This study suggests that a TPB2 piggyBac transposase has evolved to facilitate heterochromatin assembly and carry out the final DNA excision step of programmed DNA deletion in Tetrahymena thermophila. TPB2 appears to have gone through a domestication process to become a host gene and be maintained in the macronuclear genome.
Abstract
Transposons comprise large fractions of eukaryotic genomes and provide genetic reservoirs for the evolution of new cellular functions. We identified TPB2, a homolog of the piggyBac transposase gene that is required for programmed DNA deletion in Tetrahymena. TPB2 was expressed exclusively during the time of DNA excision, and its encoded protein Tpb2p was localized in DNA elimination heterochromatin structures. Notably, silencing of TPB2 by RNAi disrupts the final assembly of these heterochromatin structures and prevents DNA deletion to occur. In vitro studies revealed that Tpb2p is an endonuclease that produces double-strand breaks with four-base 5′ protruding ends, similar to the ends generated during DNA deletion. These findings suggest that Tpb2p plays a key role in the assembly of specialized DNA elimination chromatin architectures and is likely responsible for the DNA cleavage step of programmed DNA deletion.
INTRODUCTION
Developmentally programmed, large-scaled DNA rearrangements have been known to occur in organisms ranging from protozoa to vertebrates (Borst and Greaves, 1987; Coyne et al., 1996; Smith et al., 2009). These processes drastically alter chromosome structures and genomic contents and have significant implications in gene regulation and genome integrity. However, their mechanisms and biological roles are not well understood. The ciliated protozoan Tetrahymena is among the best studied organisms to carry out such rearrangement processes and has offered considerable insights. Tetrahymena thermophila contains a silent germinal micronucleus and a transcription active somatic macronucleus in each cell. Both nuclei are derived from the same genetic source during conjugation. In this sexual process, the micronucleus goes through meiosis, postmeiotic mitosis, and cross-fertilization to generate zygotic nuclei, which further divide and differentiate to form the new macro- and micronuclei in the progeny cells while the old macronucleus is destroyed (see Figure 1A). New macronuclear development involves extensive genome-wide DNA rearrangements, deleting thousands of specific DNA segments, known as internal eliminated sequences (IESs) that comprise ∼15% of the genome. The remaining DNA is fragmented at specific chromosome breakage sequences (Cbs) and endoreplicated ∼23-fold to form the somatic genome (Yao and Chao, 2005).
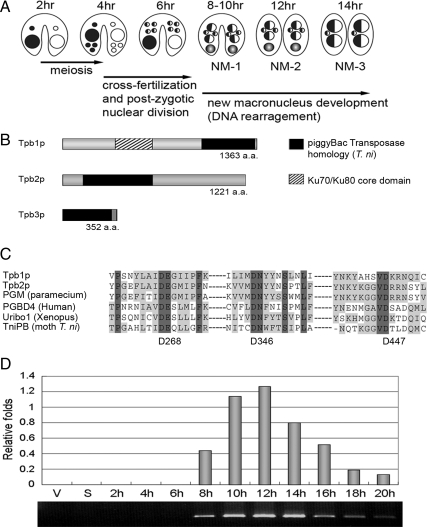
Figure 1.
Characterization of TPB. (A) Tetrahymena nuclear developmental process during conjugation. (B) Schematic representation of Tpb proteins. (C) Comparison of the catalytic DDD-motif of Tpb2p and other piggyBac-like proteins. The DDD-motif of T. ni piggyBac consisting of D268, D346, and D447 is indicated. (D) Expression of TPB2 by quantitative RT-PCR. Total RNA extracted from vegetative (V), starved (S), and conjugating cells (2, 4, 6, 8, 10, 12, 14, 16, 18, and 20 h after postmixing) were used as templates. Quantification was normalized with α-tubulin mRNA expression.
Programmed DNA deletion is an RNA-guided process (Mochizuki et al., 2002; Yao et al., 2003). Double-stranded RNA is transcribed from IES during and after meiosis and processed into 27–30-nucleotide small RNA by the Dicer-like enzyme Dcl1p (Chalker and Yao, 2001; Malone et al., 2005; Mochizuki and Gorovsky, 2005). Together with other proteins including the Argonaute protein Twi1p (Mochizuki et al., 2002), the small RNA is believed to target corresponding IESs in the developing macronucleus, leading to histone modifications (H3K27me3 and H3K9me3; Taverna et al., 2002; Liu et al., 2007) and the recruitment of chromodomain proteins (Pdd1p and Pdd3p; Madireddi et al., 1996; Nikiforov et al., 1999; Nikiforov et al., 2000) to form distinct heterochromatin structures that are eventually deleted. However, the enzymatic machinery that carries out the DNA cutting and joining reaction remains largely unknown.
Some transposase genes in eukaryotes have evolved to acquire functions that are beneficial to the host and become domesticated (Volff, 2006). Two well-known examples are RAG1 from Transib transposons in the V(D)J recombination system of vertebrate immunoglobulin genes (Agrawal et al., 1998; Kapitonov and Jurka, 2005) and Cenp-B from pogo DNA tranposons in the formation of chromosome centromere in some eukaryotes (Smit and Riggs, 1996; Casola et al., 2008). Ciliate IES could have been evolved from transposons. In Euplotes and Oxytricha, the excision of some transposons resembled the deletion of IESs (Doak et al., 1994; Klobutcher and Herrick, 1997). In Tetrahymena, most of the eliminated portion of the genome could be derived from transposons or other invading genetic elements during evolution. Studies of intermediates in the Tetrahymena DNA deletion process revealed distinct four-base 5′ protruding ends, resembling the end generated during the transposition of some transposons such as Tn7 (Saveliev and Cox, 1995, 1996).
The piggyBac transposon originally isolated from the genome of the moth Trichoplusia ni encodes a 594-amino acid transposase that mediates cut and paste excision and reinserts the transposon element into a TTAA target site in the genome (Elick et al., 1996; Fraser et al., 1996). Many piggyBac-like sequences were found in the genomes of different organisms including fungi, plants, insects, crustaceans, urochordates, amphibians, fishes, and mammals. The widespread distribution and phylogenetic analyses of piggyBac-like sequences in various organisms have led to the proposal that some of these ancient transposons might be domesticated by the host genome for cellular functions (Sarkar et al., 2003). In this study, we explored the possibility that a transposase may have evolved to participate in programmed DNA deletion in Tetrahymena. In an independent parallel study, Baudry et al. (2009) have recently provided in vivo evidence that PiggyMac (PGM), a domesticated piggyBac transposase, is required for programmed genome rearrangements in Paramecium tetraurelia. Our results here show that a piggyBac transposase gene has also been domesticated in Tetrahymena to play essential roles in programmed DNA deletion. Further more, our results revealed an essential role for this gene in the formation of specialized DNA elimination heterochromatin structures and provided evidence that it is likely responsible for the DNA cutting activity in the DNA deletion process.
MATERIALS AND METHODS
Cell Culture
Wild-type T. thermophila strains B2086 II, CU428 (Mpr/Mpr [VII, mp-s]), and CU427 (Chx/Chx [VI, cy-s]) were obtained from Peter Bruns (Cornell University, Ithaca, NY). Tetrahymena strains were maintained as previously described. Tetrahymena cells were grown in SPP medium at 30°C and prepared for mating by washing cells with 10 mM Tris-HCl (pH 7.4) buffer and incubation overnight before mixing to initiate conjugation.
