Abstract
A fluorescence confocal microscope incorporating a 1.8-mm-diam gradient-index relay lens is developed for in vivo histological guidance during resection of brain tumors. The microscope utilizes a dual-axis confocal architecture to efficiently reject out-of-focus light for high-contrast optical sectioning. A biaxial microelectromechanical system (MEMS) scanning mirror is actuated at resonance along each axis to achieve a large field of view with low-voltage waveforms. The unstable Lissajous scan, which results from actuating the orthogonal axes of the MEMS mirror at highly disparate resonance frequencies, is optimized to fully sample 500×500 pixels at two frames per second. Optically sectioned fluorescence images of brain tissues are obtained in living mice to demonstrate the utility of this microscope for image-guided resections.
Keywords: laser-scanning confocal microscopy, image-guided surgery, optical biopsy, fluorescence molecular imaging, microelectromechanical systems, microendoscopy
Surgical resection is a major component in the standard of care for the treatment of brain cancers, where clinical studies have demonstrated that improved outcomes correlate with more complete resections, especially in conjunction with adjuvant therapies such as radiochemotherapy.1, 2 Histological guidance provided by a miniaturized microscope could be used to improve the accuracy of resection at selected sites where tumor margins are difficult to identify, thereby improving survival and reducing neurological damage. We have been developing dual-axis confocal (DAC) microscopes for in vivo imaging with cellular resolution.3, 4 Unfortunately, the 10-mm-diam form factor of our current microscopes greatly exceeds their field of view (FOV) of less than 0.5 mm. The size of these instruments also surpasses that of conventional tools for tumor resection, which are typically 1 to 2 mm in diameter, rendering such a device inconvenient for guiding surgical procedures.
Here we demonstrate that the incorporation of a gradient-index (GRIN) lens at the tip of a microelectromechanical system (MEMS)-scanned DAC allows the image plane to be relayed forward by 3 cm through a 1.8-mm-diam optic (see Fig. 1). This form factor is designed to enable convenient microscopy in a clinical setting for optical biopsy and image-guided resection of questionable tumor-margin locations using fluorescent contrast agents. Previously, GRIN relay lenses have been utilized for conventional single-axis confocal5 and nonlinear microscopes.6 Here we show that two low-NA beams, with a crossing angle of ∼45 deg, can also be relayed and scanned through such an optical element with minimal aberrations.
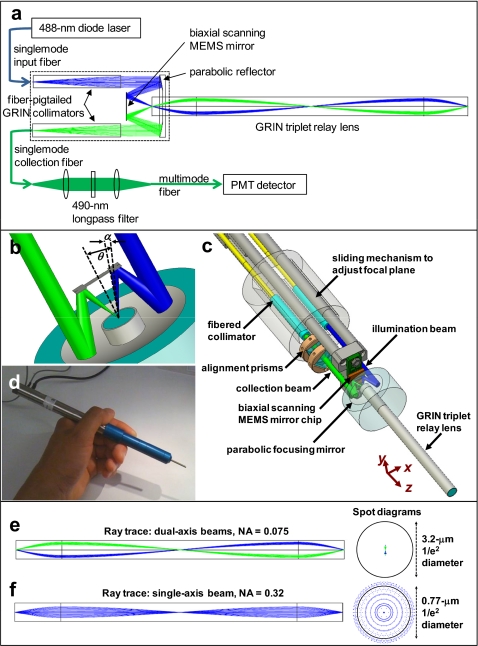
Figure 1.
The GRIN-relayed DAC. (a) A schematic of the optical circuit of the microscope. (b) A close-up view of the parabolic focusing optic and the MEMS scanning mirror. (c) The microscope scan head and relay lens. (d) A photograph of the microscope exterior. (e) Ray-trace and spot diagrams of the dual-axis illumination (blue) and collection (green) beam paths within a GRIN relay lens. (f) Ray-trace and spot diagrams of a single large-NA GRIN-relayed beam. (Color online only.)
