Molting of nematodes involves the synthesis and removal of a collagen-rich exoskeleton. We describe Caenorhabditis elegans MLT-10, which defines a large family of DUF644 and proline-rich repeat proteins. We show that MLT-10 is released from the epidermis during molting and that MLT-10 is involved in renewal of the exoskeleton and development of the epidermis.
Abstract
The molting cycle of nematodes involves the periodic synthesis and removal of a collagen-rich exoskeleton, but the underlying molecular mechanisms are not well understood. Here, we describe the mlt-10 gene of Caenorhabditis elegans, which emerged from a genetic screen for molting-defective mutants sensitized by low cholesterol. MLT-10 defines a large family of nematode-specific proteins comprised of DUF644 and tandem P-X2-L-(S/T)-P repeats. Conserved nuclear hormone receptors promote expression of the mlt-10 gene in the hypodermis whenever the exoskeleton is remade. Further, a MLT-10::mCherry fusion protein is released from the hypodermis to the surrounding matrices and fluids during molting. The fusion protein is also detected in strands near the surface of animals. Both loss-of-function and gain-of-function mutations of mlt-10 impede the removal of old cuticles. However, the substitution mutation mlt-10(mg364), which disrupts the proline-rich repeats, causes the most severe phenotype. Mutations of mlt-10 are also associated with abnormalities in the exoskeleton and improper development of the epidermis. Thus, mlt-10 encodes a secreted protein involved in three distinct but interconnected aspects of the molting cycle. We propose that the molting cycle of C. elegans involves the dynamic assembly and disassembly of MLT-10 and possibly the paralogs of MLT-10.
INTRODUCTION
The molting cycle is the hallmark of the ecdysozoan clade that encompasses more than 90% of animal species on earth (Aguinaldo et al., 1997). Molting of nematodes involves the synthesis and removal of collagen-rich extracellular matrices (ECM) and related points of cell–ECM adhesion. However, the signaling and enzymatic cascades that trigger and execute the molting process are not well understood. Dysfunction of related processes in humans contributes to the metastasis of tumors and various disorders of skin, connective tissue, and ectodermal organs. These disorders include genetic collagenopathies, certain muscular dystrophies, Marfan syndrome, and baldness (Campbell, 1995; Krause and Foitzik, 2006; Page-McCaw et al., 2007; Blank and Boskey, 2008; Ramirez and Dietz, 2009).
The exoskeleton of nematodes, also called the cuticle, is a complex, multilayered ECM secreted by underlying epidermal-like cells and syncytia (Page and Johnstone, 2007). Collagens serve as major structural components of the cuticle, along with nematode-specific proteins called cuticulins (Cox et al., 1981; Johnstone, 1994; Sapio et al., 2005). Consequently, the Caenorhabditis elegans model has been very useful for studies of collagen biosynthesis (Page and Johnstone, 2007). Less is known about the biogenesis of other cuticle components, including the glycoproteins and lipids found in the surface coat and epicuticle, respectively.
During the process of molting, a new cuticle is synthesized underneath the old one and gradually displaces the preexisting structure from the hypodermis. The outer layer of the cuticle is secreted first, and the annular furrows found there correspond to transient invaginations in the apical membrane of the hypodermis (Page and Johnstone, 2007). Particular macromolecules are depleted from the old cuticle during the process of molting, and some scavenged components may, in principle, be incorporated into the new exoskeleton. Lateral attachments that anchor the cuticle to the underlying muscle basement membrane (BM) are also remade during molting.
Larvae eventually escape (ecdyse) from the old cuticle using a stereotypical set of behaviors. This sequence includes regurgitation of the anterior half of the pharyngeal cuticle, rotation on the long axis, contractions, and forward thrusts, in that order (Singh and Soulston, 1978). C. elegans larvae molt four times, once every 8–10 h under standard culture conditions. Molting takes about 2 h, but ecdysis takes only a few minutes.
The rapid molting cycle of C. elegans requires precise temporal and spatial control over the production and destruction of ECM macromolecules. Accordingly, many genes required for the completion of molting encode proteases and antiproteases involved in the synthesis or degradation of collagens and other ECM proteins (Hashmi et al., 2002, 2004; Davis et al., 2004; Suzuki et al., 2004; Frand et al., 2005; Page et al., 2006; Partridge et al., 2008). Several genes in the general pathways for secretion and endocytosis are also required for shedding larval cuticles (Roberts et al., 2003; Frand et al., 2005; Roudier et al., 2005). However, the molecular mechanisms that coordinate the assembly and disassembly of extracellular matrices with the progression through ecdysis are not well understood.
The C. elegans nuclear hormone receptors NHR-23 and -25 are also required for the removal of larval cuticles (Kostrouchova et al., 1998; Asahina et al., 2000; Gissendanner and Sluder, 2000; Kostrouchova et al., 2001). NHR-23 and -25 are homologous to the ecdysone-responsive DHR3 and FTZ-F1 factors of Drosophila melanogaster and also the ROR/RZR/RevErb and SF-1 receptors of mammals, respectively. The requirement for NHR-23 and -25 suggests that steroid hormones regulate the molting cycle of nematodes, similar to how pulses of the steroid hormone ecdysone trigger molting and metamorphosis in insects (Thummel, 1996). NHR-23 and -25 are expressed in hypodermal cells and syncytia and regulate the expression of particular cuticle collagens and matrix modification enzymes (Kostrouchova et al., 2001; Davis et al., 2004; Chen et al., 2004; Silhankova et al., 2005). However, the targets of these receptors germane to the molting cycle have not been fully described in any system (Ruaud et al., 2010).
Large gene families whose members function synergistically were likely to evade detection in any previous RNAi-based screens for molting-defective larvae (Frand et al., 2005). We therefore conducted a forward genetic screen for mutants unable to fully shed old cuticles. Here, we describe the isolation and characterization of the C. elegans mlt-10 gene, which defines a large family of proteins involved in the episodic synthesis and removal of cuticles during postembryonic development.
MATERIALS AND METHODS
Genetic Analysis
The culture and genetic manipulation of C. elegans were performed using standard methods (Epstein and Shakes, 1995). Low-cholesterol (LC) nematode growth medium (NGM) was prepared with SeaKem GTG agarose (FMC, Rockland, ME) rather than agar and no added cholesterol. Escherichia coli OP50 were washed with M9 buffer before seeding LC plates. Bacterial-mediated RNA-interference (RNAi) was performed as described (Fraser et al., 2000), except that NGM was supplemented with 8 mM IPTG and 25 μg/ml carbenicillin. Table S1 describes the C. elegans strains used in this study.
To isolate mutants more sensitive to low cholesterol, GR1462 was mutagenized with ethylmethane sulfonate (EMS, Sigma, St. Louis, MO). About 15,000 embryos from the first filial (F1) generation were collected and cultured on NGM. Hatchlings from the second filial (F2) generation were collected and cultured on LCNGM at a density of roughly 200 animals per 6-cm plate. After 3 d, 664 larvae that had arrested at the L1 or L2 stage were individually transferred to NGM plates and 71 thereafter developed into adults and produced progeny. Descendents of those 71 animals were retested for growth on LCNGM. Monitoring expression of the dpy-7p::gfp reporter gene present in GR1462 enabled simultaneous screening for mutants defective in NHR-23 signaling.
The mlt-10(mg364) mutant was typically cultured on bacteria expressing mlt-10 double-strand RNA (dsRNA) and subsequently was fed OP50 for two generations before phenotypic analysis. To isolate intragenic suppressors, mlt-10(mg364RNAi) animals were mutagenized with EMS. Approximately 20,000 F1 animals were collected and fed OP50 on NGM. The F2 hatchlings were then collected and cultured at a density of 50 animals per 6-cm plate. Rare plates on which the F2 and F3 animals rapidly consumed all of the available food were identified. Picking a single worm from each plate led to the isolation of 44 suppressed lines. To test for linkage between a particular suppressor and mlt-10, the strain was out-crossed to KP3913 (nuIs163[myo-2p::gfp] II), and the F2 animals were observed for molting defects. We reiteratively out-crossed mlt-10(mg416mg364) animals to KP3913 to separate the two mutations in mlt-10. The mlt-10(ok2581), mlt-10(cxTi9515), and mlt-10(tm3331) mutations were out-crossed to KP3913 to generate ARF202, ARF201, and ARF204, respectively. Notably, we observed immobility and lethality in adults of the original mlt-10(ok2581) strain RB1962 but not the out-crossed strain ARF202.
The arrays mgEx701 and mgEx699 were made by microinjection of mlt-10(mg364) (5 ng/μl) or mlt-10 (10 ng/μl) DNA, respectively, along with a sur-5::gfp plasmid (50 ng/μl; Yochem et al., 1998) and pBS (40 ng/μl) into wild-type adults. To ameliorate the toxicity associated with increased dosage of mlt-10, transgenic nematodes were cultured on bacteria expressing mlt-10 dsRNA. Similar methods were used to make arrays containing mltn-4/W02B8.4 and mltn-7/Y39D8B.1. To generate aaaEx16, the mlt-10::mCherry plasmid (5 ng/μl) was microinjected into pha-1(e2123) animals along with the pha-1(+) plasmid pBX (5 ng/μl), the myo-2p::gfp plasmid pPD118.33 (10 ng/μl), and pBS (80 ng/μl). To generate aaaEx19, the gfp::mlt-10 plasmid (10 ng/μl) was microinjected into N2 animals along with pBS (80 ng/μl) and a myo-2p::rfp plasmid (10 ng/μl) provided by Cheryl Van Buskirk (California Institute of Technology).