Construction of Hairpin RNA Expression Vectors and Green Fluorescent Protein Fusion Vector
The targeted region was amplified by PCR and the product (∼500 base pairs) was cloned into the PCRII-I vector using two sets of primers with restriction enzyme cloning sites, one set with the ApaI-XhoI site and the other with the PmeI-SmaI site to generate the hairpin cassette. This hairpin cassette was then removed from the PCRII-I backbone by digestion with PmeI and ApaI and was ligated into the pIBF rDNA vector (Howard-Till and Yao, 2006). TPB2 hairpin RNA was expressed under the control of a CdCl2-inducible MTT1 promoter. The full-length TPB2 genomic DNA PCR product (4475 base pairs) was cloned into the PIGF-1 vector and fused to green fluorescent protein (GFP) at its N-terminus using the XhoI and ApaI cloning site. The sequences of the primers are listed in Oligo DNAs (see Supplemental Information).
Production of Tpb2p Antisera
Rabbits were immunized with a synthetic peptide “KQEHRSDQKKKNSY” corresponding to amino acid sequence from 1003 aa to 1016 aa of Tpb2p (commercial custom-made antibody from ProSci, Poway, CA).
RT-PCR
RNA samples were prepared using a RNA isolation kit (Roche, Indianapolis, IN). First strand cDNA was synthesized using Transcriptor reverse transcriptase with anchored-oligo(dT)18 primer. It was then followed by either conventional PCR or quantitative PCR analysis. The sequences of the primers are listed in the “Oligo DNAs” section (see Supplemental Information). The quantitative-PCR analysis was performed by LightCycler Carousel-Based PCR System with the LightCycler FastStart DNA Masterplus SYBR Green kit (Roche). Relative quantification was normalized with α-tubulin mRNA expression as an internal control.
Northern Blotting
Total RNA samples were prepared using TRIzol reagent (Invitrogen). RNA samples were mixed with RNA glyoxal reaction mixture loading dye and analyzed by electrophoresis in a 1.2% agarose gel. RNA was transferred to IMMOBILON-NY+ nylon membrane (Millipore), UV cross-linked and hybridized with probes overnight at 42°C in a hybridization buffer (Roche). Probes were made by random prime labeling reactions with PCR products amplified from genomic DNA. After hybridization, membrane was washed several times with 2–0.5× SSC and 0.1% SDS washing buffer and exposed to x-ray film. Bands were quantified and normalized by using Quantity One software (Bio-Rad, Richmond, CA).
Evaluation of TPB2 Hairpin RNA Silencing Phenotype
Two different mating types of TPB2 hairpin transformants were starved and mated with each other in 10 mM Tris-HCl (pH 7.4) buffer without or with 0.05 μg/ml CdCl2 to silence the TPB2 gene during conjugation. To test for their abilities to produce viable progeny, 300 individual mating pairs were cloned to drops of growth medium for 48 h and tested for growth and drug resistance phenotypes that distinguish parents from progeny. To determine developmental stages, conjugating cells (14, 20, and 30 h after mating initiation) were stained with DAPI (4′,6′-diamidino-2-phenylindole; 1 μg/ml) to reveal their nuclear stages. To examine the IES elimination and chromosome breakage by using PCR assay, genomic DNAs were isolated from the late conjugating cell pools (30 h after mating initiation) and used to perform PCR analysis using primers that were specific for IES elimination (M element, R element, Cam element, and Tlr element; Coyne et al., 1999; Aronica et al., 2008) and chromosome breakage (Cbs819, 5-2, and 4L-2 and the telomere primer; Coyne et al., 1999).
Immunofluorescence Analysis
Conjugating cells were fixed with 2% paraformadehyde, immobilized on slides, incubated with the primary antibodies, anti-Pdd1p (1:1000, Abcam, Cambridge, MA; ab5338) and anti-Tpb2p (1:500), respectively, and washed with 0.3% Triton X-100 in PBS buffer. The secondary antibodies were Cy5-conjugated AffiniPure F(ab′)2 fragment goat anti-rabbit IgG (1:500, Jackson ImmunoResearch, West Grove, PA; 111-176-003). Digital images were performed and analyzed using Zeiss Axio Imager system (Thornwood, NY) and Applied Precision Deltavision system (Issaquah, WA) fluorescence microscopy.
Chromatin Immunoprecipitation
Cells were cross-linked in 1% paraformadehyde, harvested, and washed as previously described (Dedon et al., 1991). Cells were then resuspended at 5 × 104 cells/ml in 0.1% SDS lysis buffer (Chadee et al., 1999) and sonicated for 4 min in 30-s bursts. Cell lysates were used to immunoprecipitate with antibodies, anti-Pdd1p (Abcam, ab5338), anti-H3K27me3 (Millipore, Bedford, MA; 07-449), and anti-H3K4me2 (Millipore, 07-030), respectively. The immunoprecipitated complexes were collected by protein A agarose (Millipore, 16-157) and then incubated in proteinase K buffer overnight. The cross-links were reversed at 65°C for 6 h, and DNA was extracted with phenol/chloroform and ethanol precipitation. DNA isolated from the lysate without antibody immunoprecipitation was used as an input control. The primer sets used are as previously described (Liu et al., 2007). Quantitative-PCR analysis was performed as description above.
Production of Recombinant Proteins
To produce recombinant Tpb2p, DNA (TPB2Ec) encoding Tpb2p that had been optimized for codon usage in Escherichia coli was synthesized (GenScript, Piscataway, NJ). The sequence of TPB2Ec is shown in Supplemental Information. TPB2Ec was cloned into EcoRI and XhoI sites of pGEX4T1-TEV (gift from Tim Clausen, Institute of Molecular Pathology, Vienna) to make pGEX-TPB2. To generate pGEX-TPB2-CD, expressing a catalytic dead Tpb2p, D297 to L, D379 to L and D495 to L mutations were introduced into pGEX-TPB2 using the QuikChange II site-directed mutagenesis Kit (Stratagene, La Jolla, CA), and DNA oligos Tpb2pD297L fw/rv, Tpb2pD379L fw/rv, and Tpb2pD495L fw/rv. Sequences of the oligo DNAs used for the mutagenesis are listed in Oligo DNAs (see Supplemental Information). GST-Tpb2p and GST-Tpb2p-CD were expressed in E. coli strain BL21(DE3). After cultivation at 37°C to an A600 of ∼0.8, cells were incubated with 0.5 mM IPTG for 8 h at 18°C. Cells were lysed in 500 mM NaCl, 80 mM Tris-HCl, pH 8.0, 0.5% Triton X-100, 0.2 mM PMSF, and 1× complete proteinase inhibitor cocktail (Roche). The cell lysate was incubated with glutathione Sepharose 4B (Amersham Biosciences, Piscataway, NJ) at 4°C. After washing with 500 mM NaCl, 80 mM Tris-HCl, pH 8.0, and 0.1% Triton X-100, the glutathione S-transferase (GST) fusion proteins were eluted in 160 mM reduced glutathione, 500 mM NaCl, and 80 mM Tris-HCl, pH 8.0, and subsequently the buffer was replaced with dialysis buffer (20 mM Tris-HCl, pH 7.5, 100 mM NaCl, 4 mM MnSO4, and 10% glycerol).