Studies have demonstrated the ability of a DAC to effectively reject out-of-focus scattered light for high-contrast optical sectioning in turbid media.7, 8, 9 The DAC utilizes low-power diode lasers along with simple low-NA optics to achieve deep tissue imaging. The long working distance afforded by the low-NA focused beams allows for postobjective beam scanning with a biaxial MEMS scanner.3, 4 This eliminates the need for compound lenses to mitigate the off-axis aberrations that result from scanning a beam through a focusing objective.
Our MEMS scanning device contains a total of four electrostatic actuators: a pair of actuators for each axis of the mirror to achieve tilt in opposing directions (pull-pull configuration).10 We have previously achieved raster-scanned imaging with these MEMS-scanned microscopes by actuating one axis of the mirror at its resonance frequency (∼1 to 3 kHz) and actuating the orthogonal axis with a slow sawtooth waveform at a frame rate of 4 to 30 Hz. The fast axis, because it is driven at resonance, can achieve large deflection angles (±3 deg or more) with relatively low-voltage waveforms (∼20 V). Furthermore, only one actuator is necessary to drive an axis at its natural resonance frequency. On the other hand, relatively large voltages (∼100 to 120 V) are applied to two actuators to achieve modest deflection angles (±1–3 deg) at off-resonance frequencies. Therefore, the ability to drive both axes of the mirror at resonance, in a Lissajous scanning pattern,11 enables large deflection angles to be achieved with fewer wired connections and lower voltages.
Previous implementations of Lissajous scanning have generally utilized stable scan patterns that self-repeat at a frame rate determined by the frequency ratio of the driving waveforms along each axis.12, 13 However, with arbitrary resonance frequencies, it is difficult to achieve a stable Lissajous scan pattern that repeats a rate of 1 to 10 Hz and also provides coverage over a large FOV of 500×500 pixels. Here we demonstrate a general method to utilize an unstable (nonrepeating) “sliding” Lissajous pattern to fully sample 500×500 pixels at a frame rate of 2 Hz. A similar strategy utilizing an unstable Lissajous scan of a MEMS mirror was demonstrated previously for two-photon microscopy.14 In that work, high frame rates of up to 10 Hz were achieved with a FOV of 256×256 pixels. In our particular implementation, the scan frequencies are tuned to yield a Lissajous pattern that predictably slides at an optimized rate to ensure that each pixel is sampled. This eliminates the need for filtering or averaging to account for unsampled pixels.
Figure 1a illustrates the optical circuit of our microscope. This diagram includes a ray-trace simulation (Zemax, Bellevue, Washington) depicting the beam paths of the DAC as they are relayed through a nonmagnifying (1:1) GRIN triplet relay assembly consisting of a pair of 1∕4-pitch, 4.25-mm-long, high-NA (0.51) GRIN lenses on either side of a 1∕2-pitch, 21.85-mm-long, low-NA (0.19) GRIN lens. Figures 1b, 1c are design drawings of the basic components of the microscope. The parabolic reflector (AgilTek) insures that the two collimated beams intersect and focus to the same point, which defines the focal volume of the microscope. A pair of 0.1-deg wedged alignment (Risley) prisms, shown in Fig. 1c, are rotated to bring the collimated beams into parallel alignment with each other. Figure 1d is a photograph of a fully packaged microscope, including the piezoelectric actuator at the proximal end that translates the MEMS mirror in the axial direction for adjusting the focal depth.
