Mutation of the apolar constriction of the yeast Sec61 translocon to polar or charged residues, while retaining functionality, affected the integration of potential transmembrane segments into the lipid bilayer. This indicates that the translocon plays an active role in setting the hydrophobicity threshold for membrane integration.
Abstract
The Sec61 translocon mediates the translocation of proteins across the endoplasmic reticulum membrane and the lateral integration of transmembrane segments into the lipid bilayer. The structure of the idle translocon is closed by a lumenal plug domain and a hydrophobic constriction ring. To test the function of the apolar constriction, we have mutated all six ring residues of yeast Sec61p to more hydrophilic, bulky, or even charged amino acids (alanines, glycines, serines, tryptophans, lysines, or aspartates). The translocon was found to be surprisingly tolerant even to the charge mutations in the constriction ring, because growth and translocation efficiency were not drastically affected. Most interestingly, ring mutants were found to affect the integration of hydrophobic sequences into the lipid bilayer, indicating that the translocon does not simply catalyze the partitioning of potential transmembrane segments between an aqueous environment and the lipid bilayer but that it also plays an active role in setting the hydrophobicity threshold for membrane integration.
INTRODUCTION
Protein translocation across the endoplasmic reticulum (ER) membrane is initiated by a hydrophobic signal sequence that, mediated by signal recognition particle (SRP) and SRP receptor, is targeted to the Sec61 translocon (Osborne et al., 2005). Here, the signal is oriented to transfer one end across the membrane and to integrate itself into the membrane. The translocon provides a pore for hydrophilic polypeptide segments to pass through, while simultaneously facilitating the integration of apolar segments into the lipid bilayer.
Mutagenesis of substrate proteins showed that charged residues flanking the hydrophobic core of a signal or signal–anchor sequence are important to define its final orientation according to the “positive-inside rule” (generally positioning the more positive end on the cytoplasmic side; von Heijne, 1986; Hartmann et al., 1989; Beltzer et al., 1991). The hydrophobicity of the signal influences the orientation process (Goder and Spiess, 2003) and drives integration into the membrane and insertion of the adjacent hydrophilic segment into the pore (Kida et al., 2009). Furthermore, subsequent apolar segments integrate into the membrane depending on their hydrophobicity. Cross-linking studies led to the proposal that membrane integration is a multistep process involving intermediate binding sites (Do et al., 1996; Ismail et al., 2006). Systematic analysis of potential transmembrane segments (TM) in mammalian in vitro and in vivo systems, in bacteria, and in yeast (Hessa et al., 2005, 2007, 2009; Xie et al., 2007; Lundin et al., 2008) yielded “biological hydrophobicity scales” and suggested that membrane insertion is fundamentally a thermodynamic partitioning process. Based on this interpretation, it was proposed that the function of the Sec61p channel is to provide a site in the membrane through which TMs can equilibrate between the lipid and aqueous phases (Heinrich et al., 2000; von Heijne, 2006).
Crystal structures of the archaeal SecYEβ translocon (van den Berg et al., 2004) provided a first basis to understand these processes mechanistically. SecY/Sec61α is a compact 10-helix bundle of two halves that may open a lateral gate toward the lipid membrane between TM helices 2/3 and 7/8 (illustrated in Figure 1 for the model of the yeast Sec61 complex; Junne et al., 2006). In the idle translocon, the central pore is obstructed by a lumenal plug domain (highlighted in Figure 1A), but in addition by a central constriction (Figure 1, B and C). The latter is generated by six, almost invariably hydrophobic side chains provided by TMs 2, 5, 7, and 10. This constriction ring might be responsible for the good viability of yeast cells and the short-term survival of bacteria with full deletion of the plug domain in Sec61p and SecY, respectively (Junne et al., 2006; Maillard et al., 2007).
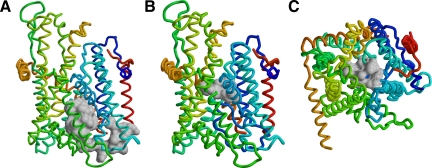
Figure 1.
Plug domain and constriction ring of Sec61p. The model of yeast Sec61 complex is shown as the polypeptide backbone (Sec61p in blue to yellow, Sbh1p in red, and Sss1p in orange) with the plug domain (residues 52-74; A) or the residues of the constriction ring (V82, I86, I181, T185, M294, and M450; B and C) in space-filling representation in gray. Views from within the membrane (A and B) or from the cytosol (C) are shown with the lateral exit gate to the front or bottom, respectively.
Mutations in the plug, in the constriction ring, as well as in the helices forming the lateral gate were found to destabilize the closed state of the translocon, resulting in a protein localization (prl) phenotype that suppresses inactivating mutations in signal sequences, both in bacteria (Emr et al., 1981; Veenendaal et al., 2004; Li et al., 2007) and in yeast (Junne et al., 2007). Such mutations in bacterial SecY produced transient channel openings in planar membrane permeability measurements (Saparov et al., 2007). In addition, prl mutants were shown to affect signal–anchor topology by premature opening of the translocation pore, before the orientation of the signal is completed (Junne et al., 2007).
To specifically analyze the importance and function of the hydrophobic constriction ring, we have here mutated all of its contributing residues to more hydrophilic or even charged amino acids, alone or in combination with a point mutation in the plug or a full plug deletion, and then we analyzed the resulting phenotypes with respect to viability, translocon assembly and stability, translocation defects, and TM integration. The translocon was found to be surprisingly tolerant to even drastic mutations in the constriction ring. Most interestingly, ring mutants were found to affect the integration of hydrophobic sequences into the lipid bilayer, indicating that the translocon does not simply catalyze the partitioning of potential TM segments between an aqueous environment and the lipid bilayer but also that it plays an active part in setting the threshold for lipid integration.
MATERIALS AND METHODS
Yeast Strains
Yeast strain VGY61 (Goder et al., 2004) corresponds to RSY1293 (matα, ura3-1, leu2-3,-112, his3-11,15, trp1-1, ade2-1, can1-100, sec61::HIS3, [pDQ1]) (Pilon et al., 1997) in which pDQ1 (i.e., YCplac111 (LEU2 CEN) containing SEC61 with codons 2–6 replaced by codons for H6RS and with its own promoter) was exchanged for YCPlac33 (URA3 CEN) with the same SEC61 gene. This made it possible to introduce mutant sec61 in YCplac111 (LEU2 CEN) by plasmid shuffling using 5-fluoro-orotic acid. The absence of wild-type SEC61 was confirmed by polymerase chain reaction (PCR) and restriction enzyme digestion of the products. VGY61 with a disruption of SSH1 was described previously (Goder et al., 2004).
