The ectodomain structure and function of the neural cell adhesion molecule L1 is shown to be regulated by the intracellular phosphorylation of a novel threonine, T1172. In pancreatic cancer cells, T1172 exhibits steady-state saturated phosphorylation, an event regulated by CKII and PKC, and which further regulates cell migration.
Abstract
Previous reports on the expression of the cell adhesion molecule L1 in pancreatic ductal adenocarcinoma (PDAC) cells range from absent to high. Our data demonstrate that L1 is expressed in poorly differentiated PDAC cells in situ and that threonine-1172 (T1172) in the L1 cytoplasmic domain exhibits steady-state saturated phosphorylation in PDAC cells in vitro and in situ. In vitro studies support roles for casein kinase II and PKC in this modification, consistent with our prior studies using recombinant proteins. Importantly, T1172 phosphorylation drives, or is associated with, a change in the extracellular structure of L1, consistent with a potential role in regulating the shift between the closed conformation and the open, multimerized conformation of L1. We further demonstrate that these distinct conformations exhibit differential binding to integrins αvβ3 and αvβ5 and that T1172 regulates cell migration in a matrix-specific manner and is required for a disintegrin and metalloproteinase-mediated shedding of the L1 ectodomain that has been shown to regulate cell migration. These data define a specific role for T1172 of L1 in regulating aspects of pancreatic adenocarcinoma cell phenotype and suggest the need for further studies to elucidate the specific ramifications of L1 expression and T1172 phosphorylation in the pathobiology of pancreatic cancer.
INTRODUCTION
L1 is a single pass type I transmembrane protein of the immunoglobulin (Ig) superfamily that contains six Ig repeats followed by five fibronectin-like (FN) repeats (Table 1). L1 regulates active neural processes, including cerebellar cell migration, neurite extension, and axon guidance (Burden-Gulley et al., 1997). L1 is also expressed in human neuroectodermal tumors and monocytic leukemias (Silletti et al., 2000b), and L1 correlates with poor prognosis and advanced disease state in uterine/ovarian carcinomas (Fogel et al., 2003a) and malignant cutaneous melanoma (Thies et al., 2002; Fogel et al., 2003b). L1 expression also correlates with stage and grade in serous ovarian neoplasms, increasing in expression with grade (Daponte et al., 2008). These findings suggest a role for L1 in the progression of tumors that express it.
Table 1.
Primers used in the construction of recombinant L1 proteins
| Primer type | Primer sequence |
|---|---|
| Mutagenesis | |
| NN T1172A | F 5′-ATGATGAAAGATGAGGCGTTCGGCGAGTACAGTG-3′ |
| R 5′-CACTGTACTCGCCGAACGCCTCATCTTTCATCAT-3′ | |
| NN T1172E | F 5′-CTGTACTCGCCGAACTCCTCATCTTTCATCATC-3′ |
| R 5′-GATGATGAAAGATGAGGAGTTCGGCGAGTACAG-3′ | |
| NN T1172F | F 5′-ATGAAAGATGAGTTCTTCGGCGAGTACAGT-3′ |
| R 5′-ACTGTACTCGCCGAAGAACTCATCTTTCAT-3′ | |
| NN Y1176F | F 5′-TTCGGCGAGTTCAGTGACAACGAGGAGAAG-3′ |
| R 5′-CTTCTCCTCGTTGTCACTGAACTCGCCGAA-3′ | |
| NN S1181A | F 5′-AGACCTTCGGCGAGTACGCTGACAACGAGGAGAAG-3′ |
| R 5′-CTTCTCCTCGTTGTCAGCGTACTCGCCGAAGGTCT-3′ | |
| N S1181A | F 5′-AGGTCCCTGGAGGCTGACAACGAGGAGAAGG-3′ |
| R 5′-CCTTCTCCTCGTTGTCAGCCTCCAGGGACCT-3′ | |
| PCR | |
| Ig 1 (14) | F 5′-CATGAATTCGAGCCACCTGTC-3′ |
| Ig 1 (115) | R 5′-GAATTCCATGAGCCGGATCT-3′ |
| Ig 1 (112, -3aa) | R 5′-AAGAATTCACCCTCGGCCAT-3′ |
| Ig 2 (116) | F 5′-GCCGAATTCGCCCCCAAGTGG-3′ |
| Ig 2 (223) | R 5′-GGGGAATTCCAGGCGCGGCTT-3′ |
| Ig 3 (216) | F 5′-GAATTCATGATTGACAGGA-3′ |
| Ig 3 (317) | R 5′-TGGGAATTCTGCAGCCAGTAC-3′ |
| Ig 4 (313) | F 5′-GTGGAATTCGCCCCCTACTGG-3′ |
| Ig 4 (404) | R 5′-CGAATTCTCACTGGACAACGTAG-3′ |
| Cyto (1144) | F 5′-CGAATTCAAGCGCAGCAAGGG-3′ |
| Cyto (1257) | R 5′-GGAATTCCTATTCTAGGGCC-3′ |
| 1168stop | R 5′-TGAATTCACGGTCGGGCCTCAG-3′ |
| Exon2-Diagnostic | F 5′-CGGTACGTGTGGCCTCTCCTCCTCTGCAG-3′ |
| Exon2-Diagnostic | R 5′-GGCTGATGTCATCTGTGGGGAAGACAACC-3′ |
| Mini exon | |
| NN 1169-1186 | F 5′-AATTCGCCCGACCGATGAAAGATGAGACCTTCGGCGAGTACAGTGACAACGAGGAGAAGG-3′ |
| R 5′-AATTCCTTCTCCTCGTTGTCACTGTACTCGCCGAAGGTCTCATCTTTCATCGGTCGGGCG-3′ | |
 a a
| |
N, neuronal isoform; NN, nonneuronal isoform.
a The domain structure of L1 is shown for reference.
Clinically, human pancreatic ductal adenocarcinoma (PDAC) is a devastating disease characterized by significant dissemination at the time of diagnosis, resulting in one of the highest mortality rates of all cancers (NCI-Pancreatic Cancer Progress Review Group, 2002). Despite this, the biology of PDAC remains poorly understood. Studies have identified key factors in the etiology of the disease, which have been incorporated into a genetic model of PDAC development (Bardeesy and DePinho, 2002). Although such a timeline is significant in the definition of factors contributing to disease onset, a similar timeline has been difficult to address with regard to disease progression. Although the majority of patients present with well differentiated (grade 1; G1) or moderately differentiated (grade 2; G2) lesions (NCI-Pancreatic Cancer Progress Review Group, 2002), patients with G1 and G2 tumors demonstrate a better overall survival rate versus those harboring poorly differentiated (grade 3; G3) or truly anaplastic/sarcomatoid lesions (Bouvet et al., 2000). Thus, the definition of factors contributing to progression to these poorly differentiated states is of fundamental importance for understanding the biology of this tumor type and may provide insights into future treatment strategies based around the differences in these grades of PDAC cells. Although a prior publication failed to detect L1 in PDAC samples (Kaifi et al., 2006), a subsequent report found L1 in 80% of G2 and 100% of G3 tumors (Muerkoster et al., 2007), with the highest expression levels in G3 tumors, similar to our findings.
As such, L1's role in regulating processes associated with migration and invasion makes it well suited for use by an aggressive tumor. Indeed, stable ectopic expression of L1 in fibroblastic and melanoma cells induces the expression of invasion and metastasis-associated genes promoting de novo integrin use and concomitant migration and invasion in vitro (Silletti et al., 2004). More recent work demonstrated that L1 is fully transforming and expressed at the invasive front of colon cancers in situ (Gavert et al., 2005), and that ectopic expression of L1 in colon cancer cells bestows a metastatic phenotype (Gavert et al., 2007). Importantly, the cytoplasmic domain (CD) of L1 was required for this effect, although the CD alone was not sufficient to drive this phenotype.
The L1 CD is highly conserved among species, and mutations in this domain cause Mental Retardation Aphasia Shuffling Gait and Adducted Thumbs (MASA) Syndrome (Fransen et al., 1995, 1997), suggesting that the L1 CD is crucial for its function. Although the role of cytoplasmic serine (S) and tyrosine (Y) phosphorylation in regulating L1 function has been studied in detail (Kamiguchi and Lemmon, 1998; Schaefer et al., 2002; Schultheis et al., 2007), little is known about threonine (T) phosphorylation of L1. Gast et al. (2008) recently demonstrated that the L1-induced invasive phenotype of ovarian carcinoma cells was abrogated by mutation of T1247 and S1248 in the L1 CD. This mutation, but not the mutation of S1248 alone, attenuated L1-mediated extracellular signal-regulated kinase (Erk) activation and the concomitant expression of malignancy-associated gene products described previously as regulated by L1 in fibroblastic and melanoma cells (Silletti et al., 2004). Interestingly, this double mutation did not affect the ability of L1 to interact with RanBPM, an Erk-activating protein that binds a site within the C-terminal 28 amino acids of L1 (Cheng et al., 2005), suggesting multiple mechanisms of Erk regulation by L1. Although these data suggest that T1247 phosphorylation might be important in regulating L1 function, the authors did not demonstrate phosphorylation of L1 at this site in their cells. A separate mass spectroscopy analysis does suggest potential phosphorylation of T1247 in L1 signal obtained from the cytoplasm and nucleus of HeLa cells (Olsen et al., 2006); however, this has yet to be corroborated by other means or investigated in other cells. Previously, we demonstrated that a novel threonine in the L1 CD, immediately upstream of the alternatively spliced neuronal exon27, can be phosphorylated in vitro (Chen et al., 2009). Here, we report that this residue exhibits steady-state saturated phosphorylation in PDAC cells, an event regulated by casein kinase II (CKII), protein kinase C (PKC) and protein phosphatase (PP)1, and that is associated with the regulation of L1 ectodomain (ECD) conformation, integrin-binding and a disintegrin and metalloproteinase (ADAM)-mediated proteolysis, and the concomitant control of cell migration in a matrix-specific manner. These findings have considerable potential significance for understanding the role of L1 in regulating aspects of the biology of cells that express it, in particular pancreatic cancer, in which L1 may contribute to the progression of this devastating and almost uniformly lethal disease.
MATERIALS AND METHODS
Cells and Transfections
Cell Lines.
CHO-K1, J558L, MIAPaCa2, and Panc1 cells are originally from American Type Culture Collection (Manassas, VA). M21 human melanoma cells were derived from the UCLA-SO-M21 cell line, which was provided by Dr. D. L. Morton (UCLA, Los Angeles, CA). M24met human melanoma cells were originally obtained from R. Reisfeld (The Scripps Research Institute, La Jolla, CA). CHO-K1, MIAPaCa2, and Panc1 cells were cultured in DMEM supplemented with 10% fetal bovine serum (FBS) and 1.5 g/l sodium bicarbonate. J558L, M21, and M24met cells were cultured in RPMI 1640 medium supplemented with 10% FBS, 1.5 g/l sodium bicarbonate, 1 mM sodium pyruvate, and 10 mM HEPES. Serum-free (SF-) medium consisted of all components except serum, as appropriate for the cell line, supplemented with 0.5% bovine serum albumin (BSA).
Transfectants.
Stable mock, L1wt, and L1-T1172A Chinese hamster ovary (CHO) transfectants were produced by Lipofectamine 2000 (Invitrogen, Carlsbad, CA) transfection of parental CHO-K1 cells with an empty pCDNA3.1 vector into which an internal ribosome entry site (IRES) had been introduced immediately upstream of a hygromycin-phosphotransferase CDS (pCDNA3.1/IRES-Hygro), or vectors into which wild-type or T1172A mutant nonneuronal L1 CDSs were inserted upstream of the IRES. J558L-L1 cells were produced essentially as described previously (Nayeem et al., 1999) by using stable transfection of neuronal L1 inserted into pCDNA3.1/IRES-Hygro. Cells were selected for hygromycin-resistance and L1 expression was confirmed.
Antibodies, Enzymes, and Reagents
Antibodies.