The single-mode fiber-pigtailed collimators (GRINTECH Gmbh, Jena, Germany, focal length=4.5 mm) used in our microscope yield a 1∕e2 beam diameter of 0.9 mm. As a result of the parabolic reflector having a 6-mm focal length and the collimators being spaced apart by 4.7 mm, the focusing half angle of each beam (1∕e2) and the intersection half angle between the beams, respectively, are α=0.075 rad and θ=22.2 deg [see Fig. 1b]. If the Gaussian illumination and collection beams are truncated (apodized) to allow 99% power transmission, diffraction theory predicts a full-width at half-maximum (FWHM) spatial resolution (in micrometers) of:7
| (1) |
where θ=22.2 deg; α=0.075 rad, n=1 (air), and λ=488 nm. The experimentally measured FWHM resolution of our microscope is: Δxd=3.7 μm, Δyd=3.6 μm, and Δzd=8.0 μm. The degradation in resolution is due to aberrations from the GRIN collimation and relay lenses, as well as alignment imperfections and diffraction noise introduced by imperfections in the MEMS mirror (beam clipping and hinge structures within the mirror surface). The spatial resolution of the GRIN-relayed DAC is degraded by less than 10% compared to the resolution measured without the GRIN relay lens (data not shown). A ray-trace simulation was performed to roughly compare the effects of aberrations due to the GRIN relay lens on our dual-axis beams versus a single large-NA beam used for conventional confocal or multiphoton microscopy. The DAC simulation reveals minimal aberrations [Fig. 1e], with various rays converging to a region well within the 3.2-μm theoretical spot size (1∕e2 diameter) of a diffraction-limited Gaussian beam with NA=0.075 (1∕e2) at 488 nm in air. In Fig. 1f, a ray trace simulation is shown for a single on-axis Gaussian beam that fills the collection NA of the GRIN relay lens (99% power transmission NA=0.5; 1∕e2 NA=0.32). The spread in the spot diagram extends beyond the 0.77-μm theoretical spot size (1∕e2 diameter) of a diffraction-limited Gaussian beam with NA=0.32 (1∕e2) at 488 nm in air. Evidently, since the DAC utilizes low-NA beam paths through the GRIN relay lens, rather than relying on the full NA of the lens, the effects of aberrations are minimized. A complete diffraction analysis, which is not presented in this work, is required to fully characterize the tradeoffs between GRIN-relayed versions of the DAC and single-axis confocal microscopes.
As mentioned previously, an unstable Lissajous scan pattern is achieved by driving our MEMS mirror at resonance along both axes, enabling large-FOV imaging with relatively low-voltage waveforms. The resulting scan pattern is described by the following parametric equations as a function of time:
| (2) |
| (3) |
where ϕx and ϕy are phase shift constants (in radians), a and b are the slow and fast frequencies, respectively, in cycles∕sec (Hz), and the FOV of the scan extends from 0 to 2A in the x direction and 0 to 2B in the y direction (in this case, A=B=250). The resonant frequencies of the two axes of our MEMS mirror are 985±10 Hz and 2220±10 Hz (FWHM bandwidth ∼20 Hz).
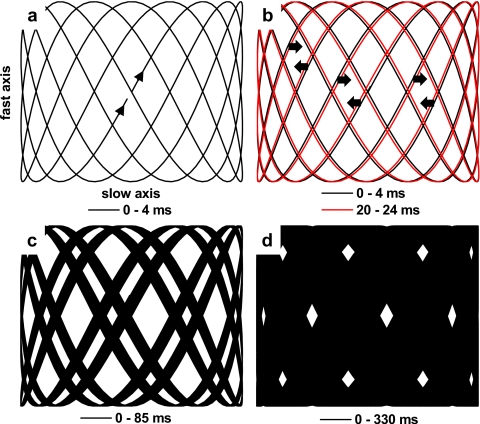
Figure 2a depicts the trajectory of a Lissajous scan pattern between t=0 and 4 ms (assuming ϕx=ϕy=0). If the frequency ratio of the fast- and slow-axis waveforms is exactly 9∕4, then a stable Lissajous pattern results, repeating once for every four cycles of the slow axis or every nine cycles of the fast axis (4∕a or 9∕b sec). If the ratio between the frequencies is slightly different than 9∕4, an unstable Lissajous pattern is generated that appears to slide in time in the horizontal direction [Fig. 2b]. The sliding speed, determined by the frequency ratio, is optimally chosen such that the Lissajous pattern shifts by a distance of less than 1 pixel for every nearly repeated cycle (4∕a or 9∕b sec), ensuring that no pixels are missed over time. Figures 2c, 2d illustrate how an entire image is sampled over time by the sliding Lissajous pattern. Taking the derivative of Eq. 3 with respect to time reveals that the maximum velocity of the fast-axis scan is given by 2π⋅b⋅B≅3.5(10)6 pixels∕sec. Thus, a data acquisition rate of 4 MHz is chosen to obviate dead (unsampled) pixels along the trajectory of the Lissajous scan.