Molecular Biology
Table S2 lists the sequence of PCR primers used in this study. To sequence mlt-10/C09E8.3, genomic DNA was amplified from worm extracts and sequenced using primers AFP6 through AFP31. To genotype tm3331, ok2581, and cxTi9515, genomic DNA was amplified with primer sets AFP15/AFP26, AFP46/AFP31, and AFP45/AFP23, respectively. Genomic DNA was amplified from RB1962, cloned into pCR-XL-TOPO (Invitrogen, Carlsbad, CA), and sequenced to determine the expanse of the ok2581 lesion. Toward the production of high-copy arrays, mlt-10 was amplified from genomic DNA using primers AFP1 and AFP3 and the Expand Long Template PCR kit (Roche, Indianapolis, IN). The mltn-4/W02B8.4 and mltn-7/Y39D8B.1 genes were amplified from genomic DNA using primer sets AFP41/AFP42 and AFP43/AFP44, respectively. Toward RNAi of mltn-10/F19H8.5, mltn-1/F32A11.7, mltn-5/W02B8.6, and mltn-7/Y39D8B.1, genomic DNA was amplified using primer sets AFP32/AFP33, AFP34/AFP35, AFP36/AFP37, and AFP38/AFP39, respectively. Each PCR product was cloned into pCR-XL-TOPO and subsequently into pPD129.36 (Fire Lab, Stanford University School of Medicine) and E. coli strain HT115(DE3).
The mlt-10p::gfp-pest fusion gene was generated as previously described (Frand et al., 2005). To construct translational fusion genes, the mlt-10 gene and regulatory sequences were amplified from N2 genomic DNA using primers AFP47 and AFP48 and Phusion High-Fidelity Polymerase (Finnzymes). The PCR product was cloned into pCR-Blunt II-TOPO (Invitrogen). The NotI site in the vector was filled using Klenow (NEB, Ipswich, MA). The resulting plasmid (pAF599) contained a single C to A transition, at the base corresponding to nucleotide 10407 of cosmid C09E8. A new NotI site was generated in pAF599 using the Phusion Site-Directed Mutagenesis kit (Finnzymes, Espoo, Finland) with primers AFP49 and AFP50. The site was inserted in-frame between the last coding codon and the stop codon of mlt-10. A NotI cassette containing the mCherry gene was cloned into the resulting vector from KP1272 (a gift from Lars Drier, University of California, Los Angeles). Separately, an NotI site was generated in pAF599 in-frame between the codons specifying Ala30 and Val31 of MLT-10. An NotI cassette containing the gfp gene was isolated from pPD114.35 (Fire Lab) and cloned into the resulting vector.
RNA was isolated from C. elegans as described (Pasquinelli et al., 2000). We used the TURBO kit (Ambion, Austin, TX) to remove any contaminating genomic DNA and the RETROscript kit (Ambion) with random primers to synthesize cDNA. The mlt-10 cDNAs were amplified using primers sets AFP14/AFP27 or AFP15/AFP30. The ama-1 cDNAs were amplified using primers ama-1-262 and ama-1-263, kindly provided by Eyleen O'Rourke (Harvard Medical School). PCR products were separated by agarose-gel electrophoresis and stained with ethidium bromide.
Microscopy and Cell Biology
Nematodes were anesthetized using sodium azide and mounted on 2% agarose pads. Images were captured using a Zeiss Axioscope (Thornwood, NY) with an attached Hamamatsu Orca ER camera (Bridgewater, NJ) and Volocity software (Improvision, Lexington, MA). For confocal work, we used a Zeiss LSM 5 PASCAL microscope and Axiovision software (Zeiss). All images were prepared for publication using Adobe Photoshop and Adobe Illustrator (San Jose, CA).
Larvae were stained with Hoechst 33258 (Sigma) as previously described (Moribe et al., 2004). To detect surface glycans, animals were incubated in M9 buffer containing 50 μg/ml WGA-Alexa Fluor 488 (Invitrogen) for 40 min with gentle agitation at room temperature. Samples were washed in M9 three times to remove unbound WGA.
RESULTS
Isolation of mlt-10 in a Forward Genetic Screen
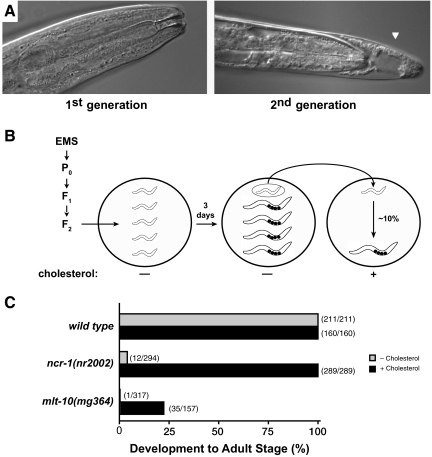
Nematodes cannot synthesize cholesterol de novo (Hieb, 1968; Chitwood, 1999). C. elegans must therefore acquire sterols from the culture medium in order to produce various steroid hormones essential for development and reproduction (Crowder et al., 2001; Merris et al., 2003; Matyash et al., 2004; Motola et al., 2006). Wild-type larvae cultured without cholesterol can rely on sterols stored by their mothers to develop to the adult stage. However, the progeny of animals cultured on low-cholesterol media arrest development as larvae, often trapped in partly shed cuticle: the molting-defective or Mlt phenotype (Figure 1A: Yochem et al., 1999). We used low cholesterol to sensitize a forward genetic screen for mutants unable to fully shed old cuticles (Figure 1B). Briefly, we isolated 17 mutants that require exogenous cholesterol, in addition to any maternal stores of cholesterol, to complete larval development. Larvae descended from 13 of the 17 mutants showed the Mlt phenotype under standard culture conditions (data not shown).
Figure 1.
Genetic screen for mutants more sensitive to low cholesterol. (A) Representative Nomarski micrographs of C. elegans grown on LCNGM for one or two generations. Left, an adult; right, a larva trapped in partly shed cuticle (arrowhead). (B) Design of the screen. (C) Growth of L1 larvae to the adult stage on LCNGM (gray) or NGM (black) after cultivation for 5 d at 20°C. Parents of the observed animals were cultured on NGM.
The mg364 allele of mlt-10 emerged from our screen. We analyzed the phenotypes associated with mg364 after out-crossing several times to remove any unlinked mutations. Only 0.3% (n = 317) of mlt-10(mg364) larvae developed to the adult stage when cultured on low-cholesterol media, compared with 100% (n = 211) of wild-type larvae (Figure 1C). As previously reported, mutations in the C. elegans ncr-1 gene also blocked development on low-cholesterol plates (Sym et al., 2000; Li et al., 2004). The ncr-1 gene is homologous to the human Niemann-Pick type C1 disease gene, which is important for the proper transport and storage of cholesterol (Smith and Levitan, 2007).
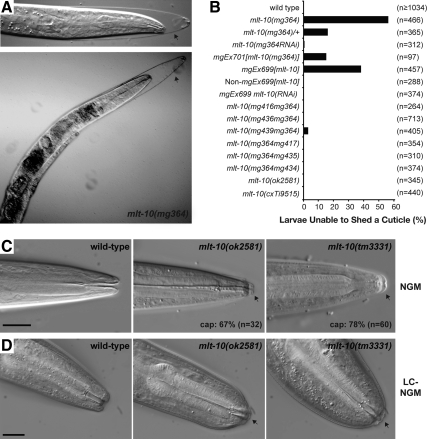
Under standard culture conditions, 56% (n = 466) of homozygous mlt-10(mg364) animals became trapped in a larval cuticle (Figure 2A). Roughly the same fraction of animals became trapped in each one of the four molts. Larvae incarcerated in a molt typically perished. An additional 22% of mg364 larvae arrested development but did not appear trapped in the cuticle upon inspection by light microscopy. The surviving mg364 animals developed slowly, with a generation time of ∼90 h at 20°C, compared with 65 h for wild-type animals. The Mlt phenotype was also observed in 16% (n = 365) of heterozygous mlt-10(mg364)/mlt-10(+) animals, indicating that mg364 is semidominant. Together, these results show that mlt-10(mg364) causes a strict blockade of the molting cycle.
Figure 2.
Mutations of mlt-10 hinder the removal of larval cuticles. (A) Representative Nomarski micrographs of mlt-10(mg364) mutants. Top, a midstage larva; bottom, an adult encased in the L4-stage cuticle (arrow). (B) Animals of each genotype were collected as embryos, cultured on NGM for 3–5 d at 20°C, and inspected using a dissecting microscope. Graph shows the percent of animals trapped in a larval cuticle. Not all genotypes were tested concurrently. (C and D) Representative Nomarski micrographs show animals of the indicated genotypes. (C) Animals were collected as embryos, cultured on NGM for 45 h at 20°C, and inspected using a compound microscope. Arrows indicate the buccal caps. (D) Hatchlings were collected and cultured on LCNGM for 3 d at 20°C. Arrows indicate unshed cuticles from the L4 larval stage. Scale bars, 10 μm.
The mlt-10 Gene Corresponds to C09E8.3
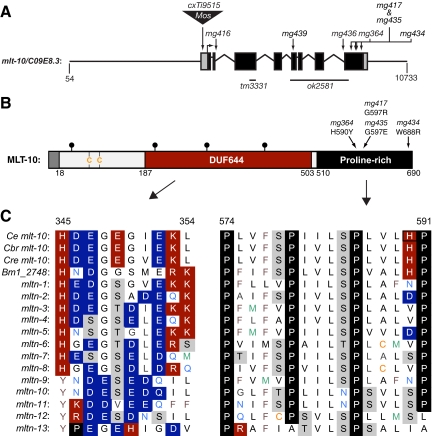
To identify the mlt-10 gene, we mapped the mg364 mutation to a 3.4 map unit region on the left end of chromosome II by standard methods (Wicks et al., 2001). Because the mg364 mutation was semidominant, we reasoned that inactivation of mlt-10 might restore the ability of mg364 mutants to shed old cuticles. We therefore systematically and individually inactivated the annotated genes in the map interval by bacterial-mediated RNAi (Timmons et al., 2001; Kamath et al., 2003). After the inactivation of C09E8.3, the vast majority of mlt-10(mg364) larvae developed into healthy adults (Figure 2B). Sequencing C09E8.3 identified a C-to-T transition in mg364 mutants that specifies the substitution H590Y in the predicted MLT-10 protein. In addition, an extrachromosomal array containing C09E8.3 amplified from mg364 mutants (mgEx701, Figure S1) rendered wild-type larvae unable to shed old cuticles. To confirm the identity of the gene, we isolated and characterized six additional alleles of mlt-10 by screening for intragenic suppressors of mg364. The new alleles included four distinct missense mutations and two splice-site mutations in C09E8.3 (Table S3). Together, these results show that mlt-10 corresponds to C09E8.3 and that mg364 is a gain-of-function (GOF) allele.