The α-human L1 monoclonal antibody (mAb) 5G3 has been described previously (Mujoo et al., 1986) and was generously provided by M. Just (eBioscience, San Diego, CA). The αL1 carboxy terminus goat polyclonal antibody (pAb) (C20) was from Santa Cruz Biotechnology (Santa Cruz, CA). Anti-CD171 mAb UJ127 has been described previously (Patel et al., 1991) and was from Neomarkers/Labvision (Fremont, CA). An α-phospho-S1181 pAb (αP-S1181) specific for the neuronal isoform of L1 and the 2C2 mAb specific for the L1 cytoplasmic domain were from Abcam (Cambridge, MA). The Neuro4 mAb was provided by BD Technologies (Durham, NC). L1-ECD is a 6xHis-tagged secreted protein produced in human embryonic kidney (HEK)293 cells (Stallcup, 2000). This protein and a pAb (αECD) generated against and affinity-purified using the L1-ECD fusion protein were generously provided by W. Stallcup (The Burnham Institute, La Jolla, CA). A rabbit α-phospho-T1172 (αP-T1172) pAb was generated for us by ProSci, (Poway, CA), by using a phosphorylated 7aa antigen (KDEpT1172FGE) coupled to KLH through an N-terminal cysteine as immunogen. Rabbit serum was affinity depleted of nonphospho-T1172-dependent species on a nonphospho-KDETFGE column, and nonbound antibody was further purified on a phospho-KDETFGE column. Antibody eluted from the nonphospho-T1172 column (αT1172-IND) recognizes the KDETFGE sequence in a manner independently of T1172 phosphorylation (i.e., it demonstrates nearly equal signal on phospho- and nonphospho-peptides); the αP-T1172 species purified from the KDEpTFGE column demonstrates specific recognition of the phosphorylated peptide (Chen et al., 2009). αErk2 pAb (C14), αGST pAb (110-218), and a 6xHis tag-specific pAb (H3) were from Santa Cruz Biotechnology. α-Actin and horseradish peroxidase (HRP)-labeled αbiotin BN-34 were from Sigma-Aldrich (St. Louis, MO). The α-phospho-S/T-phenylalanine (αP-S/T-F) motif-specific pAb was from Cell Signaling Technology (Danvers, MA). TrueBlot α-rabbit-HRP reagent was provided by M. Just (eBioScience). HRP- and fluorescein isothiocyanate (FITC)-conjugated donkey α-mouse and donkey α-rabbit secondary antibodies extensively cross-adsorbed for minimal cross-reactivity were from Jackson ImmunoResearch Laboratories (West Grove, PA). HRP-conjugated α-mouse IgG2a and IgG2b antibodies were from Southern Biotechnology (Birmingham, AL).
Proteins, Enzymes, and Cofactors.
Purified active CKIIα2 was from Invitrogen. PKC isoforms were from BIOMOL Research Laboratories/Enzo Life Sciences (Plymouth Meeting, PA), except for PKCα, which was from EMD Biosciences (San Diego, CA). The diacylglycerol (DAG) analog 1-oleoyl-2-acetyl-sn-glycerol (OAG) was purchased from BIOMOL Research Laboratories/Enzo Life Sciences. l-α-Phosphatidylserine was from Avanti Polar Lipids (Alabaster, AL). Purified integrin heterodimers lacking cytoplasmic domains were originally from Merck (Darmstadt, Germany) and have been described previously (Mehta et al., 1998). L1-ECD protein is described in the preceding section.
Inhibitors and Activators.
Purchased from EMD Biosciences unless otherwise indicated and are listed with the concentration at which they were used unless otherwise indicated in the text: calphostin C (CalC; EMD Biosciences; 500 nM, UV activated), bisindolylmaleimide I (BisI, Gö6850; EMD Biosciences; 5 μM), 4,5,6,7-tetrabromo-2-azabenzimidazole (TBB; EMD Biosciences; 50 μM), 2-Dimethylamino-4,5,6,7-tetrabromo-1H-benzimidazole (DMAT; EMD Biosciences; 10 μM), staurosporine (SS; EMD Biosciences; 100 nM), N-(R)-[2-(Hydroxyaminocarbonyl)methyl]-4-methylpentanoyl-L-naphthylalanyl-L-alanine, 2-aminoethyl amide (TAPI1; EMD Biosciences; 20 μM), okadaic acid (OA; MP Biomedicals, Solon, OH; 250 μM), calyculin A (CalA; Alexis Biochemicals, San Diego, CA; 5 μM), and phorbol 12-myristate 13-acetate (PMA; EMD Biosciences; 100 ng/ml).
Reagents.
Isopropyl β-d-thiogalactoside (IPTG), reduced glutathione, hematoxylin, fixatives, and other chemicals were from Thermo Fisher Scientific (Waltham, MA). The colorimetric substrate SureBlue 3,3′,5,5′-tetramethylbenzidine (TMB) was from Kirkegaard and Perry Laboratories (Gaithersburg, MD). Enhanced chemiluminescence (ECL) substrate PS-3 was from Lumigen (Southfield, MI).
Cell Assays
Inhibitor Treatments.
Cells were plated at 1.2 × 106 cells/35-mm culture dish and after 48 h received SF-media containing inhibitors at 10× concentration (for a final of 1×). At the indicated times, supernatant was removed and cells lysed on the plate in NP-40 lysis buffer (50 mM Tris, pH 7.4, 150 mM NaCl, and 1%NP-40) containing Complete Protease Inhibitor Cocktail (Roche Applied Science, Indianapolis, IN) supplemented with 10 mM phenylmethylsulfonyl fluoride (PMSF), 1 mM NaF, and 10 mM Na3VO4. For PKC washout analysis, cells were incubated for 90 min with CalC, before removal of media; two washes with SF-media; and further incubation for 5, 15, or 30 min in the presence or absence of PMA. Pervanadate treatment was as described previously (Gutwein et al., 2000).
L1-ECD Shedding Assay.
Cells were plated as described above and at 48 h received SF-media containing the indicated inhibitors. After 2 h, the supernatant was removed and clarified by centrifugation, and cells lysed on the plate with NP-40 lysis buffer. Equal volumes of supernatant and equal amounts of cell lysate were prepared for reducing immunoblot.
Cell Aggregation Assay.
J558L-L1 aggregation assays were performed as described previously (Nayeem et al., 1999). In brief, cells were washed with SF media and then incubated in SF-media for 24 h before another SF-media wash and application of 1.5 × 105 cells/well of a 24-well plate in SF-media containing 50 μg/ml 5G3 or Neuro4 mAbs, or an equal volume of phosphate-buffered saline (PBS; control). The plate was rotated for 30 min at 60 rpm on a horizontal orbital shaker at 37°C in a humidified chamber before photography and manual enumeration.
Cell Adhesion Assay.
Adhesion was as described previously (Silletti et al., 2000a).
Cell Migration Assay.
Haptotactic migration of CHO-K1 stable transfectants using 5-μm pore Transwell inserts was essentially as described previously (Mechtersheimer et al., 2001).
Immunoassays
Immunohistochemistry.
Samples were obtained under approved Institutional Review Board protocol from the UCSD Department of Pathology archives. Patient demographics and tissue characterization were published previously (Bouvet et al., 2000). Sections were deparaffinized, rehydrated, and endogenous peroxidases inactivated with 1% H2O2. Slides were quenched with 50 mM glycine, blocked with 2% equine serum/5% BSA, and renatured with Target Retrieval Solution (DAKO North America, Carpinteria, CA). Renatured sections were incubated with UJ127 or 2C2 in working buffer (0.2% equine serum, 0.5% BSA, and 5 mM glycine in PBS, pH 7.4), washed, and biotinylated α-mouse secondary antibody was applied according to the manufacturer's instructions for the VectaStain Elite ABC kit (Vector Laboratories, Burlingame, CA). Sections were washed and DAB chromagen was applied. Sections were counterstained with Gill's II hematoxylin and Blueing reagent, dehydrated, and then mounted.
Immunoprecipitation (IP).
Lysates were prepared from the indicated cells and treatments using phosphate lysis buffer (PLB; 10 mM NaPO4, 100 mM NaCl, 1% Triton X-100, 0.4% deoxycholate, and 0.1% SDS, pH 7.5) containing Complete Protease Inhibitor Cocktail supplemented with 10 mM PMSF, 1 mM NaF, and 10 mM Na3VO4, and incubated with the indicated antibody (2 μg) overnight on a rotator at 4°C. Protein L-Sepharose 4B beads (20 μl) were added, and tubes were rotated 2 h at 4°C. Beads were recovered by centrifugation and the supernatant was discarded. IPs were washed and prepared for reducing SDS-polyacrylamide gel electrophoresis (PAGE) and immunoblotting with the appropriate secondary or TrueBlot reagent to avoid cross-reactivity of the detection antibody with the precipitation antibody, as needed.
Immunoblotting.
Samples were separated by SDS-PAGE under reducing conditions and electroblotted to a polyvinylidene difluoride membrane. Membranes were blocked with 10% nonfat dry milk in Tris-buffered saline (TBS) containing 0.1% Tween 20 (TBS-T) and incubated 2 h to overnight in 2% milk/TBS-T with the indicated primary antibody. Primary antibody was detected with HRP-conjugated secondary antibody, and antibody complexes were visualized by ECL.
Enzyme-linked Immunosorbent Assay (ELISA)
Direct ELISA.
Glutathione transferase (GST) proteins were immobilized on 96-well microtiter plates and blocked with 0.5% gelatin before incubation with primary antibody. Wells were washed with TBS-T and incubated with HRP-conjugated secondary antibody. Antibody complexes were detected with the peroxidase substrate TMB. The reaction was stopped with 0.2 N HCl and absorbance was read at 450 nm. For titration of 2C2, wells coated with equal quantities of wild-type or T1172A protein were incubated with a serial twofold dilution of 2C2 from 1:500 to 1:32,000.
Competition Assay.
Performed as described above except that after blocking, the proteins were incubated in the presence or absence of 10 μg/ml 5G3 or Neuro4 (blocking antibody), washed with TBS-T, and reprobed with 1 μg/ml the reciprocal (detection) antibody. Bound detection antibody was visualized with HRP-conjugated α-mouse-IgG2a (5G3) or -IgG2b (Neuro4) and TMB. Isotype-specific secondary antibodies demonstrated no signal in wells that received only the blocking antibody.
Kinase Assay.
Recombinant proteins were coated and blocked as described above and then incubated with CKIIα2 in CKII buffer (20 mM Tris, 5% glycerol, 0.01% Triton X-100, 1 mM EGTA, 0.5 mM Na3VO4, 2 mM dithiothreitol (DTT), 10 mM MgCl2, 5 mM β-glycerophosphate, 0.4 mM ATP, and 0.05 mg/ml BSA) or with PKCα, -β1, -β2, -δ, or -ε in PKC buffer (20 mM HEPES for α, β1, δ, and ε or 25 mM 3-(N-morpholino)propanesulfonic acid for β2, supplemented with 10 mM MgCl2, 0.1 mM CaCl2, 0.03% Triton X-100, and 400 μM ATP) and additional supplements as follows: PKCα, -βI, -δ, or -ε all received 25 μg/ml OAG and 35 μg/ml l-α-phosphatidylserine, and in addition PKCβII, -δ, and -ε received 250 μM DTT, and PKCβII and PKCε further received 12.5 mM β-glycerophosphate. Reactions were performed for 30 min at 30°C. The presence of phosphate was assessed with the αP-T1172 pAb, the αP-S1181 pAb, or the 2C2 mAb, followed by the appropriate HRP-conjugated secondary antibody and colorimetric detection with TMB. β-Casien (1 μg/ml) or histone-H1 (1 μg/ml) were coated and treated alongside recombinant proteins as positive controls for enzyme activity. Activity on controls was assessed with αP-S/T-F pAb.
Capture ELISA.
For detection of T1172-phosphorylated L1 with the αP-T1172 pAb, biotinylated 5G3 was coated at 3 μg/ml on preblocked streptavidin-coated multiwell plates (Pierce Chemical/Thermo Fisher Scientific, Rockville, IL), before incubation overnight at 4°C with lysate from Panc1 cells treated with calyculin A (100 μg/well). Unbound protein was washed away with TBS-T, and some wells treated with 10 U/well calf alkaline phosphatase (AP) in AP buffer provided by the manufacturer (Roche Applied Science). Half of the AP-treated wells were subsequently treated with CKIIα2 as described above. Wells were incubated with αP-T1172 pAb before washing with TBS-T, detection of primary antibody with HRP-α-rabbit, and visualization with TMB/HCl. For detection of T1172-unphosphorylated L1 with the 2C2 mAb, unlabeled αECD pAb (0.1 μg/ml) was coated onto multiwell plates that were subsequently blocked with 0.5% gelatin before incubation overnight with 100 μg/ml calyculin A-treated Panc1 lysate and further treatment with AP and/or CKIIα2 as described above. Unphosphorylated captured L1 was then detected with 2C2 and HRP-α-mouse and visualized with TMB/HCl as described above.