Tpb2p Endonuclease Assay
Sequences of the oligo DNAs used in this assay are listed in Oligo DNAs (see Supplemental Information). The oligo DNAs were 5′ end labeled by incubating 20 pmol single-stranded DNA with 10 U polynucleotide kinase (Roche) in the presence of 10 μCi [γ-32P]ATP at 37°C for 1 h. Twenty picomoles of the complementary strand was added and purified with QIAquick Nucleotide removal Kit (QIAGEN, Chatsworth, CA). The samples were heat denatured at 95°C for 5 min. Tubes were placed into 200 ml boiling water and allowed to cool to room temperature to allow the oligo DNAs to anneal. The double-stranded oligo DNA was incubated with 2.5 ng/μl recombinant GST-Tpb2p or GST-Tpb2p-CD at 30°C for 2.5 h in 10 mM MnSO4, 10 mM Tris-HCl, pH 7.5. The reactions were treated with proteinase K, extracted with phenol-chloroform, and ethanol-precipitated. An aliquot of the samples with RArtfw and RArtrv were incubated with 10 U Klenow fragment (Fermentas, Hanover, MD) and 2 nmol dNTPs at 37°C for 30 min. The samples were separated on a 15% denaturing polyacrylamide gel. The dried gel was exposed to a storage phosphor screen (GE Healthcare, Piscataway, NJ), and images were captured using Typhoon Imager (GE Healthcare).
RESULTS
piggyBac Transposase Homologues in Tetrahymena
We searched the Tetrahymena genome database and found three piggyBac transposase homolog genes, TPB1, TPB2, and TPB3 (Tetrahymena piggyBac transposase 1, 2, and 3) from the gene annotations of Tetrahymena genome database (TGD; http://www.ciliate.org/). The cDNA sequences of TPB1 and TPB2 have been submitted to DDBJ/EMBL/GenBank AB544065 and AB544066. Besides the transposase domain, Tpb1p contains a Ku70/Ku80 β-barrel domain in its N-terminal region. TPB3 is present only in the micronuclear genome, and the available sequence is truncated at the C-terminal of the piggyBac transposase homology region (Figure 1B). Tpb1p and Tpb2p share transposase catalytic DDD motif (in D268, D346, and D447 of T. ni piggyBac; Sarkar et al., 2003) with other piggyBac-like proteins (Figure 1C). In this study we focus on the analysis of TPB2 due to its essential roles in programmed DNA deletion (see below).
RT-PCR analyses were carried out to determine the expression of TPB2 mRNA at different stages of the life cycle. The results showed that the expression of TPB2 was highly up-regulated at late stages of conjugation, especially at 12–14 h after cell mating begins (Figure 1D), which coincides with the stages of DNA rearrangements in new developing macronuclei (Austerberry et al., 1984). Because the old macronuclei have largely degenerated at this stage, majority of TPB2 mRNA is likely derived from new developing macronuclei.
Silencing of TPB2 Gene by Hairpin RNA
To study the function of TPB2, we generated genetic knockout strains lacking TPB2 in their macronuclear genomes. The strains were able to grow and complete conjugation without notable phenotypic defects. Because TPB2 expression is likely from the developing macronuclei, knocking out the germline copy of TPB2 will be necessary to properly test its effects. However, repeated attempts to generate germline knockout strains have not been successful.
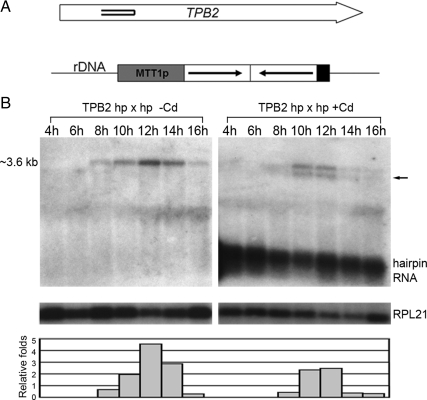
Hairpin RNA-induced gene silencing (RNAi) in Tetrahymena had been developed as an efficient method to study gene functions (Howard-Till and Yao, 2006). We used this strategy to investigate the role of TPB2 during conjugation. The region for hairpin construction in TPB2 gene is shown in Figure 2A. TPB2 hairpin RNA expression was driven from MTT1 promoter, which is induced by the presence of Cd2+ ion. We examined the effect of gene silencing during conjugation by Northern blot analysis (Figure 2B). Hairpin RNA was present and maintained at all conjugating stages in the presence of CdCl2. TPB2 mRNA expression was reduced ∼50% at the 12-h stage and more drastically (∼90%) at the 14-h stage when DNA deletion takes place. A new band likely representing the degradation products was also detected (Figure 2B, arrow). These results indicate that TPB2 mRNA expression can be efficiently down-regulated by the expression of TPB2 hairpin RNA.
Figure 2.
TPB2 gene silence by hairpin RNA expression. (A) Schematic map of hairpin RNA constructs. Open arrows represent TPB2 gene with double lines inside indicating the region targeted by the hairpin RNA. (B) TPB2 gene silencing by hairpin RNA expression. Northern blot of total RNA samples extracted from conjugating cells (4, 6, 8, 10, 12, 14, and 16 h after postmixing) transformed with the hairpin RNA construct with or without inducing with 0.05 μg/ml CdCl2 in 10 mM Tris buffer. The approximate size of TPB2 mRNA is indicated to the left of the panels, and the right arrow indicates a new band likely from the degraded RNA. The robust TPB2 hairpin RNA expression was also detected. The RPL21 mRNA was used as an internal control to quantify relative folds of the full-length TPB2 mRNA level.
TPB2 Is Essential for Nuclear Differentiation in Conjugation
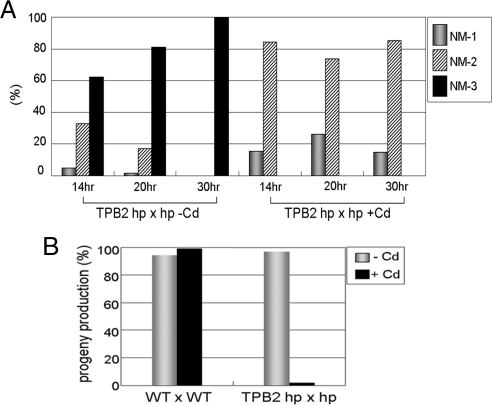
To determine whether TPB2 is essential for conjugation process, we analyzed nuclear events at different conjugating stages by DAPI staining. TPB2 silenced cells were arrested at the stage with two new Mic and two new Mac (Figure 3A). This phenotype is similar to several previously studied mutants (ΔEMA1, ΔEZL1, ΔDCL1, ΔPDD1, ΔPDD2, ΔLIA1, and ΔTWI1) that have defects in DNA rearrangements (Coyne et al., 1999; Nikiforov et al., 1999; Mochizuki et al., 2002; Malone et al., 2005; Mochizuki and Gorovsky, 2005; Liu et al., 2007; Rexer and Chalker, 2007; Aronica et al., 2008). To test for their abilities to produce progeny, single mating pairs were cloned, and their growth was analyzed. Significantly, more than 95% of the 300 individual TPB2-silenced mating cells failed to grow and eventually died (Figure 3B). Thus, we concluded that TPB2 gene is essential for the completion of conjugation and for the formation of viable progeny.
Figure 3.
Phenotype of TPB2 silencing in conjugation. (A) Late conjugating stages analysis of the mutant. Conjugating cells (14, 20, and 30 h after postmixing) of TPB2 hairpin RNA-containing strains with or without induction with 0.05 μg/ml CdCl2 were stained with DAPI to reveal their nuclear stages (Figure 1A). NM (New Mac)-1, anlagen swelled in mating cells; NM-2, mating cells separated and two macronuclei and two micronuclei were present; NM-3, two macronuclei and one micronucleus were present. (B) Viability of progeny. Wild-type and TPB2 RNA hairpin cells were mated as indicated. Individual cell mating pairs were cloned into a drop of growth media at 10 h after postmixing. Cells were examined for progeny production by viability and drug resistance, which is specific for new developing macronucleus in progeny.