Figure 3A shows the structure of the mlt-10 gene, as confirmed by the sequence of 76 distinct cDNAs curated by Wormbase (www.wormbase.org). In addition to the point mutations described above, we obtained a variety of mlt-10 alleles from public resources. These alleles included cxTi9515, tm3331, and ok2581, reagents kindly provided by Laurent Segalat (University of Lyon), the C. elegans Gene Knockout Consortium, and the National Bioresource Project of Japan, respectively (Table S3). The cxTi9515 allele is an insertion of the Mos1 transposon in the 5′ UTR of mlt-10 (Bessereau et al., 2001; Martin et al., 2002). The mutation severely decreased the level of mlt-10 expression, as indicated by RT-PCR (Figure S2). The mlt-10(tm3331) insertion/deletion causes a frame-shift in exon 3 that introduces an early stop codon. The ok2581 insertion/deletion encompasses exons 5 through 7 and part of exon 8. Transcripts of mlt-10 were shorter and less abundant in ok2581 mutants than wild-type animals (Figure S2). Sequencing those cDNAs indicated the use of an atypical splice acceptor site for exon 8 and the related introduction of a premature stop codon. As we shall describe, we analyzed the phenotypes associated with these loss-of-function (LOF) alleles of mlt-10 after out-crossing the corresponding strains several times to remove any unlinked mutations.
Figure 3.
MLT-10 defines a large family of nematode-specific proteins. (A) Diagram of the C. elegans mlt-10 gene and flanking sequences. Black boxes represent exons, and gray boxes represent untranslated sequences. Nucleotide positions correspond to cosmid C09E8 (Accession no. gb AF077529). Table S3 further describes these mutations of mlt-10. (B) Diagram of the predicted MLT-10 protein (Accession no. gi 17531703) showing the signal sequence (gray), potential acceptor sites for N-linked glycans (●), conserved cysteine residues; DUF644 (red), and the proline-rich region (black). (C) Sequence alignment of the predicted paralogs and selected orthologues of MLT-10. Amino acid positions correspond to Ce MLT-10. Acidic and basic residues are shaded blue and red, respectively. Prolines are shaded black and hydroxy amino acids are shaded gray. The H590 residue affected by mg364 is boxed. Accession numbers for these sequences are Ce, ref NM_061354; Cbr, gi 187040316; Cre, gi 183180662; and Bm1_2748, gi 170584318. Table S4 provides additional information about the mltn genes of C. elegans.
MLT-10 Defines a Family of DUF644 and Proline-rich Repeat Proteins
MLT-10 is the first characterized member of a large family of annotated proteins comprised of the domain of unknown function (DUF) 644 and distinctive tandem repeats rich in prolines and hydroxy amino acids (Figure 3, B and C, Figures S3 and S4). DUF644 contains many lysine residues and other basic or acidic amino acids. The repeats found at the C-terminus of MLT-10 have the consensus sequence P-X2-L-(S/T)-P, where X is any hydrophobic residue except glycine. MLT-10 also contains an annotated secretory signal sequence and four putative acceptor sites for N-linked glycans, features characteristic of secreted proteins.
In principle, the biosynthesis of MLT-10 and the paralogs of MLT-10 may involve several posttranslational modifications, including but not limited to 1) disulfide bond formation, 2) the addition of O-linked glycans, 3) cleavage of dibasic sites in the nonrepetitive region by subtilisin/Kex2-like proteases, 4) cross-linking of glutamine and lysine residues by transglutaminase, and 5) the hydroxylation of some proline residues in the repetitive region. These particular modifications occur during the biosynthesis of collagens and other ECM proteins of nematodes (Fetterer and Rhoads, 1990; Lustigman et al., 1995; Thacker and Rose, 2000; Edens et al., 2001; Page et al., 2006).
We identified 13 paralogs of mlt-10 in the fully sequenced genome of C. elegans and named those genes mltn-1 through mltn-13 for (mlt-ten-related; Figure 3C and Table S4). Previous high-throughput analysis indicated that most if not all of the C. elegans mltn genes are expressed (Wormbase). The MLT-10 paralogs include mltn-1/F32A11.7, mltn-2/Y52B11A.7, mltn-11/W06G6.7, mltn-12/C53B4.8, and mltn-13/F15A8.7, as well as three genes on cosmid W02B8 (II:13,916,260–13,930,421), two genes on cosmid F19H8 (II:14,616,033–14,626,346), and three genes on cosmid Y39D8B (V:364,951–379,673). Notably, several of the gene models described in Table S4 include exons not depicted in the current release of Wormbase (WS204). In particular, the gene annotated as Y39D8B.1 in WS204 represents an in silico fusion of the predicted paralogs mltn-6 and mltn-7, formerly annotated as Y39D8B.2 and Y39D8B.1, respectively, in WS100. The fused gene depicted in WS204 specifies an atypical protein containing two copies of DUF644 but only one set of proline-rich repeats. The existence of three clusters of paralogs in the genome suggests that multiple gene duplication events contributed to evolution of the mlt-10 family. Many homologues of MLT-10 were also identified in the genomes of other species of nematodes and the annotated proteomes of parasitic nematodes including Onchocerca volvulus and Brugia malayi, which cause River Blindness and lymphatic filariasis in humans, respectively (Figure 3C and S4).
Figure S4 displays an alignment of the P-X2-L-(S/T)-P repeats of the predicted paralogs and selected homologues of MLT-10. Most of these proteins contain about 30 repeats interspersed with only a few charged residues, with the exception of MLTN-13/F15A8.7. Repeats similar to those of MLT-10 were found in several annotated but uncharacterized and otherwise unrelated proteins of eukaryotes, suggesting a widespread and ancient function for the [P-X2-L-(S/T)-P]n sequence. We identified such proteins in the translated genomes of Xenopus laevis (gi 47125144), Tetraodon nigroviridis (gi 47221831), Homo sapiens (ref NW_001837930: 1954604–1955196), and Candida tropicalis (gi 255731458). A similar P-X4-P sequence also occurs in the Bordetella pertussis virulence factor Pertactin (gi 6730300; Emsley et al., 1996). Proteins with DUF644 were not readily identified by standard TblastN searches (Altschul et al., 1990) of the fully sequenced genomes of D. melanogaster, H. sapiens, or Saccharomyces cerevisiae. Thus, MLT-10 defines a large family of proteins that is well conserved only in nematodes.
Either the Loss or the Gain of mlt-10 Function Hinders the Removal of Larval Cuticles
We used genetic analysis to investigate the role of the mlt-10 family in development and better define the nature of the mlt-10(mg364) allele. A variety of mlt-10(lof) mutants developed from the L1 to the adult stage more slowly than wild-type animals, but did not display a terminal Mlt phenotype (Figure 2B). We therefore closely examined larvae partially synchronized around the time of the L2-to-L3 molt, using a compound microscope to detect buccal caps. The caps are comprised of old cuticle and temporarily seal the buccal cavity during the process of molting. Caps were observed on the majority of mlt-10(tm3331) and mlt-10(ok2581) mutants (Figure 2C), compared with 20% (n = 76) of wild-type larvae. When cultured on low-cholesterol media, 70% (n = 13) of mlt-10(tm3331) mutants and 92% (n = 12) of mlt-10(ok2581) mutants failed to shed the fourth larval cuticle, compared with 29% (n = 14) of wild-type animals (Figure 2D). Together, these results suggest that mlt-10 is needed for the efficient removal of larval cuticles, even though mlt-10 is not essential for development under standard culture conditions.
To further evaluate the function of the mlt-10 family, we used RNAi to systematically and individually knock down the 13 annotated paralogs of mlt-10 in wild-type larvae; mlt-10(lof) mutants; and rrf-3 mutants, which are more sensitive to dsRNA (Simmer et al., 2002). None of these animals became terminally trapped in cuticle (Table S5). Also, the Mlt phenotype was not associated with gk766, a deletion in the mltn-13/F15A8.7 gene. Thus, the paralogs of mlt-10 appear individually dispensable for the removal of larval cuticles, possibly due to functional redundancy among the large gene family. A thorough investigation of any such redundancy awaits the availability of null alleles in all thirteen paralogs of mlt-10.
As a complementary approach to investigate the mlt-10 family, we used high-copy arrays to increase the dosage of mlt-10 and selected paralogs of mlt-10. The array mgEx699[mlt-10] rendered 38% (n = 457) of larvae unable to shed old cuticles, and RNAi of mlt-10 suppressed that phenotype (Figure 2B). As expected, wild-type mlt-10 appeared less toxic than mlt-10(mg364) after the microinjection of equivalent amounts of DNA (data not shown). High-copy arrays that contained mltn-4 and mltn-7 also conferred the Mlt phenotype. High-copy arrays that contained mltn-3 and mltn-11 caused embryonic and larval lethality prohibitive to the propagation of transgenic nematodes (data not shown). Thus, increased expression of mlt-10, mltn-4, or mltn-7 can prevent the removal of larval cuticles. Activities of the other mltn genes will be tested in future work.
Our screen for intragenic suppressors of mlt-10(mg364) identified additional mutations that affect the proline-rich repeats of MLT-10. As previously stated, mg364 specifies the substitution H590Y. H590 is conserved among the annotated orthologues of MLT-10 and is one of only five charged residues present in the repetitive region (Figure S4). The intragenic suppressors mg417, mg434, and mg435 specified the substitutions G597R, W688R, and G597E, respectively. None of these missense mutations blocked expression of the mlt-10(mg364) gene (Figure S2). However, each of the corresponding substitutions introduced a charged residue into the repetitive region. The mechanism of suppression may therefore involve the rectification of MLT-10(H590Y) or destabilization of the otherwise toxic protein. The mg416 and mg436 mutations affected splice sites of mlt-10 and reduced expression of the gene (Figure S2). The sequence of mlt-10(mg416) cDNAs indicated correct splicing. In contrast, the sequence of mlt-10(mg436mg364) cDNAs indicated the use of an atypical splice acceptor site for exon 8 and the related introduction of a premature stop codon upstream of the proline-rich repeats (Table S3). In this case, diminished expression of the repetitive region likely accounts for the suppression of mg364. Taken together, these findings indicate that dysfunction of the repetitive region of MLT-10 blocks the removal of larval cuticles. Moreover, the data suggest that electrostatic interactions involving the P-X2-L-(S/T)-P repeats influence the utility of MLT-10. As we shall discuss, we hypothesize that MLT-10(H590Y) interferes with the function of multiple members of the MLT-10 family, as does increased expression of wild-type MLT-10.