Integrin-Binding Assay
Purified αvβ3 and αvβ5 integrin heterodimers were adsorbed at 1–5 μg/ml in TBS supplemented with 0.4 mM MnCl2 (TBS/Mn) onto 96-well plates. Wells were blocked with 0.5% BSA in TBS. Purified L1-ECD protein, His-FN3, or biotinylated vitronectin was added at 1–2 μg/ml in TBS/Mn containing 0.5% BSA to the coated wells and incubated for 90 min at 37°C. Bound L1-ECD was detected with 10 μg/ml 5G3, αECD pAb, or αFL pAb. Bound FN3 domain was detected with αHis, and bound vitronectin was detected with α-biotin-HRP. Primary antibodies were detected with HRP-conjugated secondary and visualized with TMB. Data are corrected for background binding of antibodies to integrin-coated wells (lacking L1-ECD, FN3-His, or biotinylVN) or to L1-ECD or biotinylVN in wells lacking integrin, whichever was greater.
Fluorescence-activated Cell Sorting (FACS)
FACS analysis was performed on a FACScalibur (BD Biosciences Discovery Labware, Bedford, MA) at the Moores UCSD Cancer Center Flow Cytometry Shared Resource. Cells were harvested with 0.1% trypsin/versene, inactivated with 0.1% soybean trypsin inhibitor, and resuspended in FACS buffer (1 mM MgCl2, 1 mM CaCl2, 0.1% NaN3, and 0.5% BSA in PBS, pH 7.4) and sequentially labeling with primary and FITC-labeled secondary antibodies in the dark and on ice. Gates were set with secondary alone, and 5 μg/ml propidium iodide was included to exclude dead and dying cells.
Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
cDNA was synthesized from 1 μg of total RNA from cells in standard culture using oligo(dT) primer. PCR was performed on 1 μl of total cDNA. PKC isoform transcript analysis was performed using previously described primers (Holden et al., 2008). L1 analysis was performed with primers specific for the region flanking alternatively spliced exon 2 (Table 1). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) primers were from Stratagene (San Diego, CA) and served as internal controls.
Construction and Expression of L1 Fusion Proteins
It is important to address the fact that there is some discord in the literature regarding the use of bacterial versus eukaryotic sources of recombinant proteins for the study of L1. Although the limitations of bacterial proteins are obvious (i.e., lack of eukaryotic post-translational modification), bacterial Ig1-6 actually functioned better than CHO-derived Ig1-6 previously (Holm et al., 1995), and Haspel et al. (2001) found that their inability to reproduce data from bacterial proteins reported by Zhao and Siu (1995) was due to the inappropriate folding of their L1 constructs containing less than the first four Ig domains. Therefore, our use of bacterial proteins for mapping of the Ig domains is warranted and probably relevant to the native molecule. Moreover, our mapping studies are assessed with reference to studies using the L1-ECD protein produced in HEK293 cells, as well as studies of L1 expressed by cells.
GST and 6xHis-tagged proteins were described previously or were produced as described previously (Silletti et al., 2000a). All but three new recombinant L1 fusion proteins were generated by PCR amplification of necessary coding sequences using the appropriate combination of oligonucleotide primers. Primer sequences have been published previously (Silletti et al., 2000a) or are shown in Table 1. pGEX GST fusion vectors or pProEX 6xHis-tag vectors were selected based upon required reading frame and restriction sites. The vector encoding GST/L11144-1186 was created by restriction digestion of the pGEX-6P1/nonneuronal L1 CD vector with StuI (internal to insert) and SmaI (3′ on the vector) to remove the region encoding L1 1187-1257. Religation of the blunt-cut vector yielded the appropriate coding sequence. The vector encoding GST/L11144-1168 was created by PCR of pGEX-6P1/L1NN by using the 5′pGEX sequencing primer and L1-1168stop(R). The product was digested with EcoRI and inserted into the EcoRI site of pGEX-6P1. GST/L11169-1186 was created by ligation of annealed and phosphorylated mini-exon primers (NN 1169-1186) into the EcoRI site of pGEX 6P-1. Site-directed mutagenesis was as described previously (Silletti et al., 2000a). Mutagenesis primer sequences and the mutations they introduce are shown in Table 1. All constructs were confirmed by dideoxy sequencing at the Moores UCSD Cancer Center DNA Sequencing Shared Resource.
Image Acquisition and Manipulation
Images of ethidium bromide-stained agarose gels were captured with Quantity One software on a Gel Doc XR (Bio-Rad Laboratories, Hercules, CA) by using the appropriate filter and transmitted UV light. Chemiluminescence-exposed films and printouts of agarose gels were scanned on an EpsonPerfection 4490Photo flatbed scanner. Images were imported into Photoshop (Adobe Systems, Mountain View, CA) for removal of unused levels and cropping. Minimal alterations to brightness and contrast were used for a subset of images, to improve the visual nature of the image. Nonlinear adjustments were not used. Immunohistochemical images were acquired as 24-bit RGB (.tif) and phase-contrast images as 8-bit grayscale (.tif) by using SpotBasic with TE6000-S or TE300 microscopes (Nikon, Tokyo, Japan), respectively, fitted with Model 3.2.0 charge-coupled device cameras (Diagnostic Instruments, Sterling Heights, MI) and used at the Moores UCSD Cancer Center Microscopy Shared Resource. Final images were compiled in InDesign (adobe Systems), rasterized, and converted to jpeg format using Photoshop (Adobe Systems) at a minimum resolution of 300 dpi.
Statistics
Assays were repeated at least two times. Data shown is mean ± SD unless otherwise indicated. Antibody binding and cell aggregation inhibition differences were analyzed by two-tailed Student's t test.
RESULTS
The αL1 mAb 2C2 Does Not Recognize L1 Detected by UJ127 in PDAC Cells
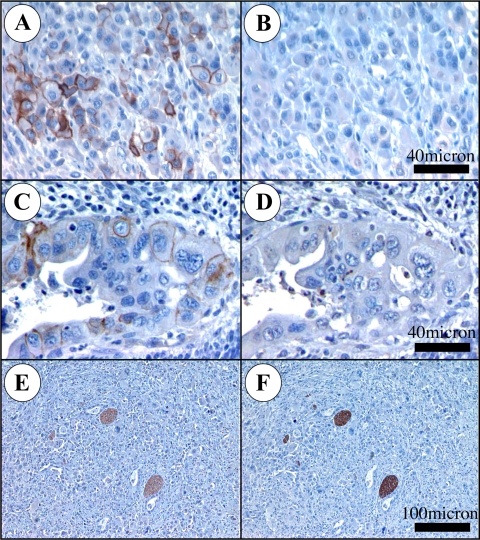
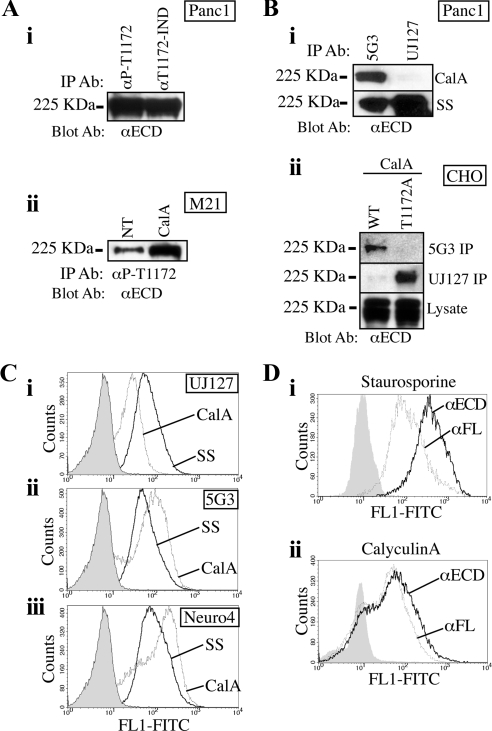
L1 expression in PDAC has been reported as absent (Kaifi et al., 2006) and high (Muerkoster et al., 2007). This latter report found L1 in G2 and G3 tumors exclusively. We analyzed 92 human patient samples with the UJ127 mAb and found that L1 is detectable in poor and undifferentiated PDAC cells (Figure 1, A and C). However, immunohistochemistry of UJ127-positive PDAC tumor sections (N = 8) using the 2C2 mAb directed toward the L1 cytoplasmic domain (CD) (Figure 1, B and D) demonstrated that 2C2 did not recognize the PDAC L1 detected by UJ127 (Figure 1, A and C), even though it was capable of detecting UJ127-positive nerves in the same sections (Figure 1, E and F). In vitro, we noted a similar lack of 2C2 reactivity in L1-positive MIAPaCa2 and Panc1 PDAC cells (Figure 2A). In contrast, PC12 cells differentiated with nerve growth factor into somatic neuron-like cells demonstrated strong 2C2 signal, as did M21 and M24met human melanoma cells.
Figure 1.
αL1 mAb 2C2 does not detect L1 expressed by PDAC cells in situ. Sections were stained with UJ127 (A, C, and E) or 2C2 (B, D, and F) (brown). Very poorly differentiated (anaplastic) PDAC cells are UJ127 positive (A), 2C2-negative (B). Poorly differentiated (G3) cells of a malignant duct are UJ127 positive (C), 2C2 negative (D). Nerve bundles from the same section as A and B are UJ127 positive (E), 2C2 positive (F).
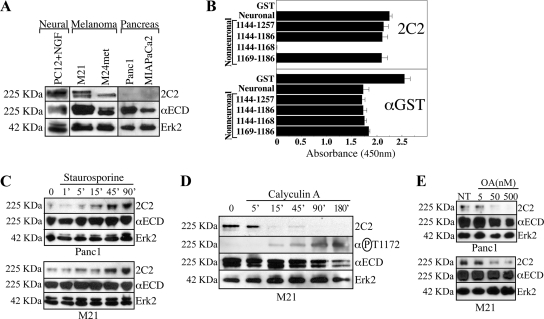
Figure 2.
Mapping the 2C2 epitope. (A) Immunoblot of the indicated cells with 2C2, αECD, and αErk2. (B) ELISA of GST proteins encoding the indicated sequences with 2C2 or αGST. (C) Cells were treated with SS and immunoblotted with 2C2, αECD, and αErk2. (D) M21 cells treated with CalA were immunoblotted with 2C2, αECD, and αErk2. The blot was reprobed with the αP-T1172 pAb. (E) Cells were treated for 90 min with the indicated concentrations of OA and immunoblotted with 2C2, αECD, and αErk2.
L1 contains two alternatively spliced small exons, one exon N-terminal and one exon in the CD. Both exons are present in the neuronal isoform and absent from the nonneuronal isoform (Hlavin and Lemmon, 1991). RT-PCR and sequencing verified the expression of the nonneuronal isoform by Panc1 and MIAPaCa2 cells, and sequencing also verified that the L1 CD coding sequence was intact (data not shown). Therefore, because PC12 cells are neural differentiated and melanoma cells are neural crest derived, and because 2C2 detects nerves in situ, the possibility existed that 2C2 only recognizes the neuronal isoform of L1. Using recombinant proteins, we found that 2C2 binds the neuronal and nonneuronal L1 CD equivalently, and in addition that the epitope for 2C2 lies between amino acids 1169 and 1186 (Figure 2B). Because the L1 CD does not contain consensus motifs for intracellular proteases (e.g., calpain), we reasoned that 2C2's lack of reactivity was probably due to a posttranslational modification such as phosphorylation. L11169-1186 contains one threonine (T1172), one tyrosine (Y1176), and one serine (S1181). Although cellular phosphorylation of Y1176 and S1181 has been demonstrated previously (Wong et al., 1996; Schaefer et al., 2002), T1172 phosphorylation has not. Previously, we demonstrated that mutation of Y1176 and S1181 does not affect 2C2 binding (Chen et al., 2009); however, a size-enhancing (T1172F) or negative charge-imparting (T1172E) mutation at 1172 abrogated 2C2 binding, whereas alanine substitution had only partial effect (Chen et al., 2009). Thus, although T1172 may not be an integral component of the 2C2 epitope, our data indicate that the addition of a large, charged phosphate to T1172 would prevent 2C2 binding to L1.