IES Elimination and Chromosome Breakage Are Severely Affected in TPB2 Silencing Cells
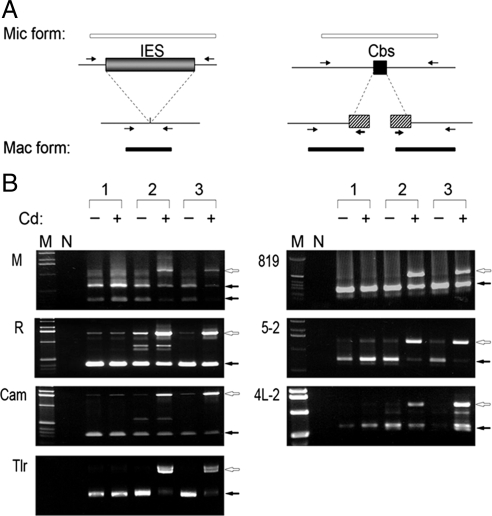
To determine the importance of TPB2 in DNA rearrangements, we examined IES elimination at four regions (M-, R-, Cam-, and Tlr element) and chromosome breakage at three sites (Cbs819, 5-2, and 4L-2) by PCR, using whole cell DNA isolated from pools of mating cells at the terminal stage of conjugation (30 h after mating initiation) as templates. In wild-type mating pools, the macronuclear form (Mac form) of PCR products was the majority compared with the micronuclear form (Mic form) at all regions studied. These ratios of the Mac/Mic form partly reflected the relative DNA amounts of the polyploid (45C) macronucleus and the diploid micronucleus. Significantly, in TPB2 silenced mating pools, the Mic form was much more abundant than the Mac form (Figure 4B), indicating the failure of DNA rearrangements. The small amounts of Mac form were most likely derived from nonmating cells that were also present in the pool. These results demonstrate that IES elimination and chromosome breakage are severely affected in TPB2-silenced cells. We concluded that TPB2 is essential for the programmed genome rearrangements in Tetrahymena.
Figure 4.
DNA elimination and chromosome breakage of TPB2 silencing in conjugation. (A) Schematic drawing of the IES elimination and chromosome breakage PCR assay. The open box represented DNA region retained in the macronucleus, and the filled box represented IES, Cbs, or telomere regions. Arrows indicate the positions of PCR primers. The relative lengths of the expected products were shown at top (Mic form) and bottom (Mac form). (B) The result of IES elimination and chromosome breakage PCR assay. The genomic DNA were isolated from pools of conjugating cells (30 h after postmixing) of wild type (1) and TPB2 hairpin RNA-containing strains (2 and 3) with or without induction with 0.05 μg/ml CdCl2. The open arrows indicate Mic form PCR product, and the filled arrows indicate Mac form PCR product.
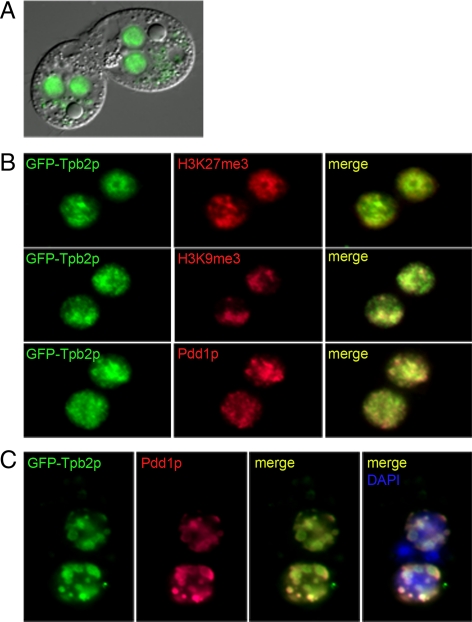
GFP-Tpb2p Is Colocalized with Heterochromatin Markers and Pdd1p-containing Structures
To further understand the role of Tpb2p, its localization was analyzed by expressing Tpb2p fused with GFP at its N-terminus in an rDNA vector and then transformed into the macronuclear genome as high copy plasmids (Yu and Blackburn, 1989; Godiska and Yao, 1990). The overexpressed GFP-Tpb2p was localized within the developing macronuclear anlagen in conjugating cells (Figure 5A). We performed immunofluorescence analysis of H3K27me3, H3K9me3, and Pdd1p in GFP-Tpb2p–overexpressed conjugating cells (∼10 h). The results indicated that GFP-Tpb2p has similar localizations with H3K27me3 and H3K9me3, two markers known to be associated with heterochromatin, and Pdd1p, which is a chromodomain-containing protein involved in the establishment of DNA elimination heterochromatin structures (Figure 5B). GFP-Tpb2p colocalized with Pdd1p-containing nuclear structures at late stages of conjugation (∼14 h; Figure 5C). These cytological studies indicate that the function of Tpb2p might be involved in chromatin modification process.
Figure 5.
Localization of GFP-Tpb2p. (A) Overexpression of the N-terminal GFP-Tpb2p fusion protein. The GFP-Tpb2p fusion protein localized within the developing macronuclear anlagen in living conjugating cells. (B) Colocalization of GFP-Tpb2p and heterochromatin marker (H3K27me3, H3K9me3, and Pdd1p). Ten-hour conjugating cells with GFP-Tpb2p (green) were processed for immunofluorescence staining with the indicated antibodies (red). (C) Colocalization of GFP-Tpb2p and Pdd1p in the DNA elimination heterochromatin structure. Fourteen-hour conjugating cells expressing GFP-Tpb2p (green) were processed for immunofluorescence staining with an anti-Pdd1p antibody (red) and stained with DAPI for DNA (blue).
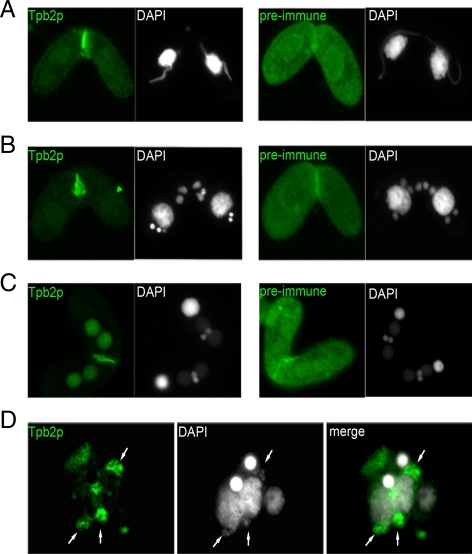
To support the GFP-Tpb2p localization results, we also generated antibodies against Tpb2p and performed immunofluorescence analysis. The results show that the endogenous Tpb2p was presented specifically within the developing macronuclear anlagen in conjugating cells (Figure 6, A–C) and especially colocalized with DNA elimination structures at the nuclear periphery similar to Pdd1p (see next section for more details; Madireddi et al., 1994, 1996; Figure 6D). These results indicate that Tpb2p is targeted to the heterochromatic DNA elimination structure just like the overexpressed GFP-Tpb2p, further supporting its direct role in DNA deletion.
Figure 6.
Localization of endogenous Tpb2p by antibody immunofluorescence staining. Cells were processed for immunofluorescence staining (green) and DAPI staining (white) using an anti-Tpb2p antibody and preimmune serum at different developmental stages in conjugation. (A) “Crescent” stage in micronuclei meiosis. (B) postmeiotic mitosis stage; (C) new developing macronuclei stage at early mating pair; (D) at the late conjugating stage after pair separation.