The mlt-10 Gene Is Expressed in the Hypodermis during Larval Development
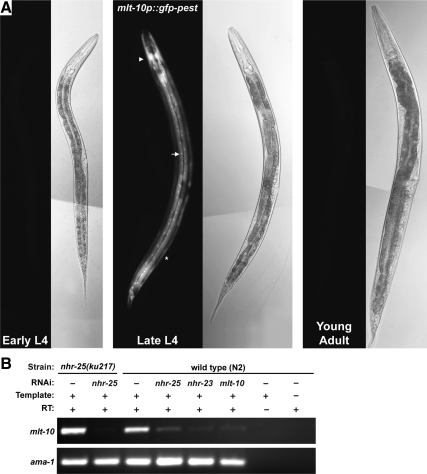
To determine the spatial and temporal expression pattern of mlt-10, we fused the promoter of mlt-10 to the gfp-pest gene, which encodes a variant of green fluorescent protein (GFP) that is rapidly degraded in vivo (Li et al., 1998; Frand et al., 2005). The mlt-10p::gfp-pest fusion gene was expressed in the major body hypodermal syncytium (Hyp7), the dorsal and ventral ridges of the hypodermis, hypodermal cells in the head and tail, and the pharyngeal myoepithelium, but not the lateral seam cells (Figure 4A). In L4-stage animals cultured at 25°C, GFP was first detected in the anterior hypodermis ∼3.5 h before ecdysis. The fluorescence spread throughout Hyp7 and intensified for about 3 h. The fluorescence dissipated at the end of lethargus and was barely detectable 1 h after ecdysis (Figure 4A). A similar pulse of expression of GFP accompanied all four molts and faithfully recapitulated expression of the endogenous mlt-10 gene, as previously described (Frand et al., 2005). GFP was also expressed in epidermal cells of pretzel-stage embryos, which synthesize the cuticle for the first larval stage (data not shown). We conclude that most hypodermal cells and syncytia express mlt-10 whenever a new exoskeleton is made.
Figure 4.
Spatial and temporal expression pattern of mlt-10. (A) Representative fluorescence and Nomarski micrographs show expression of the mlt-10p::gfp-pest fusion gene. GFP was detected in the major body hypodermal syncytium (arrow), the dorsal and ventral ridges of the hypodermis (asterisk), and the anterior hypodermis (arrowhead) of the late L4 stage larva. All fluorescence images were acquired with an exposure time of 187 ms. (B) Detection of mlt-10 transcripts by RT-PCR. Larvae were cultured on bacteria that expressed the indicated dsRNAs for 40 h at 25°C. Animals were harvested at the typical time of the L4-to-adult molt. Detection of ama-1 transcripts controlled for the quality of RNA samples and RT-PCR reactions.
We previously reported that NHR-25 and -23 drive supernumerary bouts of expression of mlt-10 in let-7 family mutants that continue molting after the fourth larval stage (Hayes et al., 2006). Here, we asked if nhr-25 and -23 also regulate expression of mlt-10 during larval development. We used RNAi to knockdown nhr-25 or -23 in mgIs49[mlt-10p::gfp-pest] larvae. The hypomorphic allele nhr-25(ku217) was used to sensitize animals to RNAi of nhr-25 (Chen et al., 2004). Sets of larvae were repeatedly monitored for expression of GFP over an 8-h time period encompassing the L4-to-adult molt. The mid-L4-stage larvae selected for this experiment were active at the start, but became lethargic and attempted to shed the L4-stage cuticle by the end. Only 40% (n = 49) of nhr-25(ku217RNAi) animals expressed the GFP reporter during this time, compared with 100% (n = 22) of control animals. Moreover, the fluorescence associated with GFP was dim and ephemeral in nhr-25(ku217RNAi) mutants, compared with wild-type animals. None (n = 24) of the nhr-23(RNAi) animals expressed any detectable GFP, as previously reported (Frand et al., 2005). RNAi of nhr-25 or -23 also greatly reduced the abundance of mlt-10 messages in late L4 larvae, as indicated by RT-PCR (Figure 4B). Thus, NHR-25 and -23 promote expression of mlt-10 during the larval molting cycle.
To detect the MLT-10 protein in vivo, we constructed two distinct, full-length translational fusion genes between mlt-10 and fluorescent reporters (Figure S1). The mCherry tag was inserted at the C-terminus of MLT-10. GFP was inserted downstream of the predicted signal sequence. The corresponding arrays conferred many of the same phenotypes as mgEx699[mlt-10], confirming expression of the MLT-10 fusion proteins.
During the process of molting, the MLT-10::mCherry and GFP::MLT-10 fusion proteins were detected in vesicle-like objects near the apical surface of the epidermis (Figure 5, A and B). Cherry was also readily detected in other structures, whereas GFP was not, possibly due to proteolytic processing of the N-terminal fusion protein.
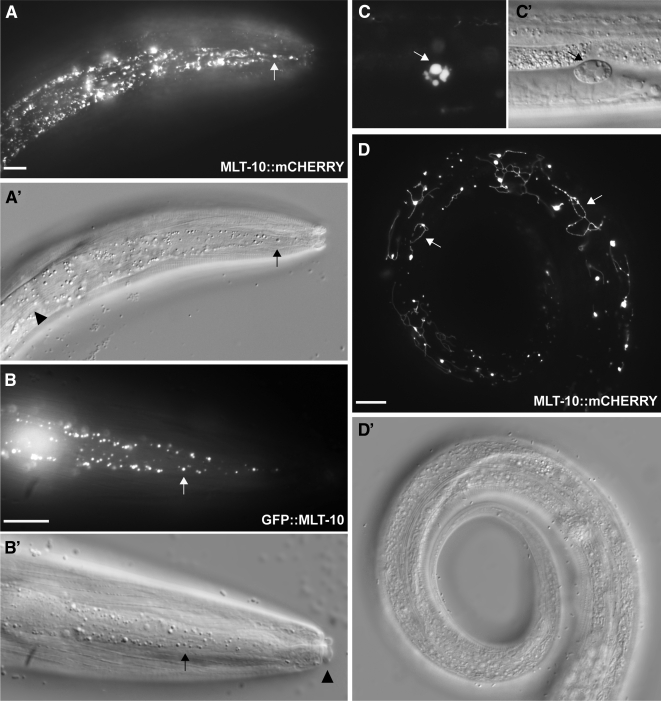
Figure 5.
Localization of the MLT-10::mCherry and GFP::MLT-10 fusion proteins. (A–D) Representative fluorescence and Nomarski micrographs show expression of the fusion proteins. (A) MLT-10::mCherry detected in vesicle-like objects (arrow) in the lateral epidermis of a late L4 larva molting to the adult stage. Alae are visible on the underlying cuticle (arrowhead). (B) GFP::MLT-10 detected in vesicles (arrow) in the lateral epidermis of a molting larva. A double cuticle covers the mouth (arrowhead). (C) Cherry detected in a coelomocyte (arrow) of a young adult. (D) MLT-10::mCherry found in strands (arrows) near the surface of a young adult. Scale bars, 10 μm.
In young adults, the MLT-10::mCherry fusion protein was also detected in coelomocytes, distinctive scavenger cells that endocytose material from the pseudocoelom (Grant and Sato, 2006). Cherry was detected in the coelomocytes of ∼22% (n = 176) of aaaEx16[mlt-10::mCherry] animals. Fusion proteins secreted from the apical surface of the hypodermis may have mixed with coelomic fluids in this particular background, because the mlt-10::mCherry array was associated with gaps in the syncytial epidermis, as we shall describe. Alternatively, some MLT-10::mCherry may have been released from the basolateral surface of Hyp7 or the pharyngeal myoepithelium. In either case, uptake by coelomocytes verified that MLT-10::mCherry was secreted, as coelomocytes do not express the mlt-10 gene. Collectively, these observations suggest that MLT-10 is released from the hypodermis to the surrounding matrices and fluids during the process of molting.
The MLT-10::mCherry fusion protein was also detected in strands and loops near the surface of transgenic animals (Figure 5D). These structures were observed in 27% (n = 123) of mixed-stage larvae, but were most prominent in animals completing the fourth molt. The strands ranged from 5 to 30 μm in length and appeared to be positioned above the hypodermis. The formal possibility that these structures result from the nonspecific aggregation of mCherry cannot be eliminated at this time. Nonetheless, these observations are consistent with the model that MLT-10 assembles into oligomeric complexes in vivo. A complete description of any such complexes awaits the availability of anti-MLT-10 antibodies.
Either the Loss or the Gain of mlt-10 Function Impinges on the Exoskeleton
The molting cycle involves both the synthesis and the removal of cuticles, and these processes are likely interconnected. We therefore examined several aspects of the exoskeleton to more fully define the phenotypes associated with particular mutations in mlt-10. To determine the effect of MLT-10(H590Y) in the absence of unshed cuticles, we examined mlt-10(mg416mg364) double mutants, rather than mlt-10(mg364) single mutants, in many experiments.