Phosphorylation of T1172 of L1 Is Responsible for Lack of 2C2 Reactivity
Panc1 PDAC or M21 melanoma cells were treated with the S/T kinase inhibitor SS, which caused a time-dependent increase in 2C2 reactivity in both cell types (Figure 2C). Because Panc1 cells demonstrate low basal 2C2 reactivity, M21 cells were reciprocally treated with the S/T phosphatase inhibitor CalA (Figure 2D). CalA caused a time-dependent loss of 2C2 signal, consistent with increased T1172 phosphorylation resulting from phosphatase blockade. Pervanadate inhibition of tyrosine phosphatases had no effect (data not shown). To demonstrate that phosphorylation of T1172 is indeed responsible for the loss of 2C2 signal, we produced a new rabbit α-phospho-T1172 pAb (αP-T1172). This affinity-purified antibody demonstrated a reciprocal pattern to 2C2 (Figure 2D), demonstrating that the loss of 2C2 signal in these samples indeed seems to be the result of T1172 phosphorylation. To elucidate the phosphatase responsible for T1172 dephosphorylation in these cells, M21 and Panc1 cells were treated with OA (Figure 2E), which differentially inhibits PP2A (IC50 = 100 pM), PP1 (IC50 = 10–15 nM), and PP2B (IC50 = 5 μM) (see Calbiochem Inhibitor Sourcebook, teachline.ls.huji.ac.il/72682/Booklets/CALBIOCHEM-Inhibitors.pdf, p. 36). Based on the spectrum of effect (50 nM), PP1 seems to be the main phosphatase responsible for dephosphorylating T1172 in both cell types.
The PhosphoMotif Finder program of the Human Protein Reference Database (www.HPRD.org) identified three kinases with a consensus motif appropriate for the region flanking T1172 (KDET1172FGE): CKII, protein kinase A (PKA), and the PKC family. Consistent with a role for PKC in regulating this event in PDAC cells, the PKC inhibitor CalC promoted an increase in 2C2 reactivity exclusively in Panc1 cells (Figure 3A). The CalC concentration used in these studies would inhibit all conventional (α, βI, βII, and γ) and novel (δ, ε, μ, and η) PKC isozymes through competitive binding of their DAG-binding sites (see Calbiochem Inhibitor Sourcebook, teachline.ls.huji.ac.il/72682/Booklets/CALBIOCHEM-Inhibitors.pdf, pp. 20–24). However, the SS concentration used previously would have inhibited only PKCα, -βI, -βII, -γ, -δ, and -ε (Calbiochem Inhibitor Sourcebook, teachline.ls.huji.ac.il/72682/Booklets/ CALBIOCHEM-Inhibitors.pdf, pp. 20–24). In addition, BisI, which competitively binds the ATP-binding site of select PKC isozymes (α, βI, δ, ε, and ζ), increased 2C2 reactivity in Panc1, but not M21 cells (Figure 3B), demonstrating that the effect of CalC is not artificial and that PKC-dependent regulation of T1172 phosphorylation in Panc1 cells is mediated by a subset of conventional (α and βI) or novel (δ and ε) PKC isoforms, because the BisI concentration used in these studies would not affect PKCζ (see Calbiochem Inhibitor Sourcebook, teachline.ls.huji.ac.il/72682/Booklets/CALBIOCHEM-Inhibitors.pdf, pp. 20–24). In support of this contention, 2C2 reactivity is lost more quickly and completely after washout of CalC in Panc1 cells treated with PMA (Figure 3C), which stimulates conventional and novel PKC isoforms by binding to their DAG-binding sites.
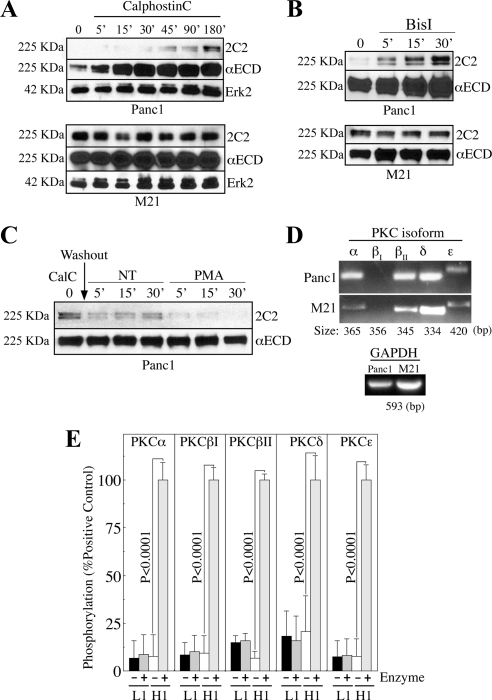
Figure 3.
PKC is involved in regulating 2C2 epitope availability but is incapable of phosphorylating T1172 of recombinant L1 CD in isolation. (A and B) Cells were treated with the PKC inhibitors CalC (A) or BisI (B) and immunoblotted with 2C2, αECD, and αErk2. (C) Cells were treated with CalC for 90 min before washout and replenishing of basal media or media containing the PKC-activating phorbol ester PMA. Lysates were immunoblotted with 2C2 and αECD. (D) RT-PCR with PKC isoform-specific primers. GAPDH, control. (E) GST-L1 nonneuronal CD (L1) or histone-H1 were incubated with PKC isoforms. T1172 phosphorylation and H1 phosphorylation were detected with αP-T1172 and αP-S/T-F pAbs, respectively.
These data demonstrate the potential involvement of PKC in the regulation or direct catalytic modification of T1172 of L1 in Panc1 cells. However, T1172 phosphorylation was not affected by PKC blockade in M21 cells. Therefore, because PKC expression is cell type dependent, the possibility existed that differential isoform expression by Panc1 and M21 cells could be responsible for the differential inhibitor sensitivity observed, thus providing a potential means to identify the PKC isoform involved in this modification in Panc1 cells. Surprisingly, RT-PCR demonstrated that all relevant Panc1 PKC isoforms are expressed at similar transcript levels in M21 cells (Figure 3D). To assess the ability of the implicated PKC isozymes to phosphorylate T1172 in vitro, we performed kinase assays with recombinant L1 nonneuronal CD or histone-H1 as control. Purified active PKCα, -βI, -βII, -δ, and -ε isozymes failed to phosphorylate T1172, as measured by lack of binding of the αP-T1172 pAb (Figure 3E). 2C2 binding was also unaffected by kinase treatment (data not shown), confirming the αP-T1172 pAb data. All kinases phosphorylated histoneH1 on the same plate, however, demonstrating that kinase activity per se is not responsible for the lack of T1172 phosphorylation observed.
Somewhat surprisingly, the CKII inhibitor DMAT also caused a time-dependent increase in 2C2 reactivity in Panc1 cells (Figure 4A), commensurate with dephosphorylation of T1172. Similar results were obtained with the DMAT analog TBB (data not shown). Analogous to the effect of PKC inhibitors, however, neither DMAT (Figure 4A) nor TBB (data not shown) significantly affected 2C2 reactivity in M21 cells; although an effect on 2C2 reactivity was noted at some time points in some experiments, a consistent and reproducible effect could not be achieved after numerous attempts including combination of TBB and DMAT, and the effects observed seem to instead represent fluctuations in the levels of 2C2 signal in control cells. Indeed, even the combination of DMAT and BisI did not affect 2C2 reactivity in M21 cells (data not shown). Therefore, we concluded that CKII is probably not involved in regulating T1172 phosphorylation in M21 cells. In contrast to the lack of effect of the purified PKC isoforms in an in vitro kinase assay, CKII promoted significant αP-T1172 pAb signal on both neuronal and nonneuronal L1 CDs (Figure 4B). Alanine substitution of T1172 abrogated CKII-mediated αP-T1172 binding to both isoforms (Figure 4B), and CKII also attenuated 2C2 binding to both isoforms (Figure 4C), confirming the phosphorylation of T1172 in this format and demonstrating the negative effect of T1172 phosphorylation on 2C2 binding. Because CKII promoted a more than threefold higher signal increase on the nonneuronal isoform of L1, the integrity of the neuronal isoform was verified by measuring S1181 phosphorylation in parallel, by using an αP-S1181 pAb, and abrogation of signal by S1181A mutation (Figure 4B). Furthermore, to directly demonstrate the negative effect of T1172 phosphorylation on 2C2 binding, maximally phosphorylated L1 from CalA-treated Panc1 cell lysates was captured on immobilized anti-L1 antibodies. This immunocaptured L1 was then treated with alkaline phosphatase with or without subsequent treatment with purified active CKII. As expected, untreated captured L1 demonstrates low 2C2 signal and strong reactivity with the αP-T1172 pAb (Figure 4D). In contrast, phosphatase treatment promotes a significant increase in 2C2 reactivity commensurate with an almost complete eradication of αP-T1172 pAb binding. This pattern is reversed by subsequent treatment of the immunocaptured L1 with CKII (Figure 4D).
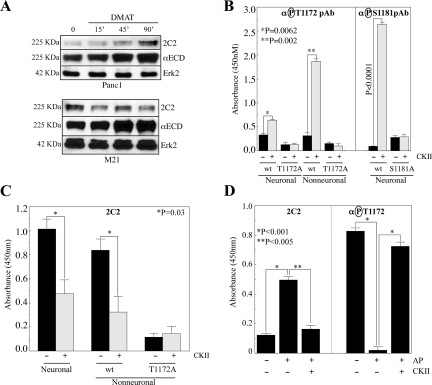
Figure 4.
CKII phosphorylates T1172 of the L1 CD and phosphorylation of T1172 is responsible for loss of 2C2 signal. (A) Cells treated with the CKII inhibitor DMAT were immunoblotted with 2C2, αECD, and αErk2. (B and C) GST-L1 CD proteins were incubated with CKII. T1172 phosphorylation was detected with αP-T1172 (B) or 2C2 (C). T1172A, phosphorylation control. Neuronal protein integrity was verified by assessing S1181 phosphorylation with αP-S1181 (B). (D) L1 from CalA-treated Panc1 cell lysates was captured on immobilized anti-L1 antibodies before treatment with alkaline phosphatase with or without subsequent treatment with CKII as described in Materials and Methods. Bound L1 was then detected with 2C2 or the αP-T1172 pAb.
L1 Phosphorylation State Is Representative of Distinct L1 Conformations on the Cell Surface
To verify both the presence of T1172-phosphorylated L1 in Panc1 cells in culture and the regulation of this phosphorylation event by inhibitor treatments, IP/immunoblotting was performed. L1 was IPed from Panc1 cells by using either the αP-T1172 pAb or the phospho-independent (αT1172-IND) pAb, which recognizes the region around T1172 irrespective of the T1172 phosphorylation state (Chen et al., 2009). Equal quantities of L1 were IPed by both antibodies (Figure 5Ai), demonstrating that the majority of L1 is T1172-phosphorylated in Panc1 cells in culture. Because Panc1 L1 is so highly T1172-phosphorylated in standard culture, M21 cells were treated with CalA to accumulate maximal S/T-phosphorylation and then subjected to IP with the αP-T1172 pAb. Although αP-T1172 pAb IPed L1 from standard culture, significantly more L1 was IPed from M21 cells treated with CalA (Figure 5Aii), demonstrating the accumulation of T1172-phosphorylated protein and corroborating the loss of 2C2 signal and reciprocal gain of αP-T1172 pAb signal shown in Figure 2D.
Figure 5.
Cytoplasmic S/T phosphorylation promotes or accompanies changes in the L1 ECD conformation. (Ai) L1 was IPed from Panc1 cells by using the αP-T1172 or T1172-independent (T1172-IND) pAb. (Aii) L1 was IPed from untreated or CalA-treated M21 cells with the αP-T1172 pAb. (B) Panc1 (Bi) or CHO cells stably expressing wild-type noneuronal L1 or L1 containing a T1172A mutation (Bii) were treated with CalA or SS and processed for IP with 5G3 and UJ127. IPs were immunoblotted with αECD. (C and D) Panc1 cells were treated with CalA or SS and processed for FACS with UJ127 (Ci), 5G3 (Cii), Neuro4 (Ciii), αECD (D, i and ii), or αFL (D, i and ii). FACS controls received secondary alone (solid).