TPB2 Is Not Required for the Physical Association of Pdd1p and H3K27me3 with IESs
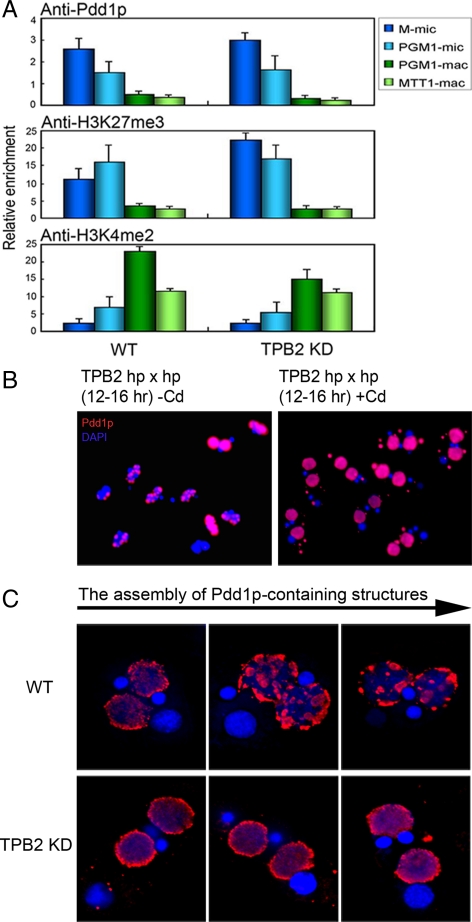
Genes involved in small RNA production, such as DCL1 and TWI1, are expressed early in conjugation and are essential for the targeted heterochromatin formation on IESs (Liu et al., 2007). Because of its late expression timing, TPB2 might not be required for IES targeting. This issue was examined by chromatin immunoprecipitation (ChIP) studies. Wild-type and TPB2 silenced cells at the 10-h stage were processed for ChIP, and two IESs, M-mic and PGM1-mic and two macronuclear genes, PGM1-mac and MTT1-mac, were analyzed by real-time PCR. In both wild-type and TPB2-silenced cells, Pdd1p and H3K27me3 were enriched in the IES regions, whereas H3K4me2, a euchromatin marker, was enriched in the macronuclear genes (Figure 7A). Thus, TPB2 is dispensable for the IES-targeting process. These results place the action of TPB2 downstream of the targeting to induce DNA elimination.
Figure 7.
TPB2 is dispensable for recruiting heterochromatin markers to IES and is essential for the assembly of heterochromatin structures. (A) Conjugating cells of wild type (WT) and TPB2 knockdown strains (TPB2 KD) were processed for ChIP using the antibodies indicated. Two micronuclear IESs (M-mic and PGM1-mic) and two macronuclear genes (PGM1-mac and MTT1-mac) were assayed by quantitative PCR, and the relative enrichment values are shown. (B) Conjugating cells (12–16 h) of TPB2 hairpin RNA strains with or without induction with 0.05 μg/ml CdCl2 were processed for immunoflurescence staining with an anti-Pdd1p antibody (red) and DAPI staining (blue). (C) Magnified images of the assembly of Pdd1p-containing structures in WT and TPB2 KD cells.
TPB2 Is Essential for the Assembly of Pdd1p-containing Structures
Previous studies have shown that H3K27me3 and H3K9me3 accumulate on IES loci shortly after new macronuclear formation begins (∼7 h after mating) and before the assembly of large Pdd1p-containing structures. Pdd1p-containing structures first appeared homogeneously distributed in the developing macronuclei and later formed a few large spherical structures (∼14 h after mating) near the nuclear periphery. Timing of DNA elimination (12–14 h) corresponds well with the aggregation of the Pdd1p-containing structures (Austerberry et al., 1984; Madireddi et al., 1994, 1996; Chalker, 2008). We performed cytological analysis of the assembly of Pdd1p-containing structures by immunofluorescence staining using an anti-Pdd1p antibody. TPB2-silenced cells did not form large Pdd1p-containing structures, and some cells formed abnormal Pdd1p aggregates in the cytosol (Figure 7, B and C). Thus, TPB2 plays an important role in the assembly of Pdd1p-containing structures.
Tpb2p Has an Endonuclease Activity in Vitro
Because Tpb2p shares the conserved transposase catalytic DDD motif (Figure 1C), we speculated that Tpb2p might posses a nuclease activity that carries out the final DNA-cutting step in programmed DNA deletion. To test this possibility, we examined possible nuclease activities of Tpb2p in vitro. Because of the difference in genetic codon usage, the TPB2 gene sequence was modified and fused to GST to allow protein purification from E. coli.
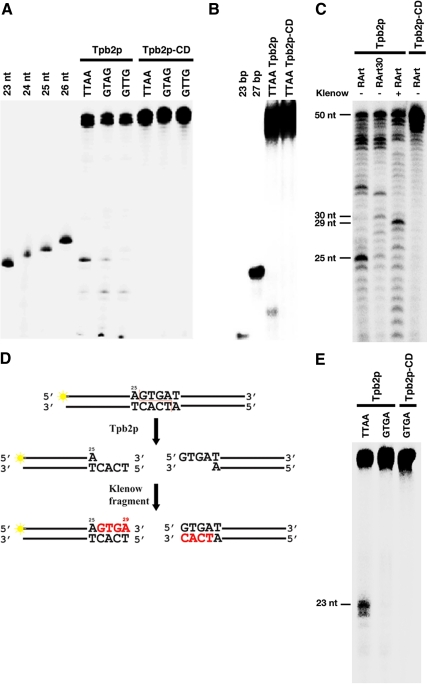
PiggyBac recognizes 5′-TTAA-3′ sequence and induces double-strand break at the bond before the first T of 5′-TTAA-3′ (Cary et al., 1989). Therefore we used a 50-base pair DNA that had 5′-TTAA-3′ sequence only once (the first T is at the 24th position). One strand of the DNA was end-labeled, incubated with recombinant GST-Tpb2p, and analyzed by denaturing gel-electrophoresis. A 23-base product was detected by autoradiography (Figure 8A, Tpb2p-TTAA), indicating that Tpb2p cuts the bond between 23rd and 24th (right before the first T of 5′-TTAA-3′) of the labeled DNA strand. Analysis of the same product in a native gel detected a ∼25-base pair product indicating that Tpb2p produces double-strand break. No cleavage product was detected when we used a Tpb2p mutant in which all three aspartic acids in the DDD motif (Figure 1C) were replaced by leucines (Figure 8, A and B, Tpb2p-CD). These results suggest that Tpb2p has DDD motif-dependent endodeoxyribonuclease activity.
Figure 8.