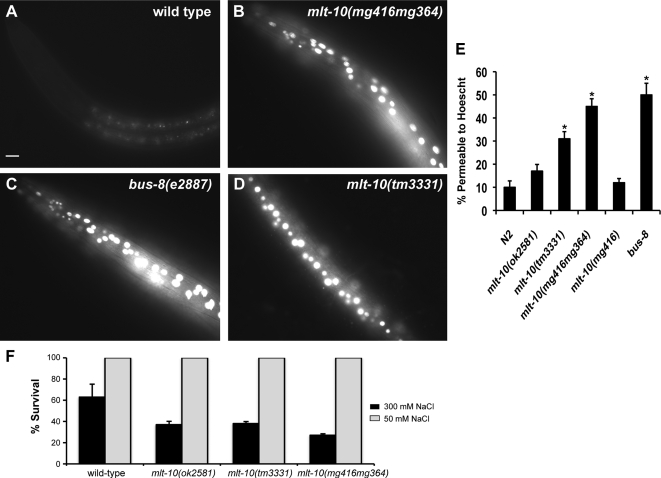
To evaluate the barrier function of the exoskeleton, we treated larvae with the DNA-binding dye Hoechst 33258, as previously described (Moribe et al., 2004). The mlt-10(tm3331), mlt-10(ok28581), mlt-10(mg364), and mlt-10(mg416mg364) mutants were all more permeable to Hoechst than wild-type larvae (Figure 6). In this regard, mlt-10(mg364) larvae resembled bus-8(e2887) mutants, which have more porous cuticles because of a defect in the glycosylation of secreted proteins (Partridge et al., 2008). The increased permeability of mlt-10(mg416mg364) mutants to Hoechst was directly attributable to expression of the MLT-10(H590Y) protein, because mlt-10(mg416) single mutants were not permeable to the dye. High-copy mlt-10(+) and mlt-10::mCherry arrays were also associated with increased permeability of the cuticle (data not shown). As a complementary approach, we tested the ability of animals to survive chronic osmotic stress. The survival rate of various mlt-10 mutants was significantly lower than that of wild-type animals on media containing 300 mM NaCl (Figure 6F). Thus, mutations of mlt-10 decrease the utility of the exoskeleton as a barrier to the environment.
Figure 6.
Increased permeability of the cuticle in mlt-10 mutants. (A–D) Representative fluorescence micrographs of larvae stained with Hoechst 33258. All images were acquired with an exposure time of 50 ms. Scale bar, 10 μm. (E) The fraction of larvae with nuclei stained by Hoechst 33258. Values represent the average of two independent experiments; error bars, SEM. Asterisks indicate a significant difference from wild-type animals (p ≤ 0.05). (F) Early L1 stage larvae of the indicated genotypes were collected and cultured on high-salt or standard NGM plates for 3 d at 20°C. Values represent the average of two independent experiments; error bars, SEM.
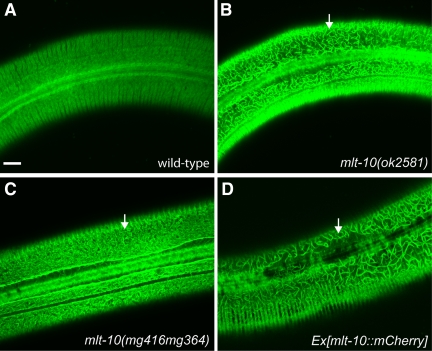
To detect a cuticle collagen in vivo, we used a translational fusion gene between col-19 and gfp, kindly provided by Anthony Page (University of Glasgow). In wild-type animals, COL-19::GFP is incorporated into the circumferential annuli and longitudinal alae of the adult exoskeleton (Thein et al., 2003). In contrast, COL-19::GFP macromolecules were disorganized in many mlt-10(tm3331) and mlt-10(ok2581) animals (Figure 7). Expression of MLT-10(H590Y) or MLT-10::mCherry also interfered with the assembly of COL-19::GFP, particularly above the lateral hypodermis (Figure S5). The severity of disorganization of COL-19::GFP varied among isogenic mlt-10 mutants, perhaps because of compensatory changes in the expression of other MLTN proteins. We conclude that both LOF and GOF mutations of mlt-10 affect the patterning of COL-19::GFP and possibly other cuticle collagens.
Figure 7.
Disorganization of a cuticle collagen in mlt-10 mutants. (A–D) Representative confocal fluorescence micrographs show COL-19::GFP in the adult exoskeleton. Arrows point to disorganized assemblies of COL-19::GFP flanking the longitudinal alae. Scale bar, 10 μm.
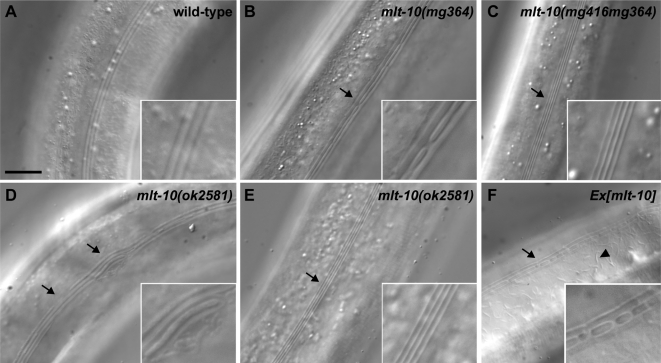
We also examined the morphology of the longitudinal alae on the adult exoskeleton. The alae of wild-type adults were continuous, straight, and comprised of three ridges (n = 28). In contrast, in many mlt-10(ok2581) and mlt-10(tm3331) mutants, segments of the alae were broken, branched, or comprised of four ridges (Figure 8). The alae were malformed in 24% (n = 80) of mlt-10(lof) mutants. The alae were similarly malformed in 72% (n = 22) of mlt-10(mg364) animals and the majority of mgEx699[mlt-10] adults. Atypical sausage-shaped structures were observed on the surface of some mlt-10(gof) mutants (Figure 8F). Thus, both LOF and GOF mutations of mlt-10 affect the morphology of the adult exoskeleton, which is synthesized during the final molt. Taken together, these observations indicate that mlt-10 is involved in the synthesis of new cuticles, as well as the removal of old ones.
Figure 8.
Malformation of the adult-specific alae in mlt-10 mutants. (A–F) Representative Nomarski micrographs show the adult exoskeleton. Arrow, abnormalities in the alae including gaps, branches, and regions with four ridges. Arrowhead, an atypical structure in the cuticle. Each region of interest was digitally magnified 2.5-fold for display in the inset. Scale bar, 10 μm.
Mutations of mlt-10 Affect Development of the Epidermis
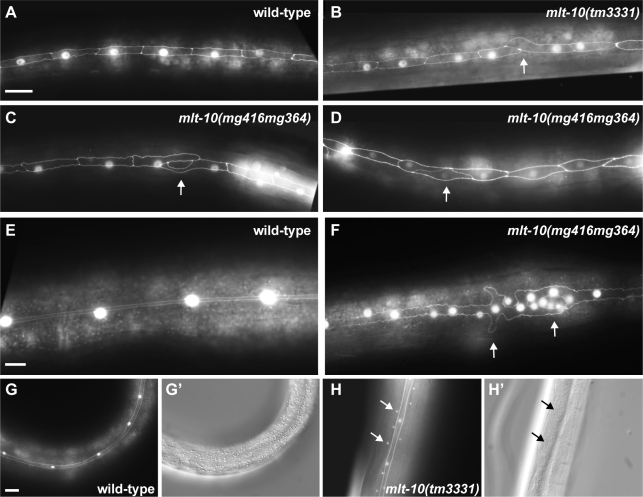
Molting requires the coordinated activity of epidermal cells and syncytia spread across the body. We therefore examined the status of the epidermis in mlt-10 mutants, focusing on the stem cell-like lateral seam cells, which produce the longitudinal alae. The seam cells divide asymmetrically early in the L1 stage (Soulston and Horvitz, 1977). Afterward, the anterior daughters fuse with the hypodermis, whereas the posterior daughters elongate and reconnect with one another. In this report, adherens junctions at the seam cell margins were detected using the AJM-1::GFP fusion protein and the seam cell nuclei were detected using a scm::gfp transcriptional fusion gene, as previously described (Hope, 1991; Mohler et al., 1998). As expected, bilateral rows of rectangular cells were observed in wild-type larvae late in the L1 stage (Figure 9A; Podbilewicz and White, 1994). In contrast, particular seam cells were misshapen and overlapped their sisters in mlt-10(tm3331) and mlt-10(ok2581) mutants (Figure 9B). Abnormal cells were observed in 6% (n = 278) of mlt-10(tm3331) mutants, but were not observed in any of 150 wild-type larvae. Oddly shaped seam cells were also present in 8.5% (n = 200) of mlt-10(mg416mg364) larvae at this stage of development (Figure 9, C and D).
Figure 9.
Abnormal development of the epidermis in mlt-10 mutants. (A–F) Fluorescence micrographs show expression of AJM-1::GFP and the nuclear scm::gfp reporter in the lateral hypodermis. The anterior end of each worm is to the right, and the ventral side is down. All images were acquired with an exposure time of 500 ms. Scale bars, 10 μm. (A–D) Hatchlings were cultured on NGM for 16 h at 25°C before imaging. Arrows, irregularly shaped seam cells in the late L1 larvae. (E and F) Larvae were imaged late in the L4 stage. Arrows, an irregularly shaped region of the syncytial seam with extra nuclei. (G and H) Representative fluorescence and Nomarski micrographs show adults. Arrows, examples of GFP detected outside of the syncytial seam. Ectopic expression of GFP was observed in 80% (n = 54) of mlt-10(tm3331) mutants, 73% (n = 44) of mlt-10(ok2581) animals, and 81% (n = 114) of mlt-10(mg416m364) adults, but was not observed in wild-type animals (n = 36).
In wild-type animals, the seam cells fuse with their sisters late in the L4 stage and thereafter cease to divide (Soulston and Horvitz, 1977). The resulting bilateral syncytia were malformed and contained extra nuclei in some mlt-10(mg416mg364) mutants, a phenotype suggestive of seam cell hyperplasia (Figure 9F). Moreover, the syncytial seam of many Ex[mlt-10::mCherry] and mlt-10(mg364) adults contained gaps lacking detectable AJM-1 at the cell margins (Figure S6 and data not shown). In addition, GFP was detected outside of the syncytial seam in the majority of mlt-10(lof) and mlt-10(416mg364) mutants expressing the scm::gfp and ajm-1::gfp fusion genes (Figure 9H). Preliminary studies using Nomarski microscopy and DAPI staining identified some sites of ectopic GFP expression as hypodermal nucleoli. Collectively, these findings show that mutations of mlt-10 affect several aspects of epidermal development, including dynamic changes in cell shape. Aberrant development of the seam cells likely contributes to the malformation of alae in mlt-10 mutants and possibly to defects in the molting cycle.