While assessing IP patterns, we noted that UJ127 and 5G3 were differentially affected by the phosphorylation state of L1. To assess the effect of phosphorylation on antibody binding, we used lysates from CalA- and SS-treated Panc1 cells, which contain maximally phosphorylated or unphosphorylated L1, respectively. The ability of UJ127 to IP L1 from CalA-treated Panc1 lysates was markedly lower than that of 5G3, even though UJ127 IPed more L1 from SS-treated Panc1 lysates than 5G3 (Figure 5Bi). These data suggest that S/T phosphorylation of the L1 CD promotes, or is associated with, a change in extracellular epitope availability, and by extension conformation, of the L1 ECD, either directly or indirectly. To assess the involvement of T1172 in this phenomenon, 5G3 and UJ127 IPs were performed from lysates of CHO-K1 cells stably expressing wild-type nonneuronal L1 (CHO/L1WT) or nonneuronal L1 containing a T1172A mutation (CHO/L1T1172A). As can be seen in Figure 5Bii, 5G3 IPs significant L1 from calA-treated CHO/L1WT cells, whereas 5G3 is largely incapable of IPing L1 from CalA-treated CHO/L1T1172A cells, which are incapable of being phosphorylated on T1172. Reciprocally, UJ127 IPed significant quantities of L1 from CalA-treated CHO/L1T1172A cells but was largely incapable of IPing L1 from CalA-treated CHO/L1WT cells, which contain maximally T1172-phosphorylated L1. To determine whether this differential epitope availability was limited to the detergent conditions of IP, live Panc1 cells were analyzed by FACS. This analysis demonstrated that UJ127 signal is dramatically higher in SS-treated cells than CalA-treated cells (Figure 5Ci), whereas 5G3 binding is reciprocally higher in CalA-treated cells than SS-treated cells (Figure 5Cii), similar to what was observed by IP. Because 5G3 reactivity is dependent on folding and the integrity of disulfide bonds, whereas UJ127 is not, we also used an αL1 mAb that recognizes a linear epitope in the membrane-distal Ig domains, Neuro4. Similar to 5G3, the Neuro4 epitope is more exposed in CalA-treated, and thus L1-phosphorylated conditions (Figure 5Ciii), thereby establishing that the differential binding of UJ127 and 5G3 are due to the location of, rather than the nature of, their respective epitopes.
To further demonstrate that this epitope masking/unmasking is not an artifact of the monospecific nature of these mAbs, we used pAbs generated against different L1 sources that recognize different L1 conformations. The αECD pAb was produced against recombinant L1 ECD lacking transmembrane and cytoplasmic domains (Stallcup, 2000). In contrast, the αFL (full length) was produced against L1 purified from human neuroblastoma cells by immunoaffinity chromatography on a 5G3 mAb column. As a result, this pAb seems to recognize the conformation recognized by the 5G3 mAb, albeit a larger number of epitopes. In this case, SS treatment promoted dramatic up-regulation of αECD reactivity over αFL reactivity (mean fluorescence intensity 444.7 vs. 149.8) (Figure 5Di). In a partially reciprocal manner, CalA reduced αECD binding to approximately the same level as the αFL pAb (Figure 5Dii). It should be noted that, due to the high level of basal T1172 in untreated Panc1 cells, results using untreated cells were similar to the CalA-treated cells in these assays (data not shown).
αL1 Epitope Specificities Identify Folding Characteristics of Distinct L1 ECD Conformations
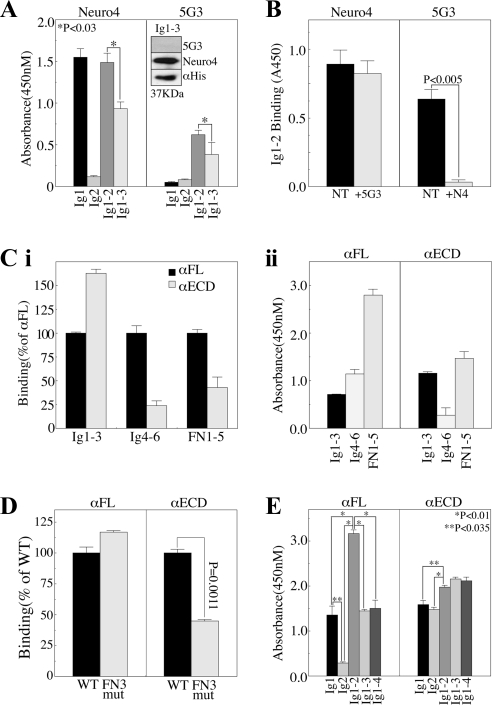
Data presented in Figure 5 suggest the presence of at least two conformations of L1 on the cell surface, and the potential regulation of ECD conformation by factors driven by, or associated with the L1 intracellular phosphorylation state in general, and that of T1172 phosphorylation in particular. Furthermore, these data demonstrate the sensitivity of αL1 antibodies to epitope masking as a result of these conformational changes. An alternative explanation would be that protein-protein interactions are responsible, in whole or in part, for the differential epitope availability noted in Figure 5. If this were the case, the binding of these antibodies should be identical in various bacterial recombinant proteins containing their respective epitopes. To test this, we performed epitope mapping studies. First, we confirmed that the epitope of 5G3 is disulfide stabilized by showing loss of reactivity against reduced antigen, and we identified the linear epitope of the Neuro4 mAb as being within the first three Ig domains of L1 (Figure 6A, inset). Further analysis demonstrated that the Neuro4 epitope is in the first Ig domain of L1 (Figure 6A). 5G3, in contrast, did not bind either Ig1 or Ig2 in isolation, but it did bind a protein encoding both domains (i.e., Ig1-2) (Figure 6A), consistent with the previously reported loss of 5G3 binding in recombinant L1 lacking the second half of Ig1 and first half of Ig2 (Dahlin-Huppe et al., 1997). Although Neuro4 also bound Ig1-2 equivalent to its binding of Ig1 alone, incorporation of the adjacent Ig3 domain caused a significant reduction of Neuro4 binding. A similar effect was observed for 5G3, whereby binding to Ig1-3 was less than that to Ig1-2. These data suggest that Ig3 exerts a steric effect on Ig1-2, partially obscuring the 5G3 and Neuro4 epitopes. Further inclusion of Ig4 did not increase this effect (data not shown). Importantly, Neuro4 did not recognize an Ig1 construct lacking the final three amino acids (AEG115) (data not shown), thus mapping its epitope to the region immediately proximal to Ig2 and directly in the middle of the putative 5G3 epitope. Therefore, the possibility existed that Neuro4 and 5G3 might have overlapping epitope requirements and/or be competitive. Indeed, pretreatment of Ig1-2 with Neuro4 completely blocked 5G3 binding, although the opposite was not true, as 5G3 pretreatment had no effect on Neuro4 binding (Figure 6B).
Figure 6.
ECD conformation regulates the availability of αL1 antibody epitopes. Immobilized GST proteins encoding the indicated domains were incubated with the indicated antibodies. (A) Neuro4 or 5G3 detection of Ig domains. Inset, reducing immunoblot of Ig1-3 with 5G3, Neuro4, or αHis. (B) Competition assay: Ig1-2 was incubated with or without 10-fold excess of Neuro4 or 5G3 and then probed with the opposite antibody and isotype-specific secondary antibody. (C) Domain mapping of αFL and αECD. (Ci) Relative binding of the αECD plotted as percent of αFL binding to the indicated fusion proteins. (Cii) Relative binding of each pAb to the indicated fusion proteins. (D) Multimerization analysis: wild-type FN3 domain (WT) or FN3 domain harboring the dibasic C-C′ loop mutation (FN3 mut) that impairs multimerization (Silletti et al., 2000a) were probed with αFL or αECD. (E) Sequential domain addition to map Ig domain interactions using αFL or αECD.
To elucidate the specifics of the αFL and αECD pAbs, we performed epitope mapping with recombinant proteins encoding Ig1-3, Ig4-6, and FN1-5. αECD recognizes Ig1-3 better than αFL, whereas αFL recognizes the other domains better than αECD (Figure 6Ci). From the standpoint of each individual antibody, αFL recognizes the FN1-5 region most strongly, with 2.5-fold more signal than that observed for Ig4-6, and 3.6-fold more signal than observed on Ig1-3 (Figure 6Cii). In contrast, αECD recognizes FN1-5 and Ig1-3 almost equally and demonstrates dramatically (6-fold) less reactivity with Ig4-6. These data clearly demonstrate that the conformation of L1 used as immunogen for these two pAbs differed significantly, leading to the presentation of different epitopes to the host immune systems.
One major conformational difference that could be responsible for obscuring large regions of the L1 ECD is the multimerization that has been described as a means of regulating integrin binding (Silletti et al., 2000a). To assess whether these pAbs recognize the monomeric and multimerized forms of L1 equivalently, we assessed their recognition of wild-type FN3 domain, or FN3 domain harboring the C-C′ loop mutation that abrogated multimerization as well as integrin binding and cell adhesion previously (Silletti et al., 2000a). Although the C-C′ loop mutation slightly increased αFL binding, αECD reactivity was dramatically (65%) reduced by this mutation (Figure 6D), consistent with the presence of monomeric L1 in the αFL immunogen, and multimerized L1 in the αECD immunogen.
Another conformational change that could affect epitope presentation is the adopting of the horseshoe structure proposed for the distal four Ig domains of L1 (Schürmann et al., 2001). To assess whether the proposed looping of the first four Ig domains affects the binding of these pAbs, ELISA was performed. αFL recognizes Ig1 individually but not Ig2 (Figure 6E). When combined (i.e., Ig1-2), the reactivity of this pAb almost triples, suggesting the exposure of cryptic epitopes by the interactions between Ig1 and Ig2. Similar to what was observed with Neuro4 and 5G3, however, the addition of Ig3 (and Ig4) actually caused a reduction in signal to the level observed with Ig1 alone. In contrast, αECD recognized both individual Ig1 and Ig2 domains equally, and combination of the domains (Ig1-2) only marginally increased signal beyond that observed with the individual domains, suggesting obscuring of epitopes exposed in the original immunogen by interactions between Ig1 and Ig2. Further addition of Ig3 or Ig4 did not increase αECD signal. These data, and those presented in Figure 6A, are consistent with a model whereby Ig1 and Ig2 interact in absentia of the other Ig domains to form a “mini-loop” that is destabilized by inclusion of adjacent Ig domains (Ig3 and Ig4).
S/T Phosphorylation Reflects the Proteolysis and Integrin-Binding Capability of the L1 ECD and Regulates Cell Migration
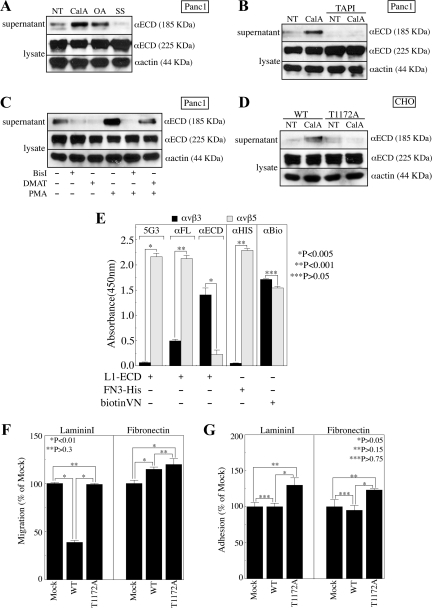
We noted a trend toward reduced cellular L1 levels in Panc1 cells treated with S/T-phosphatase inhibitors (Figure 2, D and E), and a reciprocal trend toward increased cellular L1 levels in Panc1 cells treated with S/T-kinase inhibitors (Figures 2C and 3, A and B). Densitometry verified that these changes did not fully account for the changes in 2C2 reactivity, and these changes in total L1 levels were not observed in experiments where 2C2 reactivity was not affected, establishing at least partial specificity for the effect with regard to T1172. Thus, intracellular S/T phosphorylation events including T1172 could be at least partly responsible for ECD changes that regulate proteolysis and shedding of L1 ECD from PDAC cells. In support of this contention, treatment of Panc1 cells with either CalA or OA resulted in significant shedding of the 185-kDa L1 ECD proteolysis product (Figure 7A). Reciprocal treatment of Panc1 cells with SS suppressed shedding of this fragment. Importantly, CalA-induced ECD shedding was abrogated by the matrix metalloproteinase (MMP) inhibitor TAPI1 (Figure 7B), which blocks the ADAMs-mediated release of L1 from the surface of other cells (Beer et al., 1999; Gutwein et al., 2000). It should be noted that the proteolysis described here is not responsible for the altered 5G3 and Neuro4 binding in Figure 5C, because CalA caused an increase in reactivity of these membrane distal-specific antibodies, and ADAMs proteolysis would remove essentially the entire L1 ECD. Potential proteolytic differences may stem from the fact that the shedding assays are performed in SF-media, whereas FACS treatments are performed in culture media containing FBS.
Figure 7.
Regulation of L1 proteolysis and integrin-binding by CD phosphorylation and ECD conformation. (A and B) Panc1 cells were treated with CalA, OA, or SS in the presence or absence of TAPI1 (TAPI). Conditioned media and cell lysate were immunoblotted with αECD or αactin. (C) Panc1 cells were treated with DMAT or BisI in the presence or absence of PMA. Conditioned media and cell lysate were immunoblotted with αECD or αactin. (D) CHO-K1 cells stably expressing wild type (WT) or T1172A mutant (T1172A) nonneuronal L1 were treated with CalA and immunoblotted with αECD or αactin. (E) Solid phase integrin capture assay of αvβ3 and αvβ5 binding to soluble L1-ECD, FN3 domain (FN3-His), or biotinylated vitronectin (biotinVN) as detected with 5G3, αFL, αECD, αHis, or αbiotin (αBio). (F and G) LamininI or fibronectin haptotactic migration (F) or adhesion (G) of mock-transfected (Mock) or CHO-K1 cells stably expressing wild-type nonneuronal (WT) or T1172A mutant (T1172A) nonneuronal L1.