Tpb2p has DDD-motif dependent endonuclease activities. (A) Wild-type Tpb2p and a mutant Tpb2p in which all three aspartic acids in the DDD motif were replaced by leucines (Tpb2p-CD) were expressed in E. coli and incubated with 50-base pair DNA substrates that had TTAA, GTAG, or GTTG sequence once at their middle. One of two strands was 5′-end labeled. These products were separated in a denaturing polyacrylamide gel and detected by a phosphorimager. 23-, 24-, 25-, and 26-nt oligo DNAs were used as size markers. (B) The same product from the 50-base pair DNA substrate, which has TTAA shown in A, was separated in a native polyacrylamide gel and detected by a phosphorimager. Twenty-three- and 27-base pair oligo DNAs were used as size markers. (C) Tpb2p and Tpb2p-CD were expressed in E. coli and incubated with 50-base pair DNA substrates RArt or RArt30, which had AGTGAT sequence identified at a boundary of Tetrahymena R-element at their 25th and 30th position, respectively. One of two strands was 5′-end labeled. An aliquot of the product from RArt was also treated with Klenow fragment. These products were separated in a denaturing polyacrylamide gel and detected by a phosphorimager. Positions of 25-, 29-, 30-, and 50-nt oligo DNAs used as size markers are indicated in left. (D) Schematic representation of RArt cleavage by Tpb2p and followed by the fill-in reaction by Klenow fragment. 32P-labeled end is marked with yellow stars, and nucleotides added by Klenow fragment are highlighted by red. (E) Tpb2p and Tpb2p-CD were expressed in E. coli and incubated with 50-base pair DNA substrates which had TTAA or GTGA sequence once at their middle. One of two strands was 5′-end labeled. These products were separated in a denaturing polyacrylamide gel and detected by a phosphorimager. Twenty-three-nucleotide oligo DNA was used as size markers.
We then used 50-base pair DNA that has 5′-GTAG-3′ or 5′-GTTG-3′ instead of 5′-TTAA-3′ and found that Tpb2p could utilize the 50-base pair DNA with GTAG as a substrate and produce a 23-nucleotide (nt) product, albeit the amount of the product was lower than that from 50-base pair DNA with 5′-TTAA-3′ (Figure 8A, Tpb2p-GTAG). In contrast, 50-base pair DNA with 5′-GTTG-3′ could not produce any detectable 23-nt product (Figure 8A, Tpb2p-GTTG). These results suggest that Tpb2p has some sequence preference in its cutting site.
In Tetrahymena, DNA elimination machinery is thought to recognize 5′-ANNNNT-3′ sequence to induce DNA double-strand break (Saveliev and Cox, 1996). Therefore, we next tested the 50-base pair DNA substrate RArt and RArt30, which have the 5′-AGTGAT-3′ sequence. This sequence is found at the left boundary of the Tetrahymena R-element and the first A of the 5′-AGTGAT-3′ is located at the 25th and 30th position of the labeled strand of RArt and RArt 30, respectively. The observed cleavage site during DNA elimination of the R-element in vivo (Saveliev and Cox, 1996) corresponds to the bond between the first A and the first G of the 5′-AGTGAT-3′ sequence. The products were separated in a denaturing polyacrylamide gel. Twenty-five-nucleotide products from RArt (Figure 8C, RArt Tpb2p) were detected, indicating that Tpb2p has an endonuclease activity, which prefers to cut at the in vivo cleavage site of the R-element. A 30-nt product from RArt30 was also detected (Figure 8C, RArt30 Tpb2p), although the product was less abundant than that from RArt. This was possibly because Tpb2 needs certain length of DNA at both sides to make stable Tpb2p-DNA complex for inducing efficient cleavage. Tpb2p-CD could not produce any specific cleavage products from RArt (Figure 8C, RArt Tpb2p-CD), strongly support the specificity of the endonuclease activity detected for Tpb2p. Treatment of the product from RArt with Klenow fragment results in a migration shift of the prominent band from that of 25 nt to that of 29 nt (Figure 8C, RArt Tpb2p + Klenow), demonstrating that Tpb2p cleavage leaves a four-base 5′ protruding end (Figure 8D). This is consistent with the intermediate product of the previously reported DNA elimination pathway (Saveliev and Cox, 1995, 1996), suggesting that Tpb2p has the ability to produce proper double-strand break at the endogenous IES boundary sequence to initiate DNA elimination.
The 5′-TTAA-3′ sequence in the oligo used in Figure 8A was in the context of 5′-A“TTAA”C-3′. Therefore, we speculated that the double-strand break at the 5′-AGTGAT-3′ sequence in RArt oligo (Figure 8C) was possibly mediated not by the proposed consensus 5′-ANNNNT-3′ but by the internal 5′-GTGA-3′ sequence. To test this possibility, Tpb2p was incubated with the oligo possessing 5′-GTGA-3′, which was otherwise identical to the TTAA oligo used in Figure 8A. In contrast to our prediction, no detectable 23-nt product was produced (Figure 8E). This result indicates neither internal four bases nor the first and the last base of the 5′-AGTGAT-3′ sequence is sufficient to be cleaved by Tpb2p. We need further comprehensive analysis to find out what types of sequences Tpb2p recognizes and cuts.
DISCUSSION
In this study we presented evidence that a domesticated piggyBac transposase plays a key role in programmed DNA deletion in Tetrahymena. In an independent recent study, a piggyBac transposase gene was also found to have an essential role in DNA deletion in Paramecium (Baudry et al., 2009), although its exact mode of action was unclear. Thus, the domestication of piggyBac transposase gene for DNA deletion appears to have preceded the split of Tetrahymena and Paramecium in evolution. In Tetrahymena, our studies have further demonstrated that TPB2 participates in heterochromatin organization and likely carries out the DNA cleavage reaction, which was not known from the Paramecium study. The expression timing and localization of Tpb2p suggest that the actions of Tpb2p are with the DNA elimination heterochromatin structures at late stages of conjugation. The TPB2 RNAi silencing studies further revealed its essential role in the assembly of these distinctive heterochromatin structures: their assembly was blocked without the normal expression of TPB2. In these cells the targeting of histone modifications and Pdd1p to the IES-containing chromatin occurred normally. Thus, TPB2 acts downstream of the targeting and appears to play an essential role in inducing large heterochromatin structure formation. The timing of this cytological stage coincides with that of DNA cutting, placing the action of TPB2 in this critical stage of DNA elimination. The evolutional conservation of the DDD catalytic sites in Tpb2p offered a hint for the endonuclease activity. The in vitro enzymatic assay results provided direct evidence that Tpb2p indeed has such endonuclease activities, which could have been evolved from the transposase activity. The fact that the same type of ends found in DNA deletion, a 4-nt 5′ protruding end, was generated by this protein further indicated the relevance of this endonuclease activity to DNA deletion.
In many transposons, their terminal repeat sequences are the targets of tranposases. However, IESs in Tetrahymena do not share common sequence feature at their termini, and their identities are apparently established through heterochromatin formation guided by small RNAs. Because Tpb2p colocalizes with the heterochromatin component Pdd1p (Figures 5C and 6D) and is required for the assembly of IES elimination heterochromatin structure, Tpb2p would have the ability to recognize some features of heterochromatin. We speculate that Tpb2p may be recruited to IES regions by heterochromatin structures but not by specific DNA sequences. This is reminiscent of the chromatin structure–dependent recruitment of some transposases. It is known that heterochromatin conformation enhances the recruitment of Sleeping Beauty transposases and facilitates transposition (Yusa et al., 2004; Ikeda et al., 2007), and RAG1/2 recombinases, which are domesticated transposases, can interact with H3K4me3-containing chromatin in the V(D)J recombination process (Matthews et al., 2007). Understanding how Tpb2p interacts with heterochromatin could shed light on these processes.
On the other hand, our in vitro study showed that some preference for a DNA-cutting site sequence was preserved in Tpb2p (Figure 8). Previous studies have shown that, in the R and M elements, the deletion boundaries are determined by flanking cis-acting sequences located 40–50 base pairs away, and the precise nature of the deletion are dependent on the sequences at the boundaries (Chalker and Yao, 1996; Yao, 1996). Indeed, when new deletions were induced by RNA injections, no precise boundaries were observed, presumably due to the lack of these cis-acting sequences in regions not normally targeted for deletion (Yao et al., 2003). The sequence preference of Tpb2 cutting activities we have observed in this study agrees well with those in vivo findings. Thus, deletion boundary is possibly established through an interaction between Tpb2p and the flanking and boundary sequences after it is targeted to the heterochromatin region.