We obtained additional information about the status of the seam cells in mlt-10 mutants by staining larvae with the lectin wheat germ agglutinin (WGA) conjugated to the fluorescent dye Alexa Fluor 488 (Figure S7). Similar probes are regularly used to detect the surface glycans of C. elegans (Politz et al., 1990), and mucin-type glycans are major ligands of WGA in this system (Natsuka et al., 2005). WGA bound to the margins of the seam cells in mlt-10(mg364) mutants but not in wild-type larvae (Figure S7), suggesting the unnatural exposure or accretion of particular glycans. Those glycans may influence the adhesiveness and shape of the seam cells. Widespread ligands of WGA were also detected in the shed cuticles (molts) of mlt-10 mutants and wild-type larvae. Moreover, ligands of WGA were concentrated at the anterior end of partly shed cuticles on mlt-10(lof) and mlt-10(mg364) mutants (Figure S7). Such mucus has not yet been observed on wild-type larvae, but could, in theory, provide natural lubrication at the moment of ecdysis.
Additional Phenotypes of mlt-10 Mutants
A variety of other phenotypes were associated with the mlt-10(mg364) mutation, including the presence of large, seemingly fluid-filled spaces and vacuoles in the body; aberrant shape of the body; uncoordinated movement; and improper development of the gonad and vulva. Some of these phenotypes may be attributable to abnormalities in the exoskeleton and epidermis. Notably, mlt-10(mg364) adults produced only 6 ± 10 (n = 18) progeny, whereas wild-type animals produced 262 ± 26 (n = 9) offspring. The cause of this sterility is not yet understood.
DISCUSSION
Here, we describe the isolation and characterization of the mlt-10 gene of C. elegans. MLT-10 is the first reported member of a large family of nematode-specific proteins characterized by DUF644 and tandem P-X2-L-(S/T)-P repeats. The mg364 allele of mlt-10 emerged from a genetic screen on low cholesterol. The mutation specifies the substitution H590Y in the repetitive region of MLT-10 and is thought to interfere with the function of multiple members of the MLT-10 family. Nine additional alleles of mlt-10, including two intragenic deletions and four substitution mutations, as well as high-copy mlt-10 arrays, were used to analyze the role of mlt-10 in larval development.
Our findings suggest that MLT-10 is a secreted protein involved in the removal of old cuticles as well as the synthesis of new cuticles. In review, either the loss or the gain of mlt-10 function impedes the shedding of old cuticles. Both LOF and GOF mutations of mlt-10 are also associated with abnormalities in the larval and adult exoskeletons. The synthesis and removal of cuticles are almost certainly interconnected in nematodes, as newly synthesized cuticles displace the preexisting ones during the process of molting. Consistent with that view, deformities in the ultrastructure of the underlying cuticle have been observed in particular molting-defective mutants (Hao et al., 2006). Moreover, certain enzymes involved in the biosynthesis of collagens and other ECM proteins are needed for the removal of larval cuticles, including the glycosyltransferase BUS-8, the peroxidases BLI-3 and MLT-7, and the protease BLI-5 (Edens et al., 2001; Davis et al., 2004; Frand et al., 2005; Page et al., 2006; Partridge et al., 2008; Thein et al., 2009; Stepek et al., 2010). In theory, progress through ecdysis might depend on physiological feedback on the status of the new exoskeleton. Consistent with that idea, animals forced to ecdyse before completion of a new cuticle perish, probably due to osmotic shock (Ruaud and Bessereau, 2006).
Mutations of mlt-10 also affect the development of epidermal cells and syncytia that synthesize the exoskeleton. Notably, several transcription factors required for proper development of the seam cells are also necessary to remove larval cuticles, including NHR-25 and the GATA factors ELT-5 and -6 (Asahina et al., 2000; Gissendanner and Sluder, 2000; Koh and Rothman, 2001; Chen et al., 2004; Silhankova et al., 2005). We hypothesize that some abnormalities in seam cell development observed in nhr-25 mutants relate to the deregulation of mlt-10. Further, we speculate that MLT-10 directly or indirectly affects cell–ECM interactions important for epidermal development.
Using a transcriptional fusion gene, we show that mlt-10 is transiently expressed in hypodermal cells and syncytia whenever the exoskeleton is remade. The conserved nuclear hormone receptors NHR-23 and -25 are required for the periodic expression of mlt-10. Consequently, the amount of mlt-10 expressed at any time in development probably relates to the abundance of steroid hormones that bind these receptors. Transcriptional control by NHR-23 and -25 may coordinate the expression of MLT-10 with the production of particular collagens, matrix modification enzymes, and signaling molecules involved in the molting cycle.
Two full-length, translational fusion proteins were used to evaluate the distribution of MLT-10. Both the MLT-10::mCherry and the GFP::MLT-10 fusion proteins were detected in putative vesicles in the lateral hypodermis during molting. The MLT-10::mCherry fusion protein was also detected in strands near the surface of animals. After completion of the molting cycle, Cherry was detected in coelomocytes. Collectively, these findings suggest that MLT-10 is secreted from the hypodermis to the surrounding matrices and fluids during the process of molting.
Proline-rich repeats are found in several well-characterized extracellular proteins, including collagen, elastin, and flagelliform silks (Bhattacharjee and Bansal, 2005; He et al., 2007; Matsushima et al., 2008; Savage and Gosline, 2008; Wise and Weiss, 2009). After comparing MLT-10 with these proteins, we predict that three main factors contribute to the secondary structure of the C-terminal region of MLT-10: 1) steric repulsion among proline residues, 2) attraction among hydrophobic residues, and 3) hydrogen bonding among hydroxy amino acids. One possibility is that the P-X2-L-(S/T)-P repeats of MLT-10 promote the assembly of oligomeric complexes, perhaps including other MLTN proteins. The structure of any such complexes would likely be distinct from collagen, because the P-X2-L-(S/T)-P repeats contain very few glycine residues. An alternative possibility is that the C-terminal region of MLT-10 lacks regular secondary structure and mostly provides extension or flexibility. In either case, we expect the tertiary structure of MLT-10 to allow segregation of the hydrophobic P-X2-L-(S/T)-P repeats from the hydrophilic DUF644.
We propose that the molting cycle involves the dynamic assembly and disassembly of oligomeric MLT-10 complexes. In principle, the MLT-10(H590Y) substitution mutation might interfere with intermolecular interactions among multiple members of the MLTN family, by altering the electrochemical properties of the repetitive region or forming inappropriate di-tyrosine cross-links. Interference with multiple paralogs could account for the severe phenotypes caused by the mlt-10(mg364) mutation. Excessive production of MLT-10 might also disrupt interactions between MLT-10 and the MLTN proteins. Notably, many disease-associated substitution mutations in human collagens interfere with the assembly of collagen fibrils (Blank and Boskey, 2008). Studies of these particular dominant-negative mutations in collagens have greatly enriched our understanding of ECM remodeling in human development and disease.
Our working model is that MLT-10 serves as an instructive or structural component of the cuticle and thereby influences the assembly or disassembly of collagens. Alternatively, MLT-10 might promote the trafficking of collagens through the secretory pathway of the hypodermis. A similar function has been proposed for the membrane-spanning proteins CUTI-1 and TSP-15, which are required for shedding larval cuticles (Moribe et al., 2004; Fritz and Behm, 2009). A third possibility is that MLT-10 and the paralogs of MLT-10 serve as monomeric lubricants that help dislodge old cuticles from the hypodermis. As lubricants, the MLTN proteins might also facilitate particular behaviors used to escape old cuticles, including longitudinal rotation. Further research is needed to fully define the function of MLT-10.
The increased dependence of mlt-10 mutants on exogenous cholesterol suggests a reduced capacity to acquire or utilize sterols. We therefore speculate that mutations of mlt-10 directly or indirectly reduce the function of other proteins linked to sterols and essential for shedding cuticles. Those proteins include the hedgehog-like protein QUA-1, several homologues of Patched, and the low-density lipoprotein receptor-like protein LRP-1 (Yochem et al., 1999; Zugasti et al., 2005; Hao et al., 2006). LRP-1 is expressed on the apical surface of the hypodermis (Yochem et al., 1999), which may represent an important site of sterol uptake in nematodes (Fleming and Fetterer, 1984).
Our ongoing investigation of the MLT-10 family may directly benefit the development of new drugs for filarial diseases currently affecting over 140 million people living primarily in tropical regions. The compounds currently in use target ion channels and cytoskeletal proteins that are conserved between nematodes and mammals, and these compounds can be toxic to humans. In contrast, DUF644 is well conserved only in nematodes. Surface glycoproteins also comprise major antigens of parasitic nematodes (Blaxter et al., 1992) and may include multiple homologues of MLT-10. The homologues of MLT-10 are therefore attractive targets for drug development.
Supplementary Material
ACKNOWLEDGMENTS
We thank Julie Ahringer and Marc Vidal for providing bacterial clones. We are grateful to Chris Miller, David Eisenberg, Robert Mecham, Fred Keeley, and Creg Darby for helpful discussions. We thank John Kim, Peter Edwards, and Stanford Frand for critical reading of the manuscript. We thank the C. elegans Knockout Consortium, Shohei Mitani of the Tokyo Women's Medical University, and Laurent Segalat for providing additional strains. This work was supported by funds from the David Geffen School of Medicine at UCLA; postdoctoral fellowships from the Jane Coffin Childs Memorial Foundation and the MGH Fund for Medical Discovery to A.F.; and a National Institutes of Health grant to G.R. Some strains used in this study were provided by the Caenorhabditis Genetics Center (CGC).
Abbreviations used:
- DUF
domain of unknown function
- ECM
extracellular matrix
- GFP
green fluorescent protein
- GOF
gain-of-function
- LOF
loss-of-function
- Mlt
molting-defective
- NGM
nematode growth medium
- NHR
nuclear hormone receptor
- RNAi
RNA-interference
- WGA
wheat germ agglutinin.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E08-07-0708) on March 24, 2010.