Previously, it was shown that PMA induces shedding of the L1 ECD in an ADAMs-mediated manner in melanoma cells (Beer et al., 1999), Moreover, PMA-induced ADAMs-mediated L1 ECD shedding from breast cancer cells is inhibited by SS and BisI (Gutwein et al., 2000). Therefore, to examine the relative roles of CKII and PKC in regulating the ADAMs-mediated shedding of L1 ECD from Panc1 cells, cells were treated with DMAT or BisI in the presence or absence of PMA. Both DMAT and BisI partially inhibited basal L1 ECD shedding (Figure 7C). PMA induced dramatic shedding of the L1 ECD by Panc1 cells, and this induced shedding was fully suppressed by BisI. Importantly, DMAT partially inhibited shedding from PMA-treated cells, suggesting that both enzymes are involved in this process even under the extreme PKC stimulation of PMA.
To directly assess the role of T1172 in L1 ECD shedding, we examined our stable wild-type nonneuronal L1-expressing L1WT, or T1172A mutant nonneuronal L1-expressing L1T1172A CHO-K1 cells, which have been shown previously to shed ectopically expressed neuronal L1 (Mechtersheimer et al., 2001). Although both CHO/L1WT and CHO/L1T1172A cells expressed similar levels of L1, CHO/L1WT cells shed significantly more L1 ECD in response to CalA treatment than the CHO/L1T1172A cells (Figure 7D). These data, coupled with the fact that cellular Panc1 L1 levels were only affected by inhibitor treatments that affected 2C2 reactivity, clearly demonstrate that at least part of the phospho-S/T-dependent regulation of L1 proteolysis is due to T1172.
Regulation of protein–protein interaction by extracellular conformation and proteolysis has been suggested to regulate the interactions of L1 with itself (Hall et al., 2000; Haspel et al., 2000; Gouveia et al., 2008), with heterophilic binding partners (De Angelis et al., 2001) and with integrins (Silletti et al., 2000a; Mechtersheimer et al., 2001; Thelen et al., 2002). Thus, the functional significance of the differential antibody reactivity noted in Figure 5 could extend to the regulation of protein-binding interactions on the cell surface. To investigate this possibility, a purified integrin-binding assay was performed using recombinant L1-ECD. The species of L1-ECD that is bound by immobilized integrin αvβ3 is not detectable by 5G3, whereas that bound by integrin αvβ5 is readily detectable by 5G3 (Figure 7E). Similar to this differential recognition by 5G3, the αFL pAb generated against L1 purified on a 5G3 column also detects the αvβ5-bound species more than fourfold better than the αvβ3-bound species, demonstrating that this effect is not limited to the masking of the single mAb epitope. Importantly, the αECD pAb detects the species of L1 bound by αvβ3 more than sevenfold better than that bound by αvβ5. The integrin-binding motifs of L1 are in Ig6 and FN3 (Felding-Habermann et al., 1997; Silletti et al., 2000a) and do not overlap directly with the 5G3 epitope. Therefore, these differences in epitope availability are likely due to folding parameters associated with conformational states of the L1 protein similar to those proposed as responsible for differential epitope exposure shown in Figure 5.
Previously, it was shown that integrins α5β1 and αvβ5 interact with L1 on the cell surface in a manner requiring the RGD sequence in Ig6 (Mechtersheimer et al., 2001). Therefore, because binding of the αECD pAb to purified FN3 domain was compromised by the C-C′ loop mutation shown previously to abrogate binding of the αvβ3 integrin (Figure 6D), and because the FN3 domain supports integrin binding and cell adhesion in a manner dependent on the folding of the adjacent FN domains (Silletti et al., 2000a), we questioned whether immobilized αvβ3 and αvβ5 are capable of binding L1 through the multimerized FN3 domain. Interestingly, the multimeric FN3 preparation is not bound by immobilized αvβ3, but it is significantly captured by immobilized αvβ5 (Figure 7E). Neither integrin was capable of binding the C-C′ loop mutant FN3 (data not shown), consistent with the prior demonstration that this mutation abrogates binding of soluble integrins to immobilized FN3 domain. Both integrins capture equivalent vitronectin, however, demonstrating equal coating of functional heterodimer (Figure 7E). Therefore, although soluble integrin αvβ3 is capable of binding FN3 domain adsorbed to plastic (Silletti et al., 2000a), immobilized αvβ3 is not capable of interacting with soluble FN3, which may more closely mimic the situation that would be presented in the context of FN3 present in the whole molecule in the plane of the cell membrane. Integrin αvβ3 was, however, capable of binding purified Ig6 domain in solution (data not shown).
Because binding of the ADAMs-cleaved L1 ECD to cell surface integrins has been shown to regulate cell migration (Mechtersheimer et al., 2001), and because mutation of T1172 suppressed ECD shedding in our cells, we assessed whether ectopic expression of L1 would affect CHO migration in a T1172-dependent manner. Interestingly, haptotactic migration of stable nonneuronal L1-expressing CHO cells toward lamininI was significantly lower than their mock-transfected, stably hygromycin-resistant counterparts (Figure 7F), contrary to what was reported previously in response to L1 expression in these cells (Mechtersheimer et al., 2001). However, it should be stressed that this prior report dealt with the neuronal isoform of L1, therefore this is the first demonstration of the regulation of cell migration by the nonneuronal isoform. More importantly, however, the T1172A mutation attenuated the reduced migration of these cells toward lamininI. It should further be noted that this effect was not universal, as CHO/L1WT cells demonstrated elevated migration toward fibronectin (Figure 7F), as had been observed previously with the neuronal isoform (Mechtersheimer et al., 2001), suggesting the regulation of specific integrin-mediated events by this isoform of L1 in these cells. Further establishing specificity for this effect is the fact that the T1172 mutation did not affect fibronectin migration, suggesting that the differential integrin regulation observed may be dependent upon differential conformational effects that are dependent (lamininI) or independent (fibronectin) of T1172 phosphorylation. Interestingly, we found that the adhesion of CHO/mock and CHO/L1WT cells was identical on both lamininI and fibronectin (Figure 7G), suggesting that differences in migration are not merely the result of augmented adhesion. Somewhat surprisingly, however, CHO/L1T1172A cells demonstrated a small but reproducible increase in adhesion to both lamininI and fibronectin (Figure 7G). Although consistent, this trend failed to achieve statistical significance.
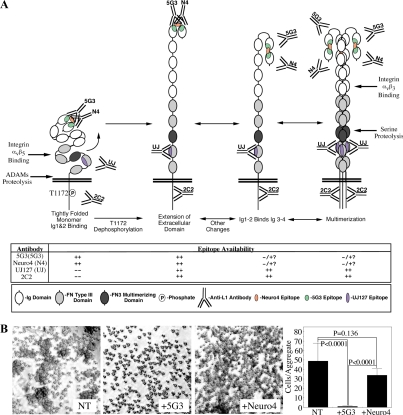
A Model of L1 Ectodomain Conformational Regulation by T1172 Phosphorylation
We have attempted to combine our data with that published in the literature to propose a working model of L1 ectodomain regulation (Figure 8A). In the first configuration, T1172-phosphorylated L1 exists as a tightly folded monomer in which Ig1 and Ig2 form a mini-loop that is highly reactive with 5G3 and Neuro4, whereas the UJ127 epitope in FN4 is largely obscured by the globular conformation of the FN domains, consistent with prior reports (Drescher et al., 1996). In response to, or concomitant with, T1172 dephosphorylation, the ECD is predicted to open up, allowing UJ127 to bind to its epitope in FN4 (second configuration). It is likely that the T1172 phosphorylation state influences interactions of the L1 CD with other cytoplasmic proteins, providing potential mechanism for these macrochanges. The mini-loop formed by Ig1 and Ig2 is probably somewhat labile as additional constraints of macrofolding are removed; thus, this second configuration may interconvert to the third configuration, adopting the canonical antiparallel horseshoe described previously (Schürmann et al., 2001). Factors not examined here may regulate this interconversion, or the protein may exist in an equilibrium state. This third configuration exhibits reduced recognition by 5G3 and Neuro4, and is potentially capable of multimerizing through the FN3 domain to the fourth configuration. These latter configurations are consistent with the previously reported lack of 5G3 reactivity by some L1-positive cells, including CD4+ T-lymphocytes (Silletti et al., 2000b). Shifting of the Ig1-2 mini-loop to the Ig1-4 horseshoe configuration may or may not be a requirement for L1 multimerization, however L1 trimerization would allow the Ig1-4/Ig1-4 trans interaction to expand from the proposed linear zipper configuration proposed for axonin1 (Freigang et al., 2000) to a two-dimensional “velcro-like” effect with significantly enhanced avidity. Indeed, Hall et al. (2000) demonstrated that trimeric L1 exhibited significantly enhanced homophilic binding capabilities. Moreover, Ig5 and Ig6 of axonin1 suppress binding through Ig1-4 by promoting a folding back of Ig1-4 onto the FN domains (Rader et al., 1996), consistent with the tightly folded conformation in Figure 8A, and suggesting that extension of the ECD might occur as a result of homophilic binding, whereas subsequent integrin-binding to Ig6 might further stabilize this extended conformation. Indeed, although Haspel et al. (2000) found Ig1-4 to be the minimal unit required for homophilic binding, this unit was less efficient than the entire ECD, and inclusion of Ig domains 5 and 6 was required to recapitulate full potency.
Figure 8.
L1 ectodomain regulation and cytoplasmic phosphorylation. (A) Putative model of L1 ectodomain conformation and its regulation by, or association with, changes in intracellular phosphorylation state. Availability of epitopes and interactions with integrins and proteases are shown. (B) Aggregation of J558L-L1 myeloma cells in the presence or absence of 50 μg/ml 5G3 or Neuro4.
This model also suggests a mechanism that may explain why pretreatment, but not posttreatment, with 5G3 blocks homotypic L1 interaction (Nayeem et al., 1999); pretreatment with 5G3 may stabilize the Ig1-2 mini-loop and prevent formation of the Ig1-4 horseshoe that is predicted to mediate L1 homophilic binding. Because Neuro4 recognizes a linear epitope within the region encompassed by the 5G3 epitope, and indeed blocks 5G3 binding, we tested whether the homotypic-blocking activity of 5G3 is due to the location of its epitope, or rather the nature of its epitope. J558L myeloma cells stably expressing L1 aggregate in an L1-dependent manner and, as demonstrated previously (Nayeem et al., 1999), pretreatment with 5G3 abrogates this aggregation (Figure 8B). Neuro4 had a much less dramatic effect on aggregation, consistent with stabilization of the “off” conformation by 5G3, rather than purely steric hindrance.
DISCUSSION
In this report, we demonstrate phosphorylation of T1172 of L1 in human PDAC and melanoma cells. The region encompassing T1172 mediates L1 interactions with the cytoskeletal linker ezrin and the clathrin adapter AP2μ2 (Dickson et al., 2002; Tyukhtenko et al., 2008). However, these interactions are restricted to the neuronal isoform (Dickson et al., 2002); therefore, the role of T1172 in cells that express nonneuronal L1 (e.g., PDAC) is unclear. Indeed, although CKII phosphorylated recombinant L1 CD in vitro, it exhibited a more than threefold preference for the nonneuronal isoform. CKII also phosphorylates S1181 of the neuronal isoform, thereby regulating L1 trafficking and axon growth (Nakata and Kamiguchi, 2007). The relevance of S1181 to nonneuronal L1 is unclear, however, although preferential CKII phosphorylation of S1181 versus T1172 in neuronal L1 may represent a regulatory mechanism based on the alternatively spliced small exon (1177RSLE) that separates these residues and recapitulates the tyrosine-based motif required for sorting of L1 to the axonal growth cone (Kamiguchi and Lemmon, 1998). Indeed, we recently demonstrated that S1181 phosphorylation is required for PKC phosphorylation of T1172 in vitro (Chen et al., 2009), suggesting that in cells CKII may be primarily responsible for S1181 phosphorylation, whereas PKC may mediate subsequent T1172 phosphorylation.