Although piggyBac transposase exclusively targets TTAA sequence by generating a double-strand break at the 5′ end of TTAA, this sequence is not commonly found at the known deletion boundaries in Tetrahymena. Instead, ANNNNT was identified as a consensus sequence at the ends of a number of IESs (Saveliev and Cox, 1996). Our in vitro study indicated that Tpb2p has ability to induce a double-strand break not only at the TTAA site (Figure 8A) but also at GTAG (Figure 8A) and AGTGAT (Figure 8C) sites. Thus, Tpb2p has probably acquired an ability to target some non-TTAA sequence. Future detailed in vitro study of Tpb2p may elucidate how targeting specificity of Tpb2p determines deletion boundaries of IESs in vivo.
TPB2 appears to have gone through a domestication process to become a host gene and be maintained in the macronucleus. Because programmed DNA deletion is a common trait of most ciliates, we speculate that TPB2 domestication occurred early in their evolution, agreeing well with the observation that piggyBac transposase homolog gene is also involved in programmed genome rearrangements in Paramecium tetraurelia (Baudry et al., 2009). However, studies of a distantly related ciliate Oxytricha trifallax have suggested the involvement of transposases encoded by a group of abundant germline transposons (TBEs) in IES deletion (Nowacki et al., 2009). Therefore, direct participation of transposons in their own removal in a programmed manner, which can be observed in O. trifallax today, could represent an ancient system in the evolution of DNA deletion, which could be followed by the domestication of one of the transposases to carry out most deletion processes like in Tetrahymena. Although several domesticated transposon proteins are known to be involved in host genome regulations in other eukaryotes, their evolutional histories are not clear. Ciliated protozoa may preserve traceable “fossil” records that help illuminate how a transposon has been domesticated to regulate eukaryotic genome.
Supplementary Material
ACKNOWLEDGMENTS
We thank all members of the Yao lab and the Mochizuki lab for helpful discussions and suggestions. This research received funding from European Research Council Starting Grant (204986) under the European Community's Seventh Framework Programme and from the Austrian Academy of Sciences to K.M., from the National Science Council (NSC97-2321-B-001-025) of Taiwan, ROC, and from Institute of Molecular Biology of Academia Sinica to M.C.Y.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-12-1079) on March 31, 2010.
REFERENCES
- Agrawal A., Eastman Q. M., Schatz D. G. Transposition mediated by RAG1 and RAG2 and its implications for the evolution of the immune system. Nature. 1998;394:744–751. doi: 10.1038/29457. [DOI] [PubMed] [Google Scholar]
- Aronica L., Bednenko J., Noto T., DeSouza L. V., Siu K. W., Loidl J., Pearlman R. E., Gorovsky M. A., Mochizuki K. Study of an RNA helicase implicates small RNA-noncoding RNA interactions in programmed DNA elimination in Tetrahymena. Genes Dev. 2008;22:2228–2241. doi: 10.1101/gad.481908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Austerberry C. F., Allis C. D., Yao M. C. Specific DNA rearrangements in synchronously developing nuclei of Tetrahymena. Proc. Natl. Acad. Sci. USA. 1984;81:7383–7387. doi: 10.1073/pnas.81.23.7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudry C., Malinsky S., Restituito M., Kapusta A., Rosa S., Meyer E., Betermier M. PiggyMac, a domesticated piggyBac transposase involved in programmed genome rearrangements in the ciliate Paramecium tetraurelia. Genes Dev. 2009;23:2478–2483. doi: 10.1101/gad.547309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borst P., Greaves D. R. Programmed gene rearrangements altering gene expression. Science. 1987;235:658–667. doi: 10.1126/science.3544215. [DOI] [PubMed] [Google Scholar]
- Cary L. C., Goebel M., Corsaro B. G., Wang H. G., Rosen E., Fraser M. J. Transposon mutagenesis of baculoviruses: analysis of Trichoplusia ni transposon IFP2 insertions within the FP-locus of nuclear polyhedrosis viruses. Virology. 1989;172:156–169. doi: 10.1016/0042-6822(89)90117-7. [DOI] [PubMed] [Google Scholar]
- Casola C., Hucks D., Feschotte C. Convergent domestication of pogo-like transposases into centromere-binding proteins in fission yeast and mammals. Mol. Biol. Evol. 2008;25:29–41. doi: 10.1093/molbev/msm221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chadee D. N., Hendzel M. J., Tylipski C. P., Allis C. D., Bazett-Jones D. P., Wright J. A., Davie J. R. Increased Ser-10 phosphorylation of histone H3 in mitogen-stimulated and oncogene-transformed mouse fibroblasts. J. Biol. Chem. 1999;274:24914–24920. doi: 10.1074/jbc.274.35.24914. [DOI] [PubMed] [Google Scholar]
- Chalker D. L. Dynamic nuclear reorganization during genome remodeling of Tetrahymena. Biochim. Biophys. Acta. 2008;1783:2130–2136. doi: 10.1016/j.bbamcr.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker D. L., Yao M. C. Non-Mendelian, heritable blocks to DNA rearrangement are induced by loading the somatic nucleus of Tetrahymena thermophila with germ line-limited DNA. Mol. Cell. Biol. 1996;16:3658–3667. doi: 10.1128/mcb.16.7.3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker D. L., Yao M. C. Nongenic, bidirectional transcription precedes and may promote developmental DNA deletion in Tetrahymena thermophila. Genes Dev. 2001;15:1287–1298. doi: 10.1101/gad.884601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne R. S., Chalker D. L., Yao M. C. Genome downsizing during ciliate development: nuclear division of labor through chromosome restructuring. Annu. Rev. Genet. 1996;30:557–578. doi: 10.1146/annurev.genet.30.1.557. [DOI] [PubMed] [Google Scholar]
- Coyne R. S., Nikiforov M. A., Smothers J. F., Allis C. D., Yao M. C. Parental expression of the chromodomain protein Pdd1p is required for completion of programmed DNA elimination and nuclear differentiation. Mol. Cell. 1999;4:865–872. doi: 10.1016/s1097-2765(00)80396-2. [DOI] [PubMed] [Google Scholar]
- Dedon P. C., Soults J. A., Allis C. D., Gorovsky M. A. Formaldehyde cross-linking and immunoprecipitation demonstrate developmental changes in H1 association with transcriptionally active genes. Mol. Cell. Biol. 1991;11:1729–1733. doi: 10.1128/mcb.11.3.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doak T. G., Doerder F. P., Jahn C. L., Herrick G. A proposed superfamily of transposase genes: transposon-like elements in ciliated protozoa and a common “D35E” motif. Proc. Natl. Acad. Sci. USA. 1994;91:942–946. doi: 10.1073/pnas.91.3.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elick T. A., Bauser C. A., Fraser M. J. Excision of the piggyBac transposable element in vitro is a precise event that is enhanced by the expression of its encoded transposase. Genetica. 1996;98:33–41. doi: 10.1007/BF00120216. [DOI] [PubMed] [Google Scholar]
- Fraser M. J., Ciszczon T., Elick T., Bauser C. Precise excision of TTAA-specific lepidopteran transposons piggyBac (IFP2) and tagalong (TFP3) from the baculovirus genome in cell lines from two species of Lepidoptera. Insect Mol. Biol. 1996;5:141–151. doi: 10.1111/j.1365-2583.1996.tb00048.x. [DOI] [PubMed] [Google Scholar]