REFERENCES
- Aguinaldo A. M., Turbeville J. M., Linford L. S., Rivera M. C., Garey J. R., Raff R. A., Lake J. A. Evidence for a clade of nematodes, arthropods and other moulting animals. Nature. 1997;387:489–493. doi: 10.1038/387489a0. [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Asahina M., Ishihara T., Jindra M., Kohara Y., Katsura I., Hirose S. The conserved nuclear receptor Ftz-F1 is required for embryogenesis, moulting and reproduction in Caenorhabditis elegans. Genes Cells. 2000;5:711–723. doi: 10.1046/j.1365-2443.2000.00361.x. [DOI] [PubMed] [Google Scholar]
- Bessereau J. L., Wright A., Williams D. C., Schuske K., Davis M. W., Jorgensen E. M. Mobilization of a Drosophila transposon in the Caenorhabditis elegans germ line. Nature. 2001;413:70–74. doi: 10.1038/35092567. [DOI] [PubMed] [Google Scholar]
- Bhattacharjee A., Bansal M. Collagen structure: the Madras triple helix and the current scenario. IUBMB Life. 2005;57:161–172. doi: 10.1080/15216540500090710. [DOI] [PubMed] [Google Scholar]
- Blank R. D., Boskey A. L. Genetic collagen diseases: influences of collagen mutations on structure and mechanical behavior. In: Fratzl P., editor. Collagen: Structure and Mechanics. New York: Springer Science; 2008. pp. 447–464. [Google Scholar]
- Blaxter M. L., Page A. P., Rudin W., Maizels R. M. Nematode surface coats: actively evading immunity. Parasitol. Today. 1992;8:243–247. doi: 10.1016/0169-4758(92)90126-m. [DOI] [PubMed] [Google Scholar]
- Campbell K. P. Three muscular dystrophies: loss of cytoskeleton-extracellular matrix linkage. Cell. 1995;80:675–679. doi: 10.1016/0092-8674(95)90344-5. [DOI] [PubMed] [Google Scholar]
- Chen Z., Eastburn D. J., Han M. The Caenorhabditis elegans nuclear receptor gene nhr-25 regulates epidermal cell development. Mol. Cell. Biol. 2004;24:7345–7358. doi: 10.1128/MCB.24.17.7345-7358.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitwood D. J. Biochemistry and function of nematode steroids. Crit. Rev. Biochem. Mol. Biol. 1999;34:273–284. doi: 10.1080/10409239991209309. [DOI] [PubMed] [Google Scholar]
- Cox G. N., Kusch M., Edgar R. S. Cuticle of Caenorhabditis elegans: its isolation and partial characterization. J. Cell Biol. 1981;90:7–17. doi: 10.1083/jcb.90.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowder C. M., Westover E. J., Kumar A. S., Ostlund R. E., Jr., Covey D. F. Enantiospecificity of cholesterol function in vivo. J. Biol. Chem. 2001;276:44369–44372. doi: 10.1074/jbc.C100535200. [DOI] [PubMed] [Google Scholar]
- Davis M. W., Birnie A. J., Chan A. C., Page A. P., Jorgensen E. M. A conserved metalloprotease mediates ecdysis in Caenorhabditis elegans. Development. 2004;131:6001–6008. doi: 10.1242/dev.01454. [DOI] [PubMed] [Google Scholar]
- Edens W. A., et al. Tyrosine cross-linking of extracellular matrix is catalyzed by Duox, a multidomain oxidase/peroxidase with homology to the phagocyte oxidase subunit gp91phox. J. Cell Biol. 2001;154:879–891. doi: 10.1083/jcb.200103132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Charles I. G., Fairweather N. F., Isaacs N. W. Structure of Bordetella pertussis virulence factor P.69 pertactin. Nature. 1996;381:90–92. doi: 10.1038/381090a0. [DOI] [PubMed] [Google Scholar]
- Epstein H. F., Shakes D. C., editors. San Diego: Academic Press; 1995. Caenorhabditis elegans: Modern Biological Analysis of an Organism. [Google Scholar]
- Fetterer R. H., Rhoads M. L. Tyrosine-derived cross-linking amino acids in the sheath of Haemonchus contortus infective larvae. J. Parasitol. 1990;76:619–624. [PubMed] [Google Scholar]
- Fleming M. W., Fetterer R. H. Ascaris suum: continuous perfusion of the pseudocoelom and nutrient absorption. Exp. Parasitol. 1984;57:142–148. doi: 10.1016/0014-4894(84)90073-0. [DOI] [PubMed] [Google Scholar]
- Frand A. R., Russel S., Ruvkun G. Functional genomic analysis of C. elegans molting. PLoS Biol. 2005;3 doi: 10.1371/journal.pbio.0030312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser A. G., Kamath R. S., Zipperlen P., Martinez-Campos M., Sohrmann M., Ahringer J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- Fritz J. A., Behm C. A. CUTI-1, a novel tetraspan protein involved in C. elegans CUTicle formation and epithelial integrity. PLoS One. 2009;4:e5117. doi: 10.1371/journal.pone.0005117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gissendanner C. R., Sluder A. E. nhr-25, the Caenorhabditis elegans ortholog of ftz-f1, is required for epidermal and somatic gonad development. Dev. Biol. 2000;221:259–272. doi: 10.1006/dbio.2000.9679. [DOI] [PubMed] [Google Scholar]
- Grant B. D., Sato M. The C. elegans Research Community, WormBook, editor. Intracellular trafficking (January 21, 2006), WormBook. 2006. doi/ 10.1895/wormbook.1.77.1, http://www.wormbook.org.
- Hao L., Mukherjee K., Liegeois S., Baillie D., Labouesse M., Burglin T. R. The hedgehog-related gene qua-1 is required for molting in Caenorhabditis elegans. Dev. Dyn. 2006;235:1469–1481. doi: 10.1002/dvdy.20721. [DOI] [PubMed] [Google Scholar]
- Hashmi S., Britton C., Liu J., Guiliano D. B., Oksov Y., Lustigman S. Cathepsin L is essential for embryogenesis and development of Caenorhabditis elegans. J. Biol. Chem. 2002;277:3477–3486. doi: 10.1074/jbc.M106117200. [DOI] [PubMed] [Google Scholar]
- Hashmi. S., Zhang J., Oksov Y., Lustigman S. The Caenorhabditis elegans cathepsin Z-like cysteine protease, Ce-CPZ-1, has a multifunctional role during the worms' development. J. Biol. Chem. 2004;279:6035–6045. doi: 10.1074/jbc.M312346200. [DOI] [PubMed] [Google Scholar]
- Hayes G. D., Frand A. R., Ruvkun G. The mir-84 and let-7 paralogous microRNA genes of Caenorhabditis elegans direct the cessation of molting via the conserved nuclear hormone receptors NHR-23 and NHR-25. Development. 2006;133:4631–4641. doi: 10.1242/dev.02655. [DOI] [PubMed] [Google Scholar]
- He D., Chung M., Chan E., Alleyne T., Ha K. C., Miao M., Stahl R. J., Keeley F. W., Parkinson J. Comparative genomics of elastin: sequence analysis of a highly repetitive protein. Matrix Biol. 2007;26:524–540. doi: 10.1016/j.matbio.2007.05.005. [DOI] [PubMed] [Google Scholar]
- Hieb W., Rothstein M. Sterol requirement for reproduction of a free-living nematode. Science. 1968;160:778–780. doi: 10.1126/science.160.3829.778. [DOI] [PubMed] [Google Scholar]
- Hope I. A. ‘Promoter trapping’ in Caenorhabditis elegans. Development. 1991;113:399–408. doi: 10.1242/dev.113.2.399. [DOI] [PubMed] [Google Scholar]
- Johnstone I. L. The cuticle of the nematode Caenorhabditis elegans: a complex collagen structure. Bioessays. 1994;16:171–178. doi: 10.1002/bies.950160307. [DOI] [PubMed] [Google Scholar]
- Kamath R. S., et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Koh K., Rothman J. H. ELT-5 and ELT-6 are required continuously to regulate epidermal seam cell differentiation and cell fusion in C. elegans. Development. 2001;128:2867–2880. doi: 10.1242/dev.128.15.2867. [DOI] [PubMed] [Google Scholar]
- Kostrouchova M., Krause M., Kostrouch Z., Rall J. E. CHR3, a Caenorhabditis elegans orphan nuclear hormone receptor required for proper epidermal development and molting. Development. 1998;125:1617–1626. doi: 10.1242/dev.125.9.1617. [DOI] [PubMed] [Google Scholar]
- Kostrouchova M., Krause M., Kostrouch Z., Rall J. E. Nuclear hormone receptor CHR3 is a critical regulator of all four larval molts of the nematode Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 2001;98:7360–7365. doi: 10.1073/pnas.131171898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause K., Foitzik K. Biology of the hair follicle: the basics. Semin. Cutan. Med. Surg. 2006;25:2–10. doi: 10.1016/j.sder.2006.01.002. [DOI] [PubMed] [Google Scholar]
- Li J., Brown G., Ailion M., Lee S., Thomas J. H. NCR-1 and NCR-2, the C. elegans homologs of the human Niemann-Pick type C1 disease protein, function upstream of DAF-9 in the dauer formation pathways. Development. 2004;131:5741–5752. doi: 10.1242/dev.01408. [DOI] [PubMed] [Google Scholar]
- Li X., Zhao X., Fang Y., Jiang X., Duong T., Fan C., Huang C. C., Kain S. R. Generation of destabilized green fluorescent protein as a transcription reporter. J. Biol. Chem. 1998;273:34970–34975. doi: 10.1074/jbc.273.52.34970. [DOI] [PubMed] [Google Scholar]
- Lustigman S., Brotman B., Huima T., Castelhano A. L., Singh R. N., Mehta K., Prince A. M. Transglutaminase-catalyzed reaction is important for molting of Onchocerca volvulus third-stage larvae. Antimicrob. Agents Chemother. 1995;39:1913–1919. doi: 10.1128/aac.39.9.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin E., Laloux H., Couette G., Alvarez T., Bessou C., Hauser O., Sookhareea S., Labouesse M., Segalat L. Identification of 1088 new transposon insertions of Caenorhabditis elegans: a pilot study toward large-scale screens. Genetics. 2002;162:521–524. doi: 10.1093/genetics/162.1.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima N., Yoshida H., Kumaki Y., Kamiya M., Tanaka T., Izumi Y., Kretsinger R. H. Flexible structures and ligand interactions of tandem repeats consisting of proline, glycine, asparagine, serine, and/or threonine rich oligopeptides in proteins. Curr. Protein Pept. Sci. 2008;9:591–610. doi: 10.2174/138920308786733886. [DOI] [PubMed] [Google Scholar]