Mutagenesis of Sec61p
Sec61p mutant strains with the mutations L63N and Δplug (residues 52-74 replaced by a glycine) have been described previously (Junne et al., 2006). Ring mutations were introduced sequentially in each of four quarters of ∼350 base pairs of the coding sequence delimited by unique restriction sites XbaI, SacI (created by a silent mutation), StuI, AccI, and EcoRI (464 base pairs after the stop codon) at nucleotide positions 27, 343, 710, 1099, and 1916 from the initiation codon, respectively. V82/I86 and I181/T185 were mutagenized simultaneously, whereas mutations of M294 and M450 were generated separately. In proximity of these four loci new silent restriction sites, Asp718, BamHI, PstI, and again BamHI were created at positions 232, 541, 892, and 1336, respectively. Mutagenesis was performed by PCR using appropriate mutagenic primers and Vent polymerase (New England Biolabs, Ipswich, MA). Ring and plug mutations were combined via Asp718. All constructs were verified by sequencing.
Growth Analysis and Sec61p Levels
For serial dilution experiments, yeast strains were grown in YPDA medium at 30°C to mid-log phase and diluted to 0.1 OD600. Aliquots of 6.6-fold serial dilutions were transferred onto YPDA plates and incubated at 15, 30, or 37°C.
To determine steady-state levels of Sec61p by immunoblot analysis, 10 OD600 equivalents of yeast cells were lysed in SDS-sample buffer with glass beads and boiled for 10 min. Aliquots of equal total protein were separated by SDS-gel electrophoresis, blotted onto nitrocellulose, and decorated with a rabbit antiserum against the C terminus of Sec61p. Antibody was detected using horseradish peroxidase-conjugated anti-mouse secondary antibody and the enhanced chemoluminescence kit (GE Healthcare, Little Chalfont, Buckinghamshire, United Kingdom). Equal protein loading was approximated based on Coomassie Blue staining of a separate gel.
To analyze the stability of Sec61p mutants in the presence of wild-type Sec61p, the sec61 coding sequences were extended by a sequence encoding a triple-hemagglutinin (HA) epitope tag, cloned with the original promoter into YCplac111 (LEU2 CEN), transformed into VGY61, and grown on SD-Leu-Ura to maintain both wild-type HA-tagged mutant copy of Sec61p. Translocons were analyzed by immunoblotting as described above using antibodies directed against the C terminus of Sec61p and against the HA epitope, respectively.
Model Proteins
The substrate proteins dipeptidyl aminopeptidase B (DPAPB), carboxypeptidase Y (CPY), and CPYΔ3 were described previously (Junne et al., 2007). In CPYΔC, the C-terminal 209 amino acids of CPY were deleted by PCR mutagenesis and fused to a triple-HA tag. To determine the effect of Sec61p mutations on membrane integration, the potential TM segments developed by Hessa et al. (2005) and shown in Table 1 were inserted into the translocated domain of DPAPB replacing codons 170–378, by PCR mutagenesis. The resulting model proteins thus consisted of an N-terminal cytoplasmic domain; a signal—anchor; a spacer sequence; the potential TM segment; and a C-terminal sequence of 29, 16, 124, 27, and 470 residues, respectively. Spacer and C-terminal sequence contain four and three potential glycosylation sites, respectively. They were expressed in pRS426 (URA3 2μ) with a glyceraldehyde-3-phosphate dehydrogenase promotor and a C-terminal triple-HA tag.
Table 1.
Potential TM segments to test membrane integration behavior of Sec61p mutants
| DPAPB-H nX/(19-n)A | Potential TM sequencea | ||
|---|---|---|---|
| 0L/19A | GGPG | AAAAAAAAAAAAAAAAAAA | GPGG |
| 1L/18A | GGPG | AAAAAAAAALAAAAAAAAA | GPGG |
| 2L/17A | GGPG | AAAALAAAALAAAAAAAAA | GPGG |
| 3L/16A | GGPG | AAAALAAAALAAAALAAAA | GPGG |
| 4L/15A | GGPG | AAAALALAALAAAALAAAA | GPGG |
| 5L/14A | GGPG | AAAALALAALAALALAAAA | GPGG |
| 6L/13A | GGPG | AAAALALALALALALAAAA | GPGG |
| 1S/18A | GGPG | AAAAAAAAASAAAAAAAAA | GPGG |
| 2S/17A | GGPG | AAAASAAAASAAAAAAAAA | GPGG |
| 3S/16A | GGPG | AAAASAAAASAAAASAAAA | GPGG |
a Essentially the same model sequences were used as had been described previously by Hessa et al. (2005, 2007, 2009), including flanking glycine/proline tetrapeptides to ″insulate″ the central 19-residue stretch from the surrounding sequence. For simplicity of construction, 2S, 2L, and 4L guest residues were left asymmetric, because position dependence had been shown to be negligible (Hessa et al., 2007). Guest residues are shown in bold.
Labeling and Immunoprecipitation
Yeast cells were in vivo pulse labeled for 5 min with 150 μCi/ml [35S]methionine/cysteine (PerkinElmer Life and Analytical Sciences, Boston, MA), and, if indicated, they were chased with 30 μg/ml each of unlabeled methionine and cysteine and 3 mM ammonium sulfate. Cells were lysed with glass beads, heated at 95°C for 5 min with 1% SDS, cleared by centrifugation, subjected to immunoprecipitation, and analyzed by SDS-gel electrophoresis and autoradiography as described previously (Junne et al., 2006). Signals were quantified by phosphorimager.
RESULTS
Sec61p Mutants with Hydrophilic or Even-charged Constriction Residues Retain Functionality
To test the importance of the hydrophobic constriction ring in Sec61p, the six amino acids constituting it, V82, I86, I181, T185, M294, and M450, were first mutated to either alanines (6A) or serines (6S). Because these mutants produced no striking growth defects, we also generated mutants in which the ring residues were replaced by glycines (6G), lacking any side chain. In addition, an opposite mutant with six tryptophans, the most bulky amino acid, was produced (6W). Finally, all six ring residues also were mutated to the charged amino acids aspartate (6D) or lysine (6K). We expected these latter mutants to be nonfunctional, because they were likely to interfere with the proper insertion of the Sec61p TM segments or because charge repulsion might prevent the formation of the helix bundle. To our surprise, however, all mutants supported growth at 30°C, even in the absence of the second, nonessential Sec61p homologue Ssh1p (Figure 2).