Importantly, although SS caused T1172 dephosphorylation in M21 melanoma cells, CKII and PKC blockade was ineffective. The reason for this is unclear; however, M21 cells express all implicated PKC isoforms, and CKII is ubiquitous; therefore, the involvement of a different kinase in these cells probably represents a regulatory difference. PKC has been implicated in L1 biology previously. PMA induced L1 ECD shedding in an ADAMs-mediated manner in melanoma (Beer et al., 1999) and breast cancer cells (Gutwein et al., 2000). More importantly, ovarian cancer cells shed L1 through two mechanisms, cleavage of intracellular L1 and secretion in exosomal vesicles, or cleavage of cell surface L1 in a manner induced by PMA (Stoeck et al., 2006). PMA drives T1172 phosphorylation and ADAMs-mediated shedding of L1 in Panc1 cells, consistent with shedding of cell membrane L1, and supporting our proposal that proteolytic shedding in PDAC cells results from ECD conformational changes driven by, or associated with, phosphorylation of cell surface L1.
Shedding of the L1 ECD is an important aspect of L1 biology. Not only is cleaved L1 ECD present in L1-positive tumors and the developing mouse brain, but the enhanced migration observed in neuronal L1-transfected CHO cells is due to binding of cleaved L1 ECD onto cell surface αvβ5 integrin (Mechtersheimer et al., 2001; Thelen et al., 2002). We actually observed decreased migration that required T1172 in nonneuronal L1-expressing CHO cells. Moreover, our current data suggest that αvβ3 binds to the open (potentially multimerized) L1 conformation, whereas αvβ5 interacts preferentially with the globular L1 configuration. Our model further predicts a potential docking interaction that may facilitate L1 proteolysis. Because immobilized αvβ5 binds soluble FN3 domain, αvβ5 that has an ADAMs protease bound to its RGD-binding site through the ADAMs disintegrin domain may coordinate cleavage of L1 by binding to FN3 through an accessory site. L1 proteolysis could then trigger release of the disintegrin domain, leaving cleaved L1 ECD attached through FN3 available to further bind αvβ5 through its RGD site in Ig6, similar to the TIMP2/MT1-MMP/αvβ3–mediated activation of MMP2 (Deryugina et al., 2001).
Multiple regions of L1 facilitate homophilic or heterophilic interactions, either acting alone, or in a cooperative manner (reviewed in Haspel and Grumet, 2003). Several lines of evidence demonstrate the Ig1-4 horseshoe configuration for L1-related molecules such as axonin-1 and hemolin (Su et al., 1998; Freigang et al., 2000). Sequence similarities allow extrapolation of such a conformation to L1, and L1 has been observed to adopt a globular conformation consistent with such a horseshoe configuration (Schürmann et al., 2001). However, Holm et al. (1995) did not observe significant affinity of Ig1-2 proteins for Ig3-4 proteins but rather demonstrated that Ig1-2 strongly homoaggregates. Indeed, Ig1-2 bound to itself better in absentia than it did to Ig1-4 or Ig1-6 and better than Ig1-4 or Ig1-6 bound to themselves. The significance of these findings is highlighted by our epitope mapping studies, which clearly demonstrate an interaction between the Ig1 and Ig2 domains, and the constraints imposed on this mini-loop by the adjacent domains (i.e., Ig3 and Ig4).
There is no predicted extradomain sequence between Ig1 and Ig2 that would impart an obvious flexibility like that facilitated by the 7aa linker separating Ig2 and Ig3 (Hlavin and Lemmon, 1991). This linker is proposed not only to allow the 180° turn that juxtaposes Ig3 with Ig2, and Ig4 with Ig1 but also to provide for rotational flexibility that allows Ig2 and Ig3 to face the same direction, instead of the alternating orientation proposed for the rest of the Ig domains (Haspel and Grumet, 2003). As such, the same degree of bending back is not expected for Ig1 and Ig2, however, a lesser degree of flexibility in this region is consistent with rotary shadow (Schürmann et al., 2001) and electron microscopy (EM) images (Drescher et al., 1996) of “extended” L1 and is clearly demonstrated by the masking of αECD epitopes available in the individual Ig1 and Ig2 domains by presentation of both domains together, and by the enhanced reactivity of the αFL pAb against Ig1-2, when only Ig1 is recognized individually (Figure 6E). The further effect of including adjacent Ig domains suggests Ig1-2 mini-loop destabilization due to the preferential formation of the Ig1-4 horseshoe configuration in the absence of the FN domains or other regulatory forces that may exist in the context of the cell membrane. This may explain why only extended and horseshoe configurations were observed in EM studies of recombinant L1 (Schürmann et al., 2001). However, these distinct configurations were not detected in the same preparations but rather were observed individually using separate techniques (rotary shadowing vs. negative staining EM), suggesting that the method of preparation affects the result with this flexible molecule. Thus, the Ig1-2 mini-loop may not be stable enough for studies using these fixation techniques. As a result, ELISA-based approaches such as those used in this study may be better suited to identifying more labile configurations of the L1 ECD.
Although the transmission of intracellular events such as phosphorylation into changes in ectodomain structure can be manifest in several ways, numerous cases of inside-out signaling have been demonstrated for cell adhesion molecules including L1 (Byzova and Plow, 1998; Hortsch et al., 1998; Mehta et al., 1998; Wang et al., 2001). We propose a model in which the ECD structure of L1 reflects the cytoplasmic phosphorylation state, with specific reference to T1172. Structural changes associated with T1172 phosphorylation regulate ADAMs-mediated cleavage of the L1 ECD and differential integrin binding as well as cell migration. Although we recognize that our model may not be a perfect representation of the real situation, it incorporates our data and is consistent with the published literature. It further helps explain a number of the nuances of L1 biology observed by many groups including our own. We feel this study is important in the further delineation of L1 activity, especially outside the nervous and immune systems, where L1-integrin interactions often overshadow measurable L1-L1 homophilic events.
ACKNOWLEDGMENTS
We thank F. Gunderson and M. Rasmussen for technical support and Dennis J. Young in the Moores UCSD Cancer Center Flow Cytometry Shared Resource for flow cytometer expertise. S. S. is an American Cancer Society Research Scholar supported by American Cancer Society RSG-05-116-01-CSM and National Institutes of Health grants CA-130104 and CA-109956. Moores UCSD Cancer Center is a National Cancer Institute-sponsored comprehensive cancer center, core equipment (including microscopes) of which is supported by UCSD Cancer Center Specialized Support grant P30 CA-23100.
Abbreviations used:
- ADAM
a disintegrin and metalloproteinase
- BisI
bisindolylmaleimide I
- CalA
calyculin A
- CalC
calphostin C
- CD
cytoplasmic domain
- CKII
casein kinase II
- ECD
ectodomain
- MMP
matrix metalloproteinase
- OA
okadaic acid
- PDAC
pancreatic ductal adenocarcinoma
- SF
serum-free
- SS
staurosporine.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-10-0900) on March 24, 2010.
REFERENCES
- Bardeesy N., DePinho R. A. Pancreatic cancer biology and genetics. Nat. Rev. 2002;2:897–909. doi: 10.1038/nrc949. [DOI] [PubMed] [Google Scholar]
- Beer S., Oleszewski M., Gutwein P., Geiger C., Altevogt P. Metalloproteinase-mediated release of the ectodomain of L1 adhesion molecule. J. Cell Sci. 1999;112:2667–2675. doi: 10.1242/jcs.112.16.2667. [DOI] [PubMed] [Google Scholar]
- Bouvet M., Gamagami R. A., Gilpin E. A., Romeo O., Sasson A., Easter D., Moossa A. R. Factors influencing survival after resection for periampullary neoplasms. Am. J. Surg. 2000;180:13–17. doi: 10.1016/s0002-9610(00)00405-0. [DOI] [PubMed] [Google Scholar]
- Burden-Gulley S. M., Pendergast M., Lemmon V. The role of cell adhesion molecule L1 in axonal extension, growth cone motility, and signal transduction. Cell Tissue Res. 1997;290:415–422. doi: 10.1007/s004410050948. [DOI] [PubMed] [Google Scholar]
- Byzova T. V., Plow E. F. Activation of αvβ3 on vascular cells controls recognition of prothrombin. J. Cell Biol. 1998;43:2081–2089. doi: 10.1083/jcb.143.7.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M. M., Leland H. A., C.-Lee Y., Silletti S. Tyrosine and serine phosphorylation regulate the conformation and subsequent threonine phosphorylation of the L1 cytoplasmic domain. Biochem. Biophys. Res. Commun. 2009;389:257–264. doi: 10.1016/j.bbrc.2009.08.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng L., Lemmon S., Lemmon V. RanBPM is an L1-interacting protein that regulates L1-mediated mitogen-activated protein kinase activation. J. Neurochem. 2005;94:1102–1110. doi: 10.1111/j.1471-4159.2005.03254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlin-Huppe K., Berglund E. O., Ranscht B., Stallcup W. B. Mutational analysis of the L1 neuronal cell adhesion molecule identifies membrane-proximal amino acids of the cytoplasmic domain that are required for cytoskeletal anchorage. Mol. Cell Neurosci. 1997;9:144–156. doi: 10.1006/mcne.1997.0608. [DOI] [PubMed] [Google Scholar]
- Daponte A., Kostopoulou E., Kollia P., Papamichali R., Vanakara P., Hadjichristodoulou C., Nakou M., Samara S., Koukoulis G., Messinis I. E. L1-CAM in ovarian serous neoplasms. Eur. J. Gynaecol. Oncol. 2008;29:26–30. [PubMed] [Google Scholar]
- De Angelis E., Brummendorf T., Cheng L., Lemmon V., Kenwrick S. Alternative use of a mini exon of the L1 gene affects L1 binding to neural ligands. J. Biol. Chem. 2001;276:32738–32742. doi: 10.1074/jbc.M105156200. [DOI] [PubMed] [Google Scholar]
- Deryugina E. I., Ratnikov B., Monosov E., Postnova T. I., DiScipio R., Smith J. W., Strongin A. Y. MT1-MMP initiates activation of pro-MMP-2 and integrin αvβ3 promotes maturation of MMP-2 in breast carcinoma cells. Exp. Cell Res. 2001;263:209–223. doi: 10.1006/excr.2000.5118. [DOI] [PubMed] [Google Scholar]
- Dickson T. C., Mintz C. D., Benson D. L., Salton S.R.J. Functional binding interaction identified between the axonal CAM L1 and members of the ERM family. J. Cell Biol. 2002;157:1105–1112. doi: 10.1083/jcb.200111076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drescher B., Spiess E., Schachner M., Probstmeier R. Structural analysis of the murine cell adhesion molecule L1 by electron microscopy and computer-assisted modelling. Eur. J. Neurosci. 1996;8:2467–2478. doi: 10.1111/j.1460-9568.1996.tb01541.x. [DOI] [PubMed] [Google Scholar]
- Felding-Habermann B., Silletti S., Mei F., Siu C. H., Yip P. M., Brooks P. C., Cheresh D. A., O'Toole T. E., Ginsberg M. H., A Montgomery M. A single immunoglobulin-like domain of the human neural cell adhesion molecule L1 supports adhesion by multiple vascular and platelet integrins. J. Cell Biol. 1997;139:1567–1581. doi: 10.1083/jcb.139.6.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fogel M., Gutwein P., Mechtersheimer S., Riedle S., Stoeck A., Smirnov A., Edler L., Ben-Arie A., Huszar M., Altevogt P. L1 expression as a predictor of progression and survival in patients with uterine and ovarian carcinomas. Lancet. 2003a;362:869–875. doi: 10.1016/S0140-6736(03)14342-5. [DOI] [PubMed] [Google Scholar]
- Fogel M., Mechtersheimer S., Huszar M., Smirnov A., Abu-Dahi A., Tilgen W., Reichrath J., Georg T., Altevogt P., Gutwein P. L1 adhesion molecule (CD 171) in development and progression of human malignant melanoma. Cancer Lett. 2003b;189:237–247. doi: 10.1016/s0304-3835(02)00513-x. [DOI] [PubMed] [Google Scholar]