Figure 2.
Calculated Lissajous scan patterns. Fast-axis frequency=2219 Hz. Slow-axis frequency=986 Hz. (a) 0 to 4 ms, where the pattern nearly repeats itself. (b) A comparison of two Lissajous patterns 20 ms apart, showing how the pattern shifts over time. (c) 0 to 85 ms, showing partial coverage over the FOV. (d) 0 to 330 ms, showing nearly full coverage over the FOV.
Sinusoidal waveforms are generated with National Instruments (Austin, Texas) boards programmed in LabVIEW and sent through a voltage amplifier (Agiloptics, Albuquerque, New Mexico) to drive our biaxial MEMS mirror. A National Instruments digitizer board is used to record signals from a PMT detector at a rate of 4 MHz. All processes are synchronized for phase-locked operation, and the phase shifts ϕx and ϕy are determined via calibration. In our implementation, a=986.0 Hz and b=2219.0 Hz. For each detector value sampled at time t, Eqs. 2, 3 are used to calculate the exact coordinates x and y of the Lissajous scan. These coordinates are rounded to the nearest integer to locate the most appropriate pixel for data entry within the 500×500 pixel bitmap image (programmed in LabVIEW). Image degradation due to rounding errors is minimal due to the fact that we oversample the image. The spatial dimension of our target FOV is approximately 500×500 μm. Therefore, with a pixel density of 500×500, and a lateral resolution of ∼4 μm, our sampling (1 pixel∕μm) exceeds the Nyquist criterion by a factor of 2. Two million points are collected per image, resulting in a frame rate of 2 Hz. Redundantly sampled pixels contain an average of the sampled values at that pixel location.
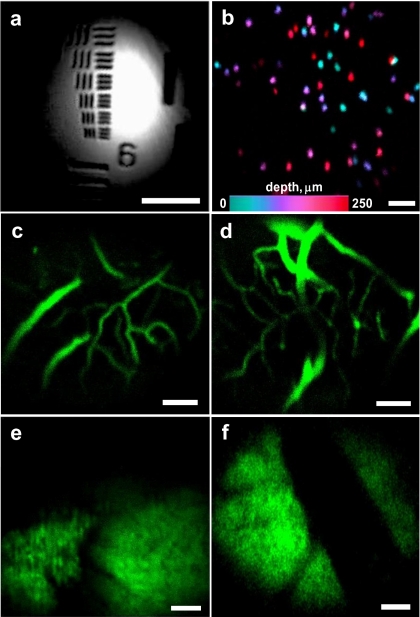
Example images acquired with our GRIN-relayed DAC are shown in Figs. 3a, 3b, 3c, 3d, 3e, 3f. Figure 3a is a reflectance image of a United States Air Force (USAF) bar target (group 7), in which the width of the smallest set of lines is 2.2 μm per line. Due to the field curvature of the microscope, only the center of the FOV is in focus for this flat reflective target. In practice, field curvature is not a major concern, since our device is designed to image at least ten microns beneath the surface of a thick tissue, rather than imaging a flat surface. Figure 3b is a depth projection (FOV=400×400 μm) of 10-μm-diam fluorescent beads embedded in Matrigel. The color map encodes depths from 0 to 250 μm. Figure 3c is an in vivo image at a depth of 50 μm (FOV=325×325 μm) of vasculature within the cerebrum of a mouse after a portion of the skull has been surgically removed (five-frame average). To highlight the vasculature in this image, fluorescein-dextran (MW=70,000) was injected into the retro-orbital capillary plexus at a dose of ∼1 mg (40-mg∕kg body weight) in a 200-μL volume. Figure 3d is of the same tissue as Fig. 3c, except that it was acquired at a depth of 100 μm (five-frame average). Figure 3e is an ex vivo image at a depth of 25 μm (FOV=350×350 μm) of cerebellum from a transgenic mouse (10 days of age) that expresses green fluorescent protein (GFP) under the Math1 promoter.15 After weaning, expression of this promoter