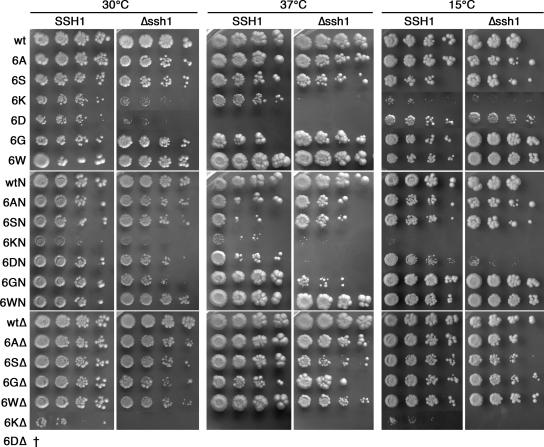
Figure 2.
Growth of yeast cells with wild-type or mutant Sec61p in the presence or absence of Ssh1p. SSH1 or Δssh1 cells expressing the indicated Sec61p mutants were plated at serial dilutions onto YPDA plates and incubated for 3 d at 30°C, 5 d at 37°C, or 11 d at 15°C.
We further constructed Sec61p mutants in which the ring mutations were combined with the L63N point mutation in the plug domain (Junne et al., 2006) named 6XN (X standing for A, S, G, W, D, or K), or with the full plug deletion (replacement of residues 52-74 by a glycine; Junne et al., 2006) named 6XΔ. As is shown in Figure 2, yeast cells with any of these mutants in place of wild-type Sec61p and in the absence of the nonessential SEC61 homologue SSH1 were viable except for cells containing 6DΔ, which could not lose the wild-type copy of SEC61. In addition, cells with 6KΔ grew so poorly that they were not yet visible after 3 d. SSH1 rescued growth of cells with 6KΔ but not with 6DΔ. Not unexpectedly, it was the charge mutants that showed the severest growth defects: 6K, 6D, 6KN, and 6KΔ had the lowest growth rates, and 6K, 6D, 6KN, 6DN, and 6KΔ showed heat and/or cold sensitivity, in some cases rescued by expression of SSH1. Ssh1p is functional only in cotranslational translocation, because it does not assemble with the Sec62–Sec63 complex essential for posttranslational translocation (Finke et al., 1996), but it is found associated with translating ribosomes (Prinz et al., 2000) and cotranslational substrate proteins (Wittke et al., 2002). Rescue of growth in the presence of Ssh1p thus suggests that cotranslational translocation was limiting. The growth behavior of the other mutants showed little, if any, deviation from wild type.
Ring Mutations Affect Translocation Efficiency
To test the functionality of mutant translocons with respect to co- and posttranslational translocation, the translocation efficiency was tested for DPAPB and CPY (Figure 3, A–C), established co- and posttranslational substrates, respectively (Ng et al., 1996). Rather modest defects were detected for cotranslational translocation of DPAPB with <30% nonintegration even for the charge mutants. However, ∼50% of CPY precursor failed to be translocated by 6K, 6D, and 6KN translocons during the 5-min labeling period, and even more by 6KΔ, whereas the other mutants showed only mild defects in comparison with the respective control (wt, wtN, or wtΔ; Figure 3, A and C).
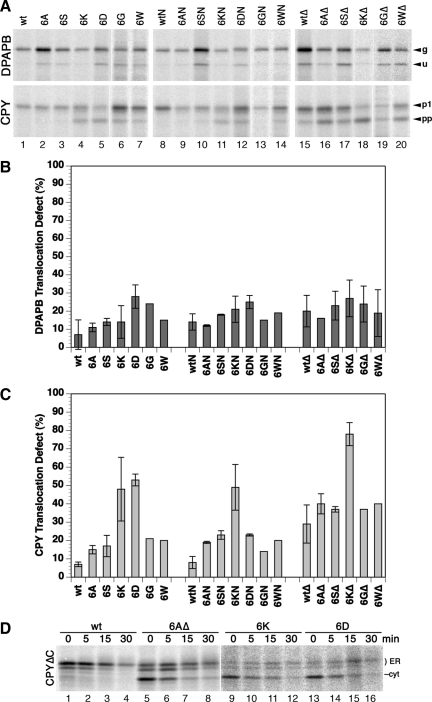
Figure 3.
Translocation efficiency of wild-type and mutant Sec61p. Integration of DPAPB as a cotranslational and of CPY as a posttranslational substrate of the Sec61 translocon was analyzed in a Δssh1 background by pulse labeling for 5 min with [35S]methionine, immunoprecipitation, gel electrophoresis, and autoradiography (A). The products correspond to glycosylated (g) and unglycosylated (u) forms of DPAPB and to the glycosylated first proform (p1) and the unglycosylated preproform (pp) of CPY. Results were quantified by phosphorimaging, and the fraction of untranslocated DPAPB (B) and CPY (C) was plotted (mean and SD of three determinations; single measurements for 6GΔ and 6WΔ in C). In D, C-terminally truncated CPYΔC was expressed in cells with the indicated wild-type and mutant translocons, pulse labeled for 5 min, and chased with unlabeled methionine for up to 30 min before immunoprecipitation, gel electrophoresis, and autoradiography to separate the translocated, two- and threefold glycosylated ER forms (ER) from cytosolic precursor (cyt).
Although unglycosylated full-length products of an obligatory cotranslational substrate directly reflect the defect in translocation, unglycosylated products of a posttranslational substrate primarily indicate a reduced rate of translocation resulting in an increased pool of cytosolic precursor. To test whether the CPY precursors not translocated after the labeling period can still be translocated later on, we performed pulse-chase experiments. With CPY, this is complicated by the fact that mature CPY, or after deglycosylation the ER and Golgi forms, comigrate with the unglycosylated precursor. For this reason, we analyzed CPYΔC, a C-terminally truncated version of CPY that cannot fold and is retained in the ER. Expressed with wild-type Sec61p, CPYΔC was almost completely glycosylated and thus translocated within the pulse period (Figure 3D, lane 1). During the chase, the signal was gradually reduced to ∼25% within 30 min by degradation (lanes 2–4). In cells with mutant translocons 6AΔ, 6K, or 6D (lanes 5–16), the signal of the glycosylated ER forms initially increased during the chase and then decreased more slowly, indicating that cytosolic precursors continued to be translocated in this period. The posttranslational defects observed by pulse-labeling in Figure 3 (A and C) reflect reduced translocation rates and not the final loss of translocated protein, which is defined by competition between the rates of translocation and cytosolic degradation.