- Fransen E., Lemmon V., van Camp G., Vits L., Coucke P., Willems P. J. CRASH syndrome: clinical spectrum of corpus callosum hypoplasia, retardation, adducted thumbs, spastic paraparesis and hydrocephalus due to mutations in one single gene, L1. Eur. J. Hum. Genet. 1995;3:273–284. doi: 10.1159/000472311. [DOI] [PubMed] [Google Scholar]
- Fransen E., Van Camp G., Vits L., Willems P. J. L1-associated diseases: clinical geneticists divide, molecular geneticists unite. Hum. Mol. Genet. 1997;6:1625–1632. doi: 10.1093/hmg/6.10.1625. [DOI] [PubMed] [Google Scholar]
- Freigang J., Proba K., Leder L., Diederichs K., Sonderegger P., Welte W. The crystal structure of the ligand binding module of axonin-1/TAG-1 suggests a zipper mechanism for neural cell adhesion. Cell. 2000;101:425–433. doi: 10.1016/s0092-8674(00)80852-1. [DOI] [PubMed] [Google Scholar]
- Gast D., et al. The cytoplasmic part of L1-CAM controls growth and gene expression in human tumors that is reversed by therapeutic antibodies. Oncogene. 2008;27:1281–1289. doi: 10.1038/sj.onc.1210747. [DOI] [PubMed] [Google Scholar]
- Gavert N., Conacci-Sorrell M., Gast D., Schneider A., Altevogt P., Brabletz T., Ben-Ze'ev A. L1, a novel target of β-catenin signaling, transforms cells and is expressed at the invasive front of colon cancers. J. Cell Biol. 2005;168:633–642. doi: 10.1083/jcb.200408051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavert N., et al. Expression of L1-CAM and ADAM10 in human colon cancer cells induces metastasis. Cancer Res. 2007;67:7703–7712. doi: 10.1158/0008-5472.CAN-07-0991. [DOI] [PubMed] [Google Scholar]
- Gouveia R. M., Gomes C. M., Sousa M., Alves P. M., Costa J. Kinetic analysis of L1 homophilic interaction: role of the first four immunoglobulin domains and implications on binding mechanism. J. Biol. Chem. 2008;83:28038–28047. doi: 10.1074/jbc.M804991200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutwein P., Oleszewski M., Mechtersheimer S., Agmon-Levin N., Krauss K., Altevogt P. Role of Src kinases in the ADAM-mediated release of L1 adhesion molecule from human tumor cells. J. Biol. Chem. 2000;275:15490–15497. doi: 10.1074/jbc.275.20.15490. [DOI] [PubMed] [Google Scholar]
- Hall H., Bozic D., Fauser C., Engel J. Trimerization of cell adhesion molecule L1 mimics clustered L1 expression on the cell surface: influence on L1-ligand interactions and on promotion of neurite outgrowth. J. Neurochem. 2000;75:336–346. doi: 10.1046/j.1471-4159.2000.0750336.x. [DOI] [PubMed] [Google Scholar]
- Haspel J., Friedlander D. R., Ivgy-May N., Chickramane S., Roonprapunt C., Chen S., Schachner M., Grumet M. Critical and optimal Ig domains for promotion of neurite outgrowth by L1/Ng-CAM. J. Neurobiol. 2000;42:287–302. [PubMed] [Google Scholar]
- Haspel J., Grumet M. The L1CAM extracellular region: a multi-domain protein with modular and cooperative binding modes. Front. Biosci. 2003;8:s1210–s1225. doi: 10.2741/1108. [DOI] [PubMed] [Google Scholar]
- Haspel J., Schürmann G., Jacob J., Erickson H. P., Grumet M. Disulfide-mediated dimerization of L1 Ig domains. J. Neurosci. Res. 2001;66:347–355. doi: 10.1002/jnr.1227. [DOI] [PubMed] [Google Scholar]
- Hlavin M. L., Lemmon V. Molecular structure and functional testing of human L1CAM: an interspecies comparison. Genomics. 1991;11:416–423. doi: 10.1016/0888-7543(91)90150-d. [DOI] [PubMed] [Google Scholar]
- Holden N. S., Squires P. E., Kaur M., Bland R., Jones C. E., Newton R. Phorbol ester-stimulated NF-kappaB-dependent transcription: roles for isoforms of novel protein kinase C. Cell Signal. 2008;20:1338–1348. doi: 10.1016/j.cellsig.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Holm J., Appel F., Schachner M. Several extracellular domains of the neural cell adhesion molecule L1 are involved in homophilic interactions. J. Neurosci. Res. 1995;42:9–20. doi: 10.1002/jnr.490420103. [DOI] [PubMed] [Google Scholar]
- Hortsch M., Homer D., Malhotra J. D., Chang S., Frankel J., Jefford G., Dubreuil R. R. Structural requirements for outside-in and inside-out signaling by Drosophila neuroglian, a member of the L1 family of cell adhesion molecules. J. Cell Biol. 1998;142:251–261. doi: 10.1083/jcb.142.1.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaifi J. T., Heidtmann S., Schurr P. G., Reichelt U., Mann O., Yekebas E. F., Wachowiak R., Strate T., Schachner M., Izbicki J. R. Absence of L1 in pancreatic masses distinguishes adenocarcinomas from poorly differentiated neuroendocrine tumors. Anticancer Res. 2006;26:1167–1170. [PubMed] [Google Scholar]
- Kamiguchi H., Lemmon V. A neuronal form of the cell adhesion molecule L1 contains a tyrosine-based signal required for sorting to the axonal growth cone. J. Neurosci. 1998;18:3749–3756. doi: 10.1523/JNEUROSCI.18-10-03749.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mechtersheimer S., et al. Ectodomain shedding of L1 adhesion molecule promotes cell migration by autocrine binding to integrins. J. Cell Biol. 2001;155:661–673. doi: 10.1083/jcb.200101099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta R. J., Diefenbach B., Brown A., Cullen E., Jonczyk A., Güssow D., Luckenbach G. A., Goodman S. L. Transmembrane-truncated αvβ3 integrin retains high affinity for ligand binding: evidence for an ‘inside-out’ suppressor? Biochem. J. 1998;330:861–869. doi: 10.1042/bj3300861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muerkoster S. S., et al. Drug-induced expression of the cellular adhesion molecule L1CAM confers anti-apoptotic protection and chemoresistance in pancreatic ductal adenocarcinoma cells. Oncogene. 2007;26:2759–2768. doi: 10.1038/sj.onc.1210076. [DOI] [PubMed] [Google Scholar]
- Mujoo K., Spiro R. C., Reisfeld R. A. Characterization of a unique glycoprotein antigen expressed on the surface of human neuroblastoma cells. J. Biol. Chem. 1986;261:10299–10305. [PubMed] [Google Scholar]
- Nakata A., Kamiguchi H. Serine phosphorylation by casein kinase II controls endocytic L1 trafficking and axon growth. J. Neurosci. Res. 2007;85:723–734. doi: 10.1002/jnr.21185. [DOI] [PubMed] [Google Scholar]
- Nayeem N., Silletti S., Yang X., Lemmon V. P., Reisfeld R. A., Stallcup W. B., Montgomery A. M. A potential role for the plasmin(ogen) system in the posttranslational cleavage of the neural cell adhesion molecule L1. J. Cell Sci. 1999;112:4739–4749. doi: 10.1242/jcs.112.24.4739. [DOI] [PubMed] [Google Scholar]
- NCI-Pancreatic Cancer Progress Review Group. PANC-PRG Implementation Plan. Bethesda, MD: National Cancer Institute; 2002. National Cancer Institute strategic plan for addressing the recommendations of the pancreatic cancer progress review group. [Google Scholar]
- Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- Patel K., Kiely F., Phimister E., Melino G., Rathjen F., Kemshead J. T. The 200/220 kDa antigen recognized by monoclonal antibody (MAb) UJ127.11 on neural tissues and tumors is the human L1 adhesion molecule. Hybridoma. 1991;10:481–491. doi: 10.1089/hyb.1991.10.481. [DOI] [PubMed] [Google Scholar]
- Rader C., Kunz B., Lierheimer R., Giger R. J., Berger P., Tittmann P., Gross H., Sonderegger P. Implications for the domain arrangement of axonin-1 derived from the mapping of its NgCAM binding site. EMBO J. 1996;15:2056–2068. [PMC free article] [PubMed] [Google Scholar]
- Schaefer A. W., Kamei Y., Kamiguchi H., Wong E. W., Rapaport I., Kirchhausen T., Beach C. M., Landreth G., Lemmon S. K., Lemmon V. L1 endocytosis is controlled by a phosphorylation-dephosphorylation cycle stimulated by outside-in signaling by L1. J. Cell Biol. 2002;157:1223–1232. doi: 10.1083/jcb.200203024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultheis M., Diestel S., Schmitz B. The role of cytoplasmic serine residues of the cell adhesion molecule L1 in neurite outgrowth, endocytosis and cell migration. Cell Mol. Neurobiol. 2007;27:11–31. doi: 10.1007/s10571-006-9113-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schürmann G., Haspel J., Grumet M., Erickson H. P. Cell adhesion molecule L1 in folded (horseshoe) and extended conformations. Mol. Biol. Cell. 2001;12:1765–1773. doi: 10.1091/mbc.12.6.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silletti S., Mei F., Sheppard D., Montgomery A. M. Plasmin-sensitive dibasic sequences in the third fibronectin-like domain of L1-cell adhesion molecule (CAM) facilitate homomultimerization and concomitant integrin recruitment. J. Cell Biol. 2000a;149:1485–1502. doi: 10.1083/jcb.149.7.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silletti S., Altevogt P., Montgomery A.M.P. L1 cell adhesion molecule (CD171). CD171 guide for the 2000 International Workshop in Human Leukocyte Differentiation Antigens and the online “Protein Reviews On The Web (PROW) ( www.ncbi.nlm.nih.gov/prow) Prow. 2000b;1:31–37. [Google Scholar]
- Silletti S., Yebra M., Perez B., Cirulli V., McMahon M., Montgomery A. M. Extracellular signal-regulated kinase (ERK)-dependent gene expression contributes to L1 cell adhesion molecule-dependent motility and invasion. J. Biol. Chem. 2004;279:28880–28888. doi: 10.1074/jbc.M404075200. [DOI] [PubMed] [Google Scholar]
- Stallcup W. B. The third fibronectin type III repeat is required for L1 to serve as an optimal substratum for neurite extension. J. Neurosci. Res. 2000;61:33–43. doi: 10.1002/1097-4547(20000701)61:1<33::AID-JNR4>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Stoeck A., Keller S., Riedle S., Sanderson M. P., Runz S., Le Naour F., Gutwein P., Ludwig A., Rubinstein E., Altevogt P. A role for exosomes in the constitutive and stimulus-induced ectodomain cleavage of L1 and CD44. Biochem. J. 2006;393:609–618. doi: 10.1042/BJ20051013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X. D., Gastinel L. N., Vaughn D. E., Faye I., Poon P., Bjorkman P. J. Crystal structure of hemolin: a horseshoe shape with implications for homophilic adhesion. Science. 1998;281:991–995. doi: 10.1126/science.281.5379.991. [DOI] [PubMed] [Google Scholar]
- Thelen K., Kedar V., Panicker A. K., Schmid R. S., Midkiff B. R., Maness P. F. The neural cell adhesion molecule L1 potentiates integrin-dependent cell migration to extracellular matrix proteins. J. Neurosci. 2002;22:4918–4931. doi: 10.1523/JNEUROSCI.22-12-04918.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thies A., Schachner M., Moll I., Berger J., Schulze H., Brunner G., Schumacher U. U. Overexpression of the cell adhesion molecule L1 is associated with metastasis in cutaneous malignant melanoma. Eur. J. Cancer. 2002;38:1708–1716. doi: 10.1016/s0959-8049(02)00105-3. [DOI] [PubMed] [Google Scholar]
- Tyukhtenko S., Deshmukh L., Kumar V., Lary J., Cole J., Lemmon V., Vinogradova O. Characterization of the neuron-specific L1-CAM cytoplasmic tail: naturally disordered in solution it exercises different binding modes for different adaptor proteins. Biochemistry. 2008;47:4160–4168. doi: 10.1021/bi702433q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Chen H., Brown E. J. L-plastin peptide activation of αvβ3-mediated adhesion requires integrin conformational change and actin filament disassembly. J. Biol. Chem. 2001;276:14474–14481. doi: 10.1074/jbc.M007324200. [DOI] [PubMed] [Google Scholar]
- Wong E. V., Schaefer A. W., Landreth G., Lemmon V. Casein kinase II phosphorylates the neural cell adhesion molecule L1. J. Neurochem. 1996;66:779–786. doi: 10.1046/j.1471-4159.1996.66020779.x. [DOI] [PubMed] [Google Scholar]
- Zhao X., Siu C. H. Colocalization of the homophilic binding site and the neuritogenic activity of the cell adhesion molecule L1 to its second Ig-like domain. J. Biol. Chem. 1995;270:29413–29421. doi: 10.1074/jbc.270.49.29413. [DOI] [PubMed] [Google Scholar]