is repressed in the cerebellum, except in medulloblastomas, a cerebellar brain tumor that spontaneously develops in this gene knock-out mouse (Ptc+∕−p53−∕− Math1-GFP). In Fig. 3f, in vivo imaging of the tumor is performed after surgical removal of a portion of the skull. This image (FOV=350×350 μm; depth=30 μm) reveals the GFP-expressing tumor in contrast to the dark normal brain tissue, and indicates that this instrument would have utility in detecting tumor margins when used in conjunction with appropriate exogenously applied fluorescent molecular probes. Figures 3e, 3f are single frames that have been filtered with a smoothing algorithm that replaces each pixel with an average of its adjacent pixels. Spatial resolution is minimally affected, since the image is oversampled.
Figure 3.
Images taken by the GRIN-relayed DAC. All scale bars are 50 μm. Illumination intensity=1 mW. (a) Reflectance image of a USAF bar target (group 7). (b) Depth projection of 10-μm-diam fluorescent beads embedded in a Matrigel matrix, imaged between 0 and 250 μm deep. The color map encodes depth. (c) In vivo image of mouse cerebrum vasculature after retro-orbital injection of fluorescein-dextran (MW=70,000). Depth=50 μm. Average of five frames. (d) Same as (c), but imaged at a depth of 100 μm. (e) Ex vivo image of cerebellum from a Math1-GFP mouse (10 days old). Depth=25 μm. (f) In vivo image of GFP-expressing medulloblastoma in a mouse. Depth=30 μm. (Color online only.)
We have presented the design and demonstration of a miniature DAC outfitted with a 1.8-mm-diam GRIN relay lens for convenient macroscopic positioning of the device at surgical sites. Our previous designs implemented raster-scanned MEMS mirrors, which were limited in their FOV because of the modest scan range attained by the nonresonant slow axis of the MEMS device. In addition, the raster-scanned MEMS mirrors required higher operating voltages that may not be preferred in clinical applications. Here we have described the use of resonant scanning along both axes of the MEMS mirror to achieve a large FOV with low actuation voltages and reduced numbers of actuators. We have developed the calibrated and synchronized instrumentation to decode the nonrepeating Lissajous pattern that results, and have demonstrated in vivo imaging of normal brain and tumor in living mice. Our goal is to use the GRIN-relayed DAC technology, along with exogenous fluorescent contrast agents, for spatially precise and molecularly specific resection of brain tumors
Acknowledgments
This work was funded, in part, by grants from the National Institutes of Health, including U54 CA105296 and U54 CA136465 (NCI). These studies were also supported by the Stanford Center for Children’s Brain Tumors (CCBT) at the Lucile Packard Children’s Hospital. Liu was supported, in part, by a Canary Foundation∕American Cancer Society postdoctoral fellowship for early cancer detection, and by an NIH Pathway to Independence Award, K99 EB008557 (NIBIB). We thank Ellis Garai for technical support, and Helen Rayburn and Matthew Scott for providing breeders for our medulloblastoma mouse colonies.