The Hydrophobic Constriction Ring Stabilizes the Closed State of the Translocon
Single point mutations in the constriction ring have been shown previously to produce a prl phenotype, i.e., the suppression of inactivating mutations in signal sequences, both in the bacterial system (Smith et al., 2005) and in yeast (Junne et al., 2007). In general, prl mutations are interpreted to destabilize specifically the closed state of the translocon by disturbing the structure of the plug, its binding site in the lumenal cavity, or lateral gate closure, and thus facilitate pore opening. As a result, even marginally hydrophobic signal sequences obtain access to the membrane that are rejected by the wild-type translocon. To test for a prl phenotype, translocation of CPYΔ3 was tested, a mutant CPY in which the signal was inactivated by deletion of three apolar residues to yield <15% translocation with wild-type Sec61p. Replacement of all six hydrophobic constriction residues by alanines, serines, or glycines showed a clear prl effect, because >50% of CPYΔ3 was translocated (Figure 4). 6W showed a smaller effect, whereas 6D and 6K did not suppress the signal mutation, even when taking into account their general translocation defect. The effect of the charge mutations and in part of 6W thus seems not to specifically destabilize the closed state in favor of the open one, but to disturb the structure in a more general manner.
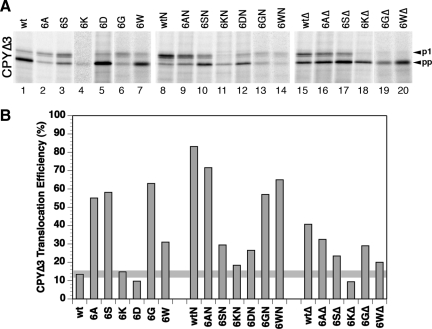
Figure 4.
prl phenotype of mutant Sec61p. (A) CPYΔ3 (CPY with a signal sequence lacking 3 apolar residues) was expressed in Δssh1 cells with wild-type (wt) or the indicated mutant Sec61p, labeled, and analyzed as described in Figure 3A. (B) Translocation efficiency was quantified by phosphorimager. The average of one to three determinations is shown. The horizontal line indicates the wild-type levels.
Ring mutations and the L63N plug mutation or the full plug deletion, which both generate a prl phenotype on their own, were not additive in suppression of the signal defect (Figure 4). The prl phenotype was always reduced, only mildly by alanines and most significantly by the charge mutations. This suggests that the effects of the combined mutations are not limited to facilitate pore opening.
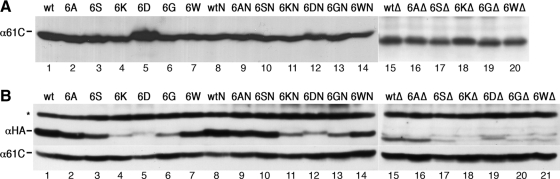
To test the stability of Sec61p mutants, their steady-state levels in cells lacking wild-type SEC61 were analyzed by immunoblot analysis. Surprisingly, charged residues replacing the hydrophobic constriction residues did not significantly reduce protein levels in comparison with the respective wild-type translocons (wt, wtN, and wtΔ; Figure 5A). In contrast, in a heterozygous situation, when coexpressed with a wild-type copy of SEC61, several of the mutant translocons (tagged with an HA-epitope for independent detection) were observed at strongly reduced levels (Figure 5B). This phenomenon was previously observed for ΔTM2 (deletion of codons 77–107; Wilkinson et al., 2000) and Δplug (Junne et al., 2006; Figure 5B, lane 15). It indicates competition of wild-type and mutant Sec61p for limiting interaction partners that are required for stability. In support of this notion, overexpression of the β and γ subunits Sbh1p and Sss1p at least partially rescued Δplug and ΔTM2 (Junne et al., 2007). Reduced levels in a heterozygous situation therefore suggest an altered protein surface with reduced binding affinity to partner molecules. Most affected were the mutations to lysines, aspartates, and glycines, whereas mutations to serines and tryptophans caused minor effects. Only the mutations to alanines showed no reduction of protein levels compared with the corresponding wild-type version of the translocon.
Figure 5.
Levels of wild-type or mutant translocons in the absence (A) or presence (B) of a second wild-type copy of Sec61p. (A) Steady-state amounts of wild-type and mutant Sec61p were determined in an SSH1 background by immunoblot analysis of total cell lysate. Equal loading was approximated based on protein determination and Coomassie staining of SDS-gels. (B) Yeast cells expressing equal amounts of wild-type Sec61p and the indicated HA-tagged mutants were analyzed by immunoblot analysis using an antiserum against the C terminus of Sec61p (α61C) and an anti-HA antibody (αHA) recognizing wild-type and mutant Sec61p, respectively. Mutation of constriction residues to aspartates consistently resulted in slightly reduced electrophoretic mobility. The asterisk indicates a background band recognized by the anti-HA antibody that also serves as a loading control. Data for SSH1 cells are shown; the same result was obtained with Δssh1 cells (unpublished data).
