a model for experiencedependent chromatin plasticity? Trends Neurosci. 2005;28:456–63. doi: 10.1016/j.tins.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Champagne FA, Weaver IC, Diorio J. Maternal care associated with methylation of the estrogen receptor-alpha1b promoter and estrogen receptor-alpha expression in the medial preoptic area of female offspring. Endocrinology. 2006;147:2909–15. doi: 10.1210/en.2005-1119. [DOI] [PubMed] [Google Scholar]
- Cervoni N, Detich N, Seo SB. The oncoprotein Set/TAF-1beta, an inhibitor of histone acetyltransferase, inhibits active demethylation of DNA, integrating DNA methylation and transcriptional silencing. J Biol Chem. 2002;277:25026–31. doi: 10.1074/jbc.M202256200. [DOI] [PubMed] [Google Scholar]
- Weaver IC, Diorio J, Seckl JR. Early environmental regulation of hippocampal glucocorticoid receptor gene expression: characterization of intracellular mediators and potential genomic target sites. Ann N Y Acad Sci. 2004;1024:182–212. doi: 10.1196/annals.1321.099. [DOI] [PubMed] [Google Scholar]
- Tremolizzo L, Carboni G, Ruzicka WB. An epigenetic mouse model for molecular and behavioral neuropathologies related to schizophrenia vulnerability. Proc Natl Acad Sci USA. 2002;99:17095–100. doi: 10.1073/pnas.262658999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver IC, Champagne FA, Brown SE. Reversal of maternal programming of stress responses in adult offspring through methyl supplementation: altering epigenetic marking later in life. J Neurosci. 2005;25:11045–54. doi: 10.1523/JNEUROSCI.3652-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaney MJ, Aitken DH, Sapolsky RM. Thyroid hormones influence the development of hippocampal glucocorticoid receptors in the rat: a mechanism for the effects of postnatal handling on the development of the adrenocortical stress response. Neuroendocrinology. 1987;45:278–83. doi: 10.1159/000124741. [DOI] [PubMed] [Google Scholar]
- Meaney MJ, Diorio J, Francis D. Postnatal handling increases the expression of cAMP-inducible transcription factors in the rat hippocampus: the effects of thyroid hormones and serotonin. J Neurosci. 2000;20:3926–35. doi: 10.1523/JNEUROSCI.20-10-03926.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante P, Diorio J, Meaney MJ. Serotonin regulates hippocampal glucocorticoid receptor expression via a 5-HT7 receptor. Brain Res Dev Brain Res. 2002;139:199–203. doi: 10.1016/S0165-3806(02)00550-3. [DOI] [PubMed] [Google Scholar]
- McCormick JA, Lyons V, Jacobson MD. 59-Heterogeneity of glucocorticoid receptor messenger RNA is tissue specific: differential regulation of variant transcripts by early-life events. Mol Endocrinol. 2000;14:506–17. doi: 10.1210/me.14.4.506. [DOI] [PubMed] [Google Scholar]
- Richardson BC. Role of DNA methylation in the regulation of cell function: autoimmunity, aging and cancer. J Nutr. 2002;132 doi: 10.1093/jn/132.8.2401S. [DOI] [PubMed] [Google Scholar]
- Cornacchia E, Golbus J, Maybaum J. Hydralazine and procainamide inhibit T cell DNA methylation and induce autoreactivity. J Immunol. 1988;140:2197–200. [PubMed] [Google Scholar]
- Scheinbart LS, Johnson MA, Gross LA. Procainamide inhibits DNA methyltransferase in a human T cell line. J Rheumatol. 1991;18:530–4. [PubMed] [Google Scholar]
- Deng C, Lu Q, Zhang Z. Hydralazine may induce autoimmunity by inhibiting extracellular signal-regulated kinase pathway signaling. Arthritis Rheum. 2003;48:746–56. doi: 10.1002/art.10833. [DOI] [PubMed] [Google Scholar]
- Quddus J, Johnson KJ, Gavalchin J. Treating activated CD4+ T cells with either of two distinct DNA methyltransferase inhibitors, 5-azacytidine or procainamide, is sufficient to cause a lupus-like disease in syngeneic mice. J Clin Invest. 1993;92:38–53. doi: 10.1172/JCI116576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balada E, Ordi-Ros J, Serrano-Acedo S. Transcript overexpression of the MBD2 and MBD4 genes in CD4+ T cells from systemic lupus erythematosus patients. J Leukoc Biol. 2007;81:1609–16. doi: 10.1189/jlb.0107064. [DOI] [PubMed] [Google Scholar]