- Godiska R., Yao M. C. A programmed site-specific DNA rearrangement in Tetrahymena thermophila requires flanking polypurine tracts. Cell. 1990;61:1237–1246. doi: 10.1016/0092-8674(90)90688-b. [DOI] [PubMed] [Google Scholar]
- Howard-Till R. A., Yao M. C. Induction of gene silencing by hairpin RNA expression in Tetrahymena thermophila reveals a second small RNA pathway. Mol. Cell. Biol. 2006;26:8731–8742. doi: 10.1128/MCB.01430-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda R., Kokubu C., Yusa K., Keng V. W., Horie K., Takeda J. Sleeping beauty transposase has an affinity for heterochromatin conformation. Mol. Cell. Biol. 2007;27:1665–1676. doi: 10.1128/MCB.01500-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov V. V., Jurka J. RAG1 core and V(D)J recombination signal sequences were derived from Transib transposons. PLoS Biol. 2005;3:e181. doi: 10.1371/journal.pbio.0030181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klobutcher L. A., Herrick G. Developmental genome reorganization in ciliated protozoa: the transposon link. Prog. Nucleic Acid Res. Mol. Biol. 1997;56:1–62. doi: 10.1016/s0079-6603(08)61001-6. [DOI] [PubMed] [Google Scholar]
- Liu Y., Taverna S. D., Muratore T. L., Shabanowitz J., Hunt D. F., Allis C. D. RNAi-dependent H3K27 methylation is required for heterochromatin formation and DNA elimination in Tetrahymena. Genes Dev. 2007;21:1530–1545. doi: 10.1101/gad.1544207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madireddi M. T., Coyne R. S., Smothers J. F., Mickey K. M., Yao M. C., Allis C. D. Pdd1p, a novel chromodomain-containing protein, links heterochromatin assembly and DNA elimination in Tetrahymena. Cell. 1996;87:75–84. doi: 10.1016/s0092-8674(00)81324-0. [DOI] [PubMed] [Google Scholar]
- Madireddi M. T., Davis M. C., Allis C. D. Identification of a novel polypeptide involved in the formation of DNA-containing vesicles during macronuclear development in Tetrahymena. Dev. Biol. 1994;165:418–431. doi: 10.1006/dbio.1994.1264. [DOI] [PubMed] [Google Scholar]
- Malone C. D., Anderson A. M., Motl J. A., Rexer C. H., Chalker D. L. Germ line transcripts are processed by a Dicer-like protein that is essential for developmentally programmed genome rearrangements of Tetrahymena thermophila. Mol. Cell. Biol. 2005;25:9151–9164. doi: 10.1128/MCB.25.20.9151-9164.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews A. G., et al. RAG2 PHD finger couples histone H3 lysine 4 trimethylation with V(D)J recombination. Nature. 2007;450:1106–1110. doi: 10.1038/nature06431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K., Fine N. A., Fujisawa T., Gorovsky M. A. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in Tetrahymena. Cell. 2002;110:689–699. doi: 10.1016/s0092-8674(02)00909-1. [DOI] [PubMed] [Google Scholar]
- Mochizuki K., Gorovsky M. A. A Dicer-like protein in Tetrahymena has distinct functions in genome rearrangement, chromosome segregation, and meiotic prophase. Genes Dev. 2005;19:77–89. doi: 10.1101/gad.1265105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikiforov M. A., Gorovsky M. A., Allis C. D. A novel chromodomain protein, pdd3p, associates with internal eliminated sequences during macronuclear development in Tetrahymena thermophila. Mol. Cell. Biol. 2000;20:4128–4134. doi: 10.1128/mcb.20.11.4128-4134.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikiforov M. A., Smothers J. F., Gorovsky M. A., Allis C. D. Excision of micronuclear-specific DNA requires parental expression of pdd2p and occurs independently from DNA replication in Tetrahymena thermophila. Genes Dev. 1999;13:2852–2862. doi: 10.1101/gad.13.21.2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowacki M., Higgins B. P., Maquilan G. M., Swart E. C., Doak T. G., Landweber L. F. A functional role for transposases in a large eukaryotic genome. Science. 2009;324:935–938. doi: 10.1126/science.1170023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rexer C. H., Chalker D. L. Lia1p, a novel protein required during nuclear differentiation for genome-wide DNA rearrangements in Tetrahymena thermophila. Eukaryot. Cell. 2007;6:1320–1329. doi: 10.1128/EC.00157-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar A., Sim C., Hong Y. S., Hogan J. R., Fraser M. J., Robertson H. M., Collins F. H. Molecular evolutionary analysis of the widespread piggyBac transposon family and related “domesticated” sequences. Mol. Genet. Genom. 2003;270:173–180. doi: 10.1007/s00438-003-0909-0. [DOI] [PubMed] [Google Scholar]
- Saveliev S. V., Cox M. M. Transient DNA breaks associated with programmed genomic deletion events in conjugating cells of Tetrahymena thermophila. Genes Dev. 1995;9:248–255. doi: 10.1101/gad.9.2.248. [DOI] [PubMed] [Google Scholar]
- Saveliev S. V., Cox M. M. Developmentally programmed DNA deletion in Tetrahymena thermophila by a transposition-like reaction pathway. EMBO J. 1996;15:2858–2869. [PMC free article] [PubMed] [Google Scholar]
- Smit A. F., Riggs A. D. Tiggers and DNA transposon fossils in the human genome. Proc. Natl. Acad. Sci. USA. 1996;93:1443–1448. doi: 10.1073/pnas.93.4.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. J., Antonacci F., Eichler E. E., Amemiya C. T. Programmed loss of millions of base pairs from a vertebrate genome. Proc. Natl. Acad. Sci. USA. 2009;106:11212–11217. doi: 10.1073/pnas.0902358106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taverna S. D., Coyne R. S., Allis C. D. Methylation of histone h3 at lysine 9 targets programmed DNA elimination in Tetrahymena. Cell. 2002;110:701–711. doi: 10.1016/s0092-8674(02)00941-8. [DOI] [PubMed] [Google Scholar]
- Volff J. N. Turning junk into gold: domestication of transposable elements and the creation of new genes in eukaryotes. Bioessays. 2006;28:913–922. doi: 10.1002/bies.20452. [DOI] [PubMed] [Google Scholar]
- Yao M. C. Programmed DNA deletions in Tetrahymena: mechanisms and implications. Trends Genet. 1996;12:26–30. doi: 10.1016/0168-9525(96)81385-0. [DOI] [PubMed] [Google Scholar]
- Yao M. C., Chao J. L. RNA-guided DNA deletion in Tetrahymena: an RNAi-based mechanism for programmed genome rearrangements. Annu. Rev. Genet. 2005;39:537–559. doi: 10.1146/annurev.genet.39.073003.095906. [DOI] [PubMed] [Google Scholar]
- Yao M. C., Fuller P., Xi X. Programmed DNA deletion as an RNA-guided system of genome defense. Science. 2003;300:1581–1584. doi: 10.1126/science.1084737. [DOI] [PubMed] [Google Scholar]
- Yu G. L., Blackburn E. H. Transformation of Tetrahymena thermophila with a mutated circular ribosomal DNA plasmid vector. Proc. Natl. Acad. Sci. USA. 1989;86:8487–8491. doi: 10.1073/pnas.86.21.8487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yusa K., Takeda J., Horie K. Enhancement of Sleeping Beauty transposition by CpG methylation: possible role of heterochromatin formation. Mol. Cell. Biol. 2004;24:4004–4018. doi: 10.1128/MCB.24.9.4004-4018.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.