Properties of the Constriction Ring Regulate Membrane Insertion
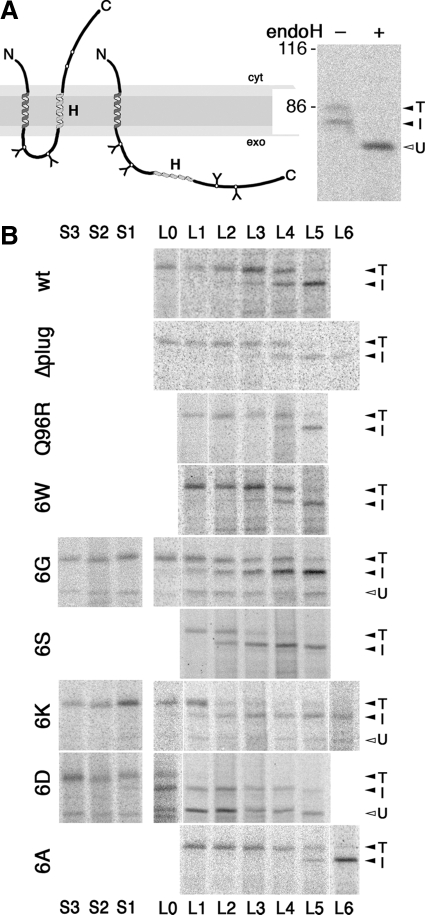
In addition to offering a passage for polypeptides through the membrane, the translocon also provides a lateral gate for the insertion of TM segments into the lipid bilayer. To analyze the effect of ring mutations on integration of TM sequences into the lipid bilayer, we tested the integration efficiency of moderately hydrophobic H-segments previously used by von Heijne and colleagues to characterize this process in mammalian in vitro and in vivo systems and in yeast (Hessa et al., 2005, 2009). They consisted of a 19-alanine host segment in which an increasing number of residues were replaced by leucines (Table 1), thus creating a series of increasing hydrophobicity. These sequences were inserted into the exoplasmic domain of DPAPB, generating a protein (DPAPB-H) with an uncleaved signal–anchor sequence for cotranslational ER targeting and translocation of its C terminus, and a potential stop-transfer sequence (as illustrated in Figure 6A). Depending upon whether this sequence is integrated into the membrane or is translocated, the protein is glycosylated only at sites between the signal–anchor and the H-sequence or also at downstream sites, respectively. The fraction of translocated to integrated H-segments can thus be determined after pulse-labeling, immunoprecipitation, gel electrophoresis, and autoradiography from the intensities of the fully and partially glycosylated forms corresponding to the translocated (T) and integrated H-segments (I), respectively. Unglycosylated products (U) generated by some of the partially defective mutant translocons were clearly separated and ignored as irrelevant to the process of membrane integration of the H-segments.
Figure 6.
Membrane insertion of H-segments of various hydrophobicities mediated by wild-type and mutant Sec61p. (A) Schematic representation of the DPAPB-H model proteins (left) and products of the DPAPB-H substrate with four leucines expressed in cells with wild-type Sec61p after [35S]methionine labeling, immunoprecipitation, incubation with (+) or without (−) endoglycosidase H (endoH), and gel electrophoresis (right). Integration of the H-segment results in a partially glycosylated double-spanning membrane protein (I), whereas its translocation yields a fully glycosylated type II protein (T). U, unglycosylated form, cyt, cytoplasmic; exo, exoplasmic. The position of molecular weight markers (in kilodaltons) is indicated. (B) SSH1 cells expressing wild-type or mutant translocons (as indicated on the left) as well as a DPAPB-H substrate (with the number of leucine or serine residues indicated above and below) were pulse labeled with[35S]methionine, and the substrate products were immunoprecipitated, separated by gel electrophoresis, and visualized by autoradiography.
Oligo-alanine H-segments with no or one leucine were fully translocated by the wild-type translocon, whereas increasing membrane integration was observed with additional leucines (Figure 6B, and quantified as the integrated fraction in Figure 7A). Fifty percent integration was obtained with approximately four leucines. Cells expressing the Δplug translocon dealt with the DPAPB-H constructs identically, indicating that the plug domain does not affect the outcome of the integration process. Similarly, the Q96R translocon, a Sec61p mutant altered at the cytoplasmic end of TM2 and affecting signal–anchor orientation (Junne et al., 2007), did not affect membrane integration. Also, the replacement of the constriction ring by six tryptophans, which are quite hydrophobic, had no effect (Figures 6B and 7B).
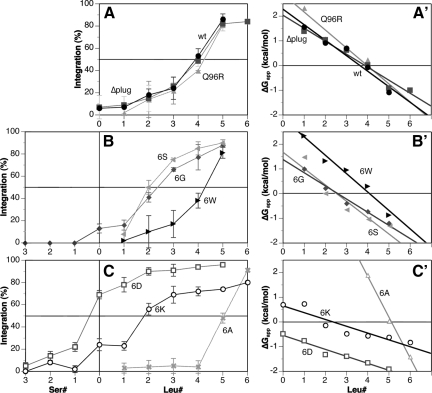
Figure 7.
Efficiency of H-segment integration by wild-type and mutant Sec61p. (A–C) The membrane-inserted fraction of two to five experiments like those shown in Figure 6 was quantified and plotted (with SDs) versus the number of leucines or serines in the H-segment. (A′–C′) The data for the H-segments containing zero to six leucines also were plotted as apparent free energies of membrane insertion, ΔGapp with straight lines determined by linear regression.
In contrast, exchange of the ring residues to hydrophilic amino acids, 6G, 6S, 6K, and 6D, clearly affected the insertion of H-segments (Figures 6B and 7, B and C). The required number of leucines to allow 50% membrane insertion was reduced to approximately two with 6S, 6G, and 6K, and the 6D translocon already mediated >60% integration for a pure 19-alanine sequence. Insertion of one to three serines into the oligo-alanine H-segment (Table 1) was necessary to prevent integration completely (Figure 6B and 7, B and C). In addition, the transition from predominantly translocated to mostly integrated H-segments occurred over a wide range of hydrophobicity for 6K and 6D, rather than within three leucines as for the other translocons. Surprisingly, mutation of the constriction residues to 6A had the opposite effects on H-domain insertion: it required five leucines for 50% insertion and the transition was completed in a range of only two additional leucines.
If the results are interpreted as an equilibration between a membrane inserted and a free state (according to Hessa et al., 2005), the ratio of integrated to translocated fractions, Kapp = fi/ft, i.e., the apparent equilibrium constant, can be used to calculate apparent free energies of membrane insertion as ΔGapp = −RTlnKapp, where R is the gas constant and T the absolute temperature of 303 K. The resulting plots (Figure 7, A′–C′) reveal a good linearity, consistent with the equilibrium assumption. The number of leucines in the oligo-alanine host sequence necessary for 50% membrane insertion with each mutant translocon was interpolated from these plots and listed in Table 2. These values and the apparent free energies of insertion ΔGapp are generally reduced with mutant translocons containing hydrophilic or charged ring residues. The amino acids lining the core of the translocation pore thus clearly influence the hydrophobicity threshold for membrane insertion, most likely by defining the polarity of the environment of the substrate sequence within the pore, i.e., of one of the two compartments between which the H-segment is partitioning. Increased polarity in the more polar compartment is expected to favor membrane integration of a moderately hydrophobic sequence. This is what we observe for the 6G, 6S, 6K, and 6D mutant translocons. The behavior of the 6A mutant, however, does not simply correlate with the increase in polarity in the ring residues and requires a different explanation.