References
- Van den Bent M. J., Stupp R., Mason W. et al. , “Impact of the extent of resection on overall survival in newly-diagnosed glioblastoma after chemo-irradiation with temozolamide: further analysis of EORTC study 26981,” Eur. J. Cancer Suppl. 3, 134 (2005). [Google Scholar]
- Riffaud L., Saikali S., Leray E., Hamlat A., Haegelen C., Vauleon E., and Lesimple T., “Survival and prognostic factors in a series of adults with medulloblastomas,” J. Neurosurg. 111, 478–487 (2009). 10.3171/2009.1.JNS081004 [DOI] [PubMed] [Google Scholar]
- Liu J. T. C., Mandella M. J., Ra H., Wong L. K., Solgaard O., Kino G. S., Piyawattanametha W., Contag C. H., and Wang T. D., “Miniature near-infrared dual-axes confocal microscope utilizing a two-dimensional microelectromechanical systems scanner,” Opt. Lett. 32, 256 (2007). 10.1364/OL.32.000256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ra H., Piyawattanametha W., Mandella M. J., Hsiung P., Hardy J., Wang T. D., Contag C. H., Kino G. S., and Solgaard O., “Three-dimensional in vivo imaging by a handheld dual-axes confocal microscope,” Opt. Express 16, 7224 (2008). 10.1364/OE.16.007224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knittel J., Schnieder L., Buess G., Messerschmidt B., and Possner T., “Endoscope-compatible confocal microscope using a gradient index-lens system,” Opt. Commun. 188, 267 (2001). 10.1016/S0030-4018(00)01164-0 [DOI] [Google Scholar]
- Jung J. C., Mehta A. D., Aksay E., Stepnoski R., and Schnitzer M. J., “In vivo mammalian brain imaging using one-and two-photon fluorescence microendoscopy,” J. Neurophysiol. 92, 3121 (2004). 10.1152/jn.00234.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. T. C., Mandella M. J., Friedland S., Soetikno R., Crawford J. H., Contag C. H., Kino G. S., and Wang T. D., “Dual-axes confocal reflectance microscope for distinguishing colonic neoplasia,” J. Biomed. Opt. 11, 054019 (2006). 10.1117/1.2363363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong L. K., Wang T. D., Mandella M. J., and Kino G. S., “Improved rejection of multiply-scattered photons in confocal microscopy using dual-axes architecture,” Opt. Lett. 32, 1674 (2007). 10.1364/OL.32.001674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J. T. C., Mandella M. J., Crawford J. H., Contag C. H., Wang T. D., and Kino G. S., “Efficient rejection of scattered light enables deep optical sectioning in turbid media with low-NA optics in a dual-axis confocal architecture,” J. Biomed. Opt. 13, 034020 (2008). 10.1117/1.2939428 [DOI] [PubMed] [Google Scholar]
- Ra H., Piyawattanametha W., Taguchi Y., Lee D., Mandella M. J., and Solgaard O., “Two-dimensional MEMS scanner for dual-axes confocal in vivo microscopy,” J. Microelectromech. Syst. 16, 969 (2007). 10.1109/JMEMS.2007.892900 [DOI] [Google Scholar]
- Lissajous J. A., “Note sur une méthode nouvelle applicable à l’étude des mouvements vibratoires,” Comptes Rendus Séances l’Academie Sci. 41, 814–817 (1855). [Google Scholar]
- Delaney P. M. and Harris M. R., “Fiberoptics in confocal microscopy,” in The Handbook of Biological Confocal Microscopy, Pawley J. B., Ed., pp. 515–523, Plenum Press, NY: (1995). [Google Scholar]
- Helmchen F., Fee M., Tank D., and Denk W., “A miniature head-mounted two-photon microscope: high-resolution brain imaging in freely moving animals,” Neuron 31, 903–912 (2001). 10.1016/S0896-6273(01)00421-4 [DOI] [PubMed] [Google Scholar]
- Hoy C. L., Durr N. J., Chen P., Piyawattanametha W., Ra H., Solgaard O., and Ben-Yakar A., “Miniaturized probe for femtosecond laser microsurgery and two-photon imaging,” Opt. Express 16, 9996–10005 (2008). 10.1364/OE.16.009996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetmore C., Eberhart D. E., and Curran T., “Loss of p53 but not ARF accelerates medulloblastoma in mice heterozygous for patched,” Cancer Res. 61, 513–516 (2001). [PubMed] [Google Scholar]