- Matyash V., Entchev E. V., Mende F., Wilsch-Brauninger M., Thiele C., Schmidt A. W., Knolker H. J., Ward S., Kurzchalia T. V. Sterol-derived hormone(s) controls entry into diapause in Caenorhabditis elegans by consecutive activation of DAF-12 and DAF-16. PLoS Biol. 2004;2 doi: 10.1371/journal.pbio.0020280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merris M., Wadsworth W. G., Khamrai U., Bittman R., Chitwood D. J., Lenard J. Sterol effects and sites of sterol accumulation in Caenorhabditis elegans: developmental requirement for 4alpha-methyl sterols. J. Lipid Res. 2003;44:172–181. doi: 10.1194/jlr.m200323-jlr200. [DOI] [PubMed] [Google Scholar]
- Mohler W. A., Simske J. S., Williams-Masson E. M., Hardin J. D., White J. G. Dynamics and ultrastructure of developmental cell fusions in the Caenorhabditis elegans hypodermis. Curr. Biol. 1998;8:1087–1090. doi: 10.1016/s0960-9822(98)70447-6. [DOI] [PubMed] [Google Scholar]
- Moribe H., Yochem J., Yamada H., Tabuse Y., Fujimoto T., Mekada E. Tetraspanin protein (TSP-15) is required for epidermal integrity in Caenorhabditis elegans. J. Cell Sci. 2004;117:5209–5220. doi: 10.1242/jcs.01403. [DOI] [PubMed] [Google Scholar]
- Motola D. L., et al. Identification of ligands for DAF-12 that govern dauer formation and reproduction in C. elegans. Cell. 2006;124:1209–1223. doi: 10.1016/j.cell.2006.01.037. [DOI] [PubMed] [Google Scholar]
- Natsuka S., Kawaguchi M., Wada Y., Ichikawa A., Ikura K., Hase S. Characterization of wheat germ agglutinin ligand on soluble glycoproteins in Caenorhabditis elegans. J. Biochem. 2005;138:209–213. doi: 10.1093/jb/mvi117. [DOI] [PubMed] [Google Scholar]
- Page A. P., Johnstone I. L. The C. elegans Research Community, WormBook, editor. WormBook. doi/ 10.1895/wormbook.1.138.1, http://www.wormbook.org. The cuticle (March 19, 2007)
- Page A. P., McCormack G., Birnie A. J. Biosynthesis and enzymology of the Caenorhabditis elegans cuticle: identification and characterization of a novel serine protease inhibitor. Int. J. Parasitol. 2006;36:681–689. doi: 10.1016/j.ijpara.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Page-McCaw A., Ewald A. J., Werb Z. Matrix metalloproteinases and the regulation of tissue remodelling. Nat. Rev. Mol. Cell Biol. 2007;8:221–233. doi: 10.1038/nrm2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge F. A., Tearle A. W., Gravato-Nobre M. J., Schafer W. R., Hodgkin J. The C. elegans glycosyltransferase BUS-8 has two distinct and essential roles in epidermal morphogenesis. Dev. Biol. 2008;317:549–559. doi: 10.1016/j.ydbio.2008.02.060. [DOI] [PubMed] [Google Scholar]
- Pasquinelli A. E., et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- Podbilewicz B., White J. G. Cell fusions in the developing epithelial of C. elegans. Dev. Biol. 1994;161:408–424. doi: 10.1006/dbio.1994.1041. [DOI] [PubMed] [Google Scholar]
- Politz S. M., Philipp M., Estevez M., O'Brien P. J., Chin K. J. Genes that can be mutated to unmask hidden antigenic determinants in the cuticle of the nematode Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 1990;87:2901–2905. doi: 10.1073/pnas.87.8.2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez F., Dietz H. C. Extracellular microfibrils in vertebrate development and disease processes. J. Biol. Chem. 2009;284:14677–14681. doi: 10.1074/jbc.R900004200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts B., Clucas C., Johnstone I. L. Loss of SEC-23 in Caenorhabditis elegans causes defects in oogenesis, morphogenesis, and ECM secretion. Mol. Biol. Cell. 2003;14:4414–4426. doi: 10.1091/mbc.E03-03-0162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roudier N., Lefebvre C., Legouis R. CeVPS-27 is an endosomal protein required for the molting and the endocytic trafficking of the low-density lipoprotein receptor-related protein 1 in Caenorhabditis elegans. Traffic. 2005;6:695–705. doi: 10.1111/j.1600-0854.2005.00309.x. [DOI] [PubMed] [Google Scholar]
- Ruaud A. F., Bessereau J. L. Activation of nicotinic receptors uncouples a developmental timer from the molting timer in C. elegans. Development. 2006;133:2211–2222. doi: 10.1242/dev.02392. [DOI] [PubMed] [Google Scholar]
- Ruaud A. F., Lam G., Thummel C. S. The Drosophila nuclear receptors DHR3 and betaFTZ-F1 control overlapping developmental responses in late embryos. Development. 2010;137:123–131. doi: 10.1242/dev.042036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapio M. R., Hilliard M. A., Cermola M., Favre R., Bazzicalupo P. The Zona Pellucida domain containing proteins, CUT-1, CUT-3 and CUT-5, play essential roles in the development of the larval alae in Caenorhabditis elegans. Dev. Biol. 2005;282:231–245. doi: 10.1016/j.ydbio.2005.03.011. [DOI] [PubMed] [Google Scholar]
- Savage K. N., Gosline J. M. The role of proline in the elastic mechanism of hydrated spider silks. J. Exp. Biol. 2008;211:1948–1957. doi: 10.1242/jeb.014225. [DOI] [PubMed] [Google Scholar]
- Silhankova M., Jindra M., Asahina M. Nuclear receptor NHR-25 is required for cell-shape dynamics during epidermal differentiation in Caenorhabditis elegans. J. Cell Sci. 2005;118:223–232. doi: 10.1242/jcs.01609. [DOI] [PubMed] [Google Scholar]
- Simmer F., Tijsterman M., Parrish S., Koushika S. P., Nonet M. L., Fire A., Ahringer J., Plasterk R. H. Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr. Biol. 2002;12:1317–1319. doi: 10.1016/s0960-9822(02)01041-2. [DOI] [PubMed] [Google Scholar]
- Singh R. N., Soulston J. E. Some observations on moulting in Caenorhabditis elegans. Nematologica. 1978;24:63–71. [Google Scholar]
- Smith M. M., Levitan D. J. Human NPC1L1 and NPC1 can functionally substitute for the ncr genes to promote reproductive development in C. elegans. Biochim. Biophys. Acta. 2007;1770:1345–1351. doi: 10.1016/j.bbagen.2007.06.004. [DOI] [PubMed] [Google Scholar]
- Soulston J. E., Horvitz H. R. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- Stepek G., McCormack G., Page A. P. The kunitz domain protein BLI-5 plays a functionally conserved role in cuticle formation in a diverse range of nematodes. Mol. Biochem. Parasitol. 2010;169:1–11. doi: 10.1016/j.molbiopara.2009.08.005. [DOI] [PubMed] [Google Scholar]
- Suzuki M., Sagoh N., Iwasaki H., Inoue H., Takahashi K. Metalloproteases with EGF, CUB, and thrombospondin-1 domains function in molting of Caenorhabditis elegans. Biol. Chem. 2004;385:565–568. doi: 10.1515/BC.2004.069. [DOI] [PubMed] [Google Scholar]
- Sym M., Basson M., Johnson C. A model for niemann-pick type C disease in the nematode Caenorhabditis elegans. Curr. Biol. 2000;10:527–530. doi: 10.1016/s0960-9822(00)00468-1. [DOI] [PubMed] [Google Scholar]
- Thacker C., Rose A. M. A look at the Caenorhabditis elegans Kex2/Subtilisin-like proprotein convertase family. Bioessays. 2000;22:545–553. doi: 10.1002/(SICI)1521-1878(200006)22:6<545::AID-BIES7>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Thein M. C., McCormack G., Winter A. D., Johnstone I. L., Shoemaker C. B., Page A. P. Caenorhabditis elegans exoskeleton collagen COL-19, an adult-specific marker for collagen modification and assembly, and the analysis of organismal morphology. Dev. Dyn. 2003;226:523–539. doi: 10.1002/dvdy.10259. [DOI] [PubMed] [Google Scholar]
- Thein M. C., Winter A. D., Stepek G., McCormack G., Stapleton G., Johnstone I. L., Page A. P. Combined extracellular matrix cross-linking activity of the peroxidase MLT-7 and the dual oxidase BLI-3 is critical for post-embryonic viability in Caenorhabditis elegans. J. Biol. Chem. 2009;284:17549–17563. doi: 10.1074/jbc.M900831200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thummel C. S. Files on steroids—Drosophila metamorphosis and the mechanisms of steroid hormone action. Trends Genet. 1996;12:306–310. doi: 10.1016/0168-9525(96)10032-9. [DOI] [PubMed] [Google Scholar]
- Timmons L., Court D. L., Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- Wicks S. R., Yeh R. T., Gish W. R., Waterston R. H., Plasterk R. H. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat. Genet. 2001;28:160–164. doi: 10.1038/88878. [DOI] [PubMed] [Google Scholar]
- Wise S. G., Weiss A. S. Tropoelastin. Int. J. Biochem. Cell Biol. 2009;41:494–497. doi: 10.1016/j.biocel.2008.03.017. [DOI] [PubMed] [Google Scholar]
- Yochem J., Gu T., Han M. A new marker for mosaic analysis in Caenorhabditis elegans indicates a fusion between hyp6 and hyp7, two major components of the hypodermis. Genetics. 1998;149:1323–1334. doi: 10.1093/genetics/149.3.1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yochem J., Tuck S., Greenwald I., Han M. A gp330/megalin-related protein is required in the major epidermis of Caenorhabditis elegans for completion of molting. Development. 1999;126:597–606. doi: 10.1242/dev.126.3.597. [DOI] [PubMed] [Google Scholar]
- Zugasti O., Rajan J., Kuwabara P. E. The function and expansion of the Patched- and Hedgehog-related homologs in C. elegans. Genome Res. 2005;15:1402–1410. doi: 10.1101/gr.3935405. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.