Table 2.
Summary of membrane integration parameters of wild-type and mutant translocons
| Sec61 | n for 50% membrane integration of nL/(19–n)Aa |
|---|---|
| Wt | 3.6 |
| Δplug | 3.8 |
| Q96R | 4.0 |
| 6W | 4.1 |
| 6G | 2.6 |
| 6S | 2.5 |
| 6K | 2.3 |
| 6D | −2.0 |
| 6A | 5.1 |
DISCUSSION
Constriction Ring Mutants Retain Translocon Functionality
The crystal structure of the idle translocon shows a pore that is closed by the lumenal plug domain and a central constriction formed by six hydrophobic residues. A likely function of this apolar constriction is thus to prevent or reduce ion permeability when the plug is out and during protein translocation. This is supported by the viability of yeast cells expressing plugless mutant translocons (Junne et al., 2006) and the fact that in Escherichia coli expression of translocons with their plug locked open, although lethal, did not lead to an immediate membrane depolarization (Harris and Silhavy, 1999). Indeed, it has recently been shown that prl mutants of SecY with single hydrophobic-to-asparagine mutations in the constriction ring or with a deletion in the plug domain caused ion conductance across inner membrane vesicles, but with strong selectivity for anions, particularly chloride (Dalal and Duong, 2009). This selectivity, which preserves the seal for protons, is also functional during protein translocation through the wild-type translocon.
Here, we have tested the effects of replacing all six constriction residues to various less hydrophobic, polar, and even charged amino acids. Only minor defects were detected for mutations to alanines, serines, tryptophans, or glycines, and significant functionality was retained even with charged residues (aspartates or lysines). Mutation to the uncharged amino acids, 6A, 6S, 6G, and 6W, produced a prl phenotype, indicating specific destabilization of the closed state and facilitated translocon opening. prl mutations specifically disturb interactions that must be overcome for preprotein insertion. Therefore, not every ring mutation causes a prl phenotype (in Sec61p I86T does, but not T185K or M450K; Junne et al., 2007), and multiple ring mutations do not necessarily have a stronger phenotype than single mutations (6S showed less suppression than I86T). The charge mutations in 6D and 6K, but also the single mutations T185K or M450K (Junne et al., 2007), caused less specific perturbations and thus no prl effect. Similarly, by cumulation of prl mutations upon combining mutations in the ring residues and in the plug domain may generally destabilize the structure and thus not enhance or even reduce suppression of signal defects.
Because the constriction ring forms part of the plug binding site, ring mutations are expected to simultaneously disturb plug insertion. This could explain that an additional point mutation or even deletion of the plug did not strongly aggravate the phenotypes. Translocon stability was not significantly compromised by the ring mutations, not even by charged residues. Only in competition with a wild-type copy of Sec61p were the levels of mutant translocons clearly reduced in the order A < S ≈ W < G < D ≈ K, suggesting altered or less stable surface binding sites for limiting interaction partners. Again, this is a surprisingly mild effect for considerable alterations in the center of the protein. The ring mutations 6A, 6S, 6G, and 6W also did not significantly affect the orientation of sensitive diagnostic constructs (as used previously in Junne et al., 2007), and 6D and 6K did not have strong and opposite effects that could be correlated with their charges (unpublished data). It suggests that the positive-inside rule is not dominated by charged residues in the core of the translocon.
The Translocon Core Regulates Membrane Integration
It is long known that hydrophobicity is the essential property of a sequence for membrane integration (Davis and Model, 1985). Cross-linking experiments with reconstituted proteoliposomes demonstrated that the translocon allows a TM domain to bypass the barrier posed by the polar head groups of the lipid bilayer and to come into contact with the hydrophobic interior of the membrane (Heinrich et al., 2000). It was proposed that Sec61 provides a site through which a TM domain can dynamically equilibrate between the lipid and aqueous phases, depending on its hydrophobicity. The systematic analyses by von Heijne and colleagues (Hessa et al., 2005, 2009) yielded a biological hydrophobicity scale for membrane integration consistent with a thermodynamic partitioning process. The observed position dependence of residues such as tryptophan or tyrosine in the H-segment reflects the symmetry of the lipid bilayer (Hessa et al., 2007) and is also consistent with equilibration. Based on this interpretation, an apparent free energy contribution (ΔGaaapp) for membrane integration could be calculated for each amino acid. Interestingly, the values, although generally similar, were different for different systems (mammalian ER in vitro and in vivo, yeast, bacteria, and biophysical measurements). From our measurements in yeast, we obtained ΔGLeuapp = −0.51 kcal/mol, ΔGAlaapp = 0.12 kcal/mol and an interpolated 3.6 leucines in a 19-residues oligo-alanine sequence for 50% integration (compared with previous measurements in yeast of −0.21 kcal/mol, 0.06 kcal/mol, and 4.4 leucines, respectively (Hessa et al., 2009).
An alternative model to partitioning is a kinetically controlled mechanism in which H-segments trigger the opening of the lateral gate for (irreversible) exit into the lipid phase. Membrane integration would reflect the probability of gate opening, as supported by the molecular modeling study by Zhang and Miller (2010). In vitro crosslinking studies with arrested nascent chains showed extended association of TM segments with translocon proteins in defined positions (e.g. McCormick et al., 2003; Pitonzo et al., 2009), arguing that the H-segments might not immediately be free in the lipid phase. If these mechanisms depend on the hydrophobicity of the H-segment, the result might be difficult to distinguish from that of a free equilibration mechanism. Here, we found that mutations in the translocon affect TM integration, seemingly in support of a kinetic model, because a pure catalyst of partitioning should not affect the equilibrium. However, the translocon not only mediates the transition between two environments but also defines the properties of one of them. The hydrophobic constriction ring provides an apolar core that has also been observed experimentally at the center of the SecYEG translocon of E. coli (Bol et al., 2007). In addition, the narrowness of the pore (particularly in the presence of a substrate) partially excludes water to create conditions less polar than in bulk solution. The choice for an H-segment is therefore not the lipid environment versus the aqueous solution, but versus a less polar pore environment. Constriction ring mutations change the conditions inside the pore and thus also the outcome of a partitioning process with the membrane. An increase in polarity inside the pore by replacing the hydrophobic constriction residues with polar amino acids reduces the hydrophobicity required for 50% membrane integration (Table 2). Replacement with serines or glycines reduced it by at least one leucine, replacement with lysines by even more. The effect was most dramatic for aspartates that resulted in predominant membrane integration even of the Ala19 H-segment. The reason is probably that the charges on the short side chains of aspartates are more concentrated whereas those on the long lysine side chains are more delocalized. Tryptophans, as relatively hydrophobic amino acids and excluding water by their size, did not alter membrane integration significantly.
Interestingly, mutation of the ring residues to alanines had a clear effect of raising the hydrophobicity threshold for membrane integration to approximately five leucines. This is contrary to the expectation for introducing less hydrophobic ring residues. Of course, the effect of the mutations on the conformation of the translocon is not known. It is conceivable that the small side chains allow the pore to contract and thus to further exclude water, resulting in conditions corresponding to increased hydrophobicity. Glycines may not have this effect, because they introduce increased conformational flexibility.
It has been shown by cysteine cross-linking that the plug can move out of its binding cavity to reach SecE and that plug movement is triggered by polypeptide translocation (Tam et al., 2005). Molecular dynamics simulations, however, suggested that the plug is not necessarily fully displaced by a translocation polypeptide, but remains positioned either toward the lateral gate for a hydrophilic substrate or toward the inner side of the pore for a hydrophobic substrate (Zhang and Miller, 2010). In our experiments, full deletion of the plug did not affect H-segment integration, suggesting that the plug does not play an active role in regulating membrane integration.
Yet, our results clearly show that the properties of the residues forming the central constriction in the Sec61 translocon adjust the hydrophobicity threshold at which translocating sequences prefer the apolar environment of the lipid bilayer and thus stop further transfer to be anchored as TM domains.
ACKNOWLEDGMENTS
We thank Drs. Simon Bernèche, Lorenza Bordoli, and Torsten Schwede (all from Biozentrum) for valuable discussions. This work was supported by grant 31003A-125423 from the Swiss National Science Foundation.
Abbreviations used:
- CPY
carboxypeptidase Y
- DPAPB
dipeptidyl aminopeptidase B
- ER
endoplasmic reticulum
- HA
hemagglutinin
- PCR
polymerase chain reaction
- TM
transmembrane.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-01-0060) on March 31, 2010.
REFERENCES
- Beltzer J. P., Fiedler K., Fuhrer C., Geffen I., Handschin C., Wessels H. P., Spiess M. Charged residues are major determinants of the transmembrane orientation of a signal-anchor sequence. J. Biol. Chem. 1991;266:973–978. [PubMed] [Google Scholar]
- Bol R., de Wit J. G., Driessen A. J. The active protein-conducting channel of Escherichia coli contains an apolar patch. J. Biol. Chem. 2007;282:29785–29793. doi: 10.1074/jbc.M702140200. [DOI] [PubMed] [Google Scholar]
- Dalal K., Duong F. The SecY complex forms a channel capable of ionic discrimination. EMBO Rep. 2009;10:762–768. doi: 10.1038/embor.2009.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis N. G., Model P. An artificial anchor domain: hydrophobicity suffices to stop transfer. Cell. 1985;41:607–614. doi: 10.1016/s0092-8674(85)80033-7. [DOI] [PubMed] [Google Scholar]
- Do H., Falcone D., Lin J., Andrews D. W., Johnson A. E. The cotranslational integration of membrane proteins into the phospholipid bilayer is a multistep process. Cell. 1996;85:369–378. doi: 10.1016/s0092-8674(00)81115-0. [DOI] [PubMed] [Google Scholar]
- Emr S. D., Hanley-Way S., Silhavy T. J. Suppressor mutations that restore export of a protein with a defective signal sequence. Cell. 1981;23:79–88. doi: 10.1016/0092-8674(81)90272-5. [DOI] [PubMed] [Google Scholar]
- Finke K., Plath K., Panzner S., Prehn S., Rapoport T. A., Hartmann E., Sommer T. A second trimeric complex containing homologs of the Sec61p complex functions in protein transport across the ER membrane of S. cerevisiae. EMBO J. 1996;15:1482–1494. [PMC free article] [PubMed] [Google Scholar]
- Goder V., Junne T., Spiess M. Sec61p contributes to signal sequence orientation according to the positive-inside rule. Mol. Biol. Cell. 2004;15:1470–1478. doi: 10.1091/mbc.E03-08-0599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goder V., Spiess M. Molecular mechanism of signal sequence orientation in the endoplasmic reticulum. EMBO J. 2003;22:3645–3653. doi: 10.1093/emboj/cdg361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris C. R., Silhavy T. J. Mapping an interface of SecY (PrlA) and SecE (PrlG) by using synthetic phenotypes and in vivo cross-linking. J. Bacteriol. 1999;181:3438–3444. doi: 10.1128/jb.181.11.3438-3444.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann E., Rapoport T. A., Lodish H. F. Predicting the orientation of eukaryotic membrane-spanning proteins. Proc. Natl. Acad. Sci. USA. 1989;86:5786–5790. doi: 10.1073/pnas.86.15.5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrich S. U., Mothes W., Brunner J., Rapoport T. A. The Sec61p complex mediates the integration of a membrane protein by allowing lipid partitioning of the transmembrane domain. Cell. 2000;102:233–244. doi: 10.1016/s0092-8674(00)00028-3. [DOI] [PubMed] [Google Scholar]
- Hessa T., Kim H., Bihlmaier K., Lundin C., Boekel J., Andersson H., Nilsson I., White S. H., von Heijne G. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature. 2005;433:377–381. doi: 10.1038/nature03216. [DOI] [PubMed] [Google Scholar]
- Hessa T., Meindl-Beinker N. M., Bernsel A., Kim H., Sato Y., Lerch-Bader M., Nilsson I., White S. H., von Heijne G. Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature. 2007;450:1026–1030. doi: 10.1038/nature06387. [DOI] [PubMed] [Google Scholar]
- Hessa T., Reithinger J. H., von Heijne G., Kim H. Analysis of transmembrane helix integration in the endoplasmic reticulum in S. cerevisiae. J. Mol. Biol. 2009;386:1222–1228. doi: 10.1016/j.jmb.2009.01.027. [DOI] [PubMed] [Google Scholar]
- Ismail N., Crawshaw S. G., High S. Active and passive displacement of transmembrane domains both occur during opsin biogenesis at the Sec61 translocon. J. Cell Sci. 2006;119:2826–2836. doi: 10.1242/jcs.03018. [DOI] [PubMed] [Google Scholar]
- Junne T., Schwede T., Goder V., Spiess M. The plug domain of yeast Sec61p is important for efficient protein translocation, but is not essential for cell viability. Mol. Biol. Cell. 2006;17:4063–4068. doi: 10.1091/mbc.E06-03-0200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junne T., Schwede T., Goder V., Spiess M. Mutations in the Sec61p channel affecting signal sequence recognition and membrane protein topology. J. Biol. Chem. 2007;282:33201–33209. doi: 10.1074/jbc.M707219200. [DOI] [PubMed] [Google Scholar]
- Kida Y., Morimoto F., Sakaguchi M. Signal anchor sequence provides motive force for polypeptide chain translocation through the endoplasmic reticulum membrane. J. Biol. Chem. 2009;284:2861–2866. doi: 10.1074/jbc.M808020200. [DOI] [PubMed] [Google Scholar]
- Li W., Schulman S., Boyd D., Erlandson K., Beckwith J., Rapoport T. A. The plug domain of the SecY protein stabilizes the closed state of the translocation channel and maintains a membrane seal. Mol. Cell. 2007;26:511–521. doi: 10.1016/j.molcel.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Lundin C., Kim H., Nilsson I., White S. H., von Heijne G. Molecular code for protein insertion in the endoplasmic reticulum membrane is similar for N(in)-C(out) and N(out)-C(in) transmembrane helices. Proc. Natl. Acad. Sci. USA. 2008;105:15702–15707. doi: 10.1073/pnas.0804842105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillard A. P., Lalani S., Silva F., Belin D., Duong F. Deregulation of the SecYEG translocation channel upon removal of the plug domain. J. Biol. Chem. 2007;282:1281–1287. doi: 10.1074/jbc.M610060200. [DOI] [PubMed] [Google Scholar]
- McCormick P. J., Miao Y., Shao Y., Lin J., Johnson A. E. Cotranslational protein integration into the ER membrane is mediated by the binding of nascent chains to translocon proteins. Mol. Cell. 2003;12:329–341. doi: 10.1016/s1097-2765(03)00304-6. [DOI] [PubMed] [Google Scholar]
- Ng D. T., Brown J. D., Walter P. Signal sequences specify the targeting route to the endoplasmic reticulum membrane. J. Cell Biol. 1996;134:269–278. doi: 10.1083/jcb.134.2.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne A. R., Rapoport T. A., van den Berg B. Protein translocation by the Sec61/SecY channel. Annu. Rev. Cell Dev. Biol. 2005;21:529–550. doi: 10.1146/annurev.cellbio.21.012704.133214. [DOI] [PubMed] [Google Scholar]
- Pilon M., Schekman R., Römisch K. Sec61p mediates export of a misfolded secretory protein from the endoplasmic reticulum to the cytosol for degradation. EMBO J. 1997;16:4540–4548. doi: 10.1093/emboj/16.15.4540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pitonzo D., Yang Z., Matsumura Y., Johnson A. E., Skach W. R. Sequence-specific retention and regulated integration of a nascent membrane protein by the endoplasmic reticulum Sec61 translocon. Mol. Biol. Cell. 2009;20:685–698. doi: 10.1091/mbc.E08-09-0902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz A., Hartmann E., Kalies K. U. Sec61p is the main ribosome receptor in the endoplasmic reticulum of Saccharomyces cerevisiae. Biol. Chem. 2000;381:1025–1029. doi: 10.1515/BC.2000.126. [DOI] [PubMed] [Google Scholar]
- Saparov S. M., Erlandson K., Cannon K., Schaletzky J., Schulman S., Rapoport T. A., Pohl P. Determining the conductance of the SecY protein translocation channel for small molecules. Mol. Cell. 2007;26:501–509. doi: 10.1016/j.molcel.2007.03.022. [DOI] [PubMed] [Google Scholar]
- Smith M. A., Clemons W. M., Jr, DeMars C. J., Flower A. M. Modeling the effects of prl mutations on the Escherichia coli SecY complex. J. Bacteriol. 2005;187:6454–6465. doi: 10.1128/JB.187.18.6454-6465.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam P. C., Maillard A. P., Chan K. K., Duong F. Investigating the SecY plug movement at the SecYEG translocation channel. EMBO J. 2005;24:3380–3388. doi: 10.1038/sj.emboj.7600804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg B., Clemons W. M., Jr, Collinson I., Modis Y., Hartmann E., Harrison S. C., Rapoport T. A. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- Veenendaal A. K., van der Does C., Driessen A. J. The protein-conducting channel SecYEG. Biochim. Biophys. Acta. 2004;1694:81–95. doi: 10.1016/j.bbamcr.2004.02.009. [DOI] [PubMed] [Google Scholar]
- von Heijne G. The distribution of positively charged residues in bacterial inner membrane proteins correlates with the trans-membrane topology. EMBO J. 1986;5:3021–3027. doi: 10.1002/j.1460-2075.1986.tb04601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Membrane-protein topology. Nat. Rev. Mol. Cell Biol. 2006;7:909–918. doi: 10.1038/nrm2063. [DOI] [PubMed] [Google Scholar]
- Wilkinson B. M., Tyson J. R., Reid P. J., Stirling C. J. Distinct domains within yeast Sec61p involved in post-translational translocation and protein dislocation. J. Biol. Chem. 2000;275:521–529. doi: 10.1074/jbc.275.1.521. [DOI] [PubMed] [Google Scholar]
- Wittke S., Dunnwald M., Albertsen M., Johnsson N. Recognition of a subset of signal sequences by Ssh1p, a Sec61p-related protein in the membrane of endoplasmic reticulum of yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 2002;13:2223–2232. doi: 10.1091/mbc.01-10-0518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie K., Hessa T., Seppala S., Rapp M., von Heijne G., Dalbey R. E. Features of transmembrane segments that promote the lateral release from the translocase into the lipid phase. Biochemistry. 2007;46:15153–15161. doi: 10.1021/bi701398y. [DOI] [PubMed] [Google Scholar]
- Zhang B., Miller T. F. A hydrophobically stabilized open state for the lateral gate of the Sec translocon. Proc. Natl. Acad. Sci. USA. 2010;107:5399–5404. doi: 10.1073/pnas.0914752107. [DOI] [PMC free article] [PubMed] [Google Scholar]