Abstract
To better understand breast cancer etiology and progression, we explored the association between promoter methylation status of three breast cancer related genes (BRCA1, APC and p16) and survival in a large cohort of women with breast cancer. About 800 archived tumor tissues were collected from women diagnosed with a first primary invasive or in situ breast cancer in 1996–1997. The vital status of the participants was followed through the end of year 2005 with a mean follow up time of 8.0 years. Promoter methylation was assessed by methylation-specific PCR (for BRCA1) and MethyLight (for APC and p16). The association of promoter methylation and breast cancer mortality was evaluated by Cox-proportional hazards models. Methylated promoters were found in 59.0%, 48.4% and 3.6% of the tumor samples for BRCA1, APC and p16, respectively. Breast cancer-specific mortality was strongly associated with promoter methylation of p16 [HR and 95% CI: 3.53(1.83–6.78)], whereas the associations with of BRCA1 and APC were less pronounced [HR and 95% CI: 1.81(1.18–2.78) and 1.46(0.98–2.17), respectively]. Similar associations were observed with all-cause mortality. As the number of methylated genes increased, the risk of breast cancer-specific mortality also increased in a dose-dependent manner (p, trend=0.01). Importantly, even with our results stratified by hormone receptor status, promoter methylation of the three genes remained predictive of mortality. Our results suggest that promoter methylation could be promising epigenetic markers to be considered for breast cancer survival.
Keywords: methylation, epigenetics, mortality, breast cancer
INTRODUCTION
During the past several years, early detection and novel treatments have improved survival rates in breast cancer [1]. However, there is still an urgent need for biomarkers that can reliably be used to predict clinical outcome of breast cancer patients [2]. Recently, evidences emerge as gene promoter methylation status could predict cancer survival. A study reported that methylation in APC was associated with an increased risk of prostate cancer–specific mortality in two cohort of patients [3].
Although the causal relationship is still being debated, evidence has shown that promoter hypermethylation is associated with silencing of many crucial genes in the neoplastic process [4]. This phenomenon has also been reported in a large panel of genes in breast cancer [5]. Breast cancer gene 1(BRCA1) encodes a multifunctional protein involved in DNA repair, control of cell-cycle checkpoints, protein ubiquitinylation and chromatin remodeling [6]. About 5–50% of familial breast cancers could be explained by inherited mutations of BRCA1 in different populations [7]. BRCA1 status may potentially be used as a prognostic marker as studies have shown that BRCA1 mutated breast cancer is associated with poor survival [8]. BRCA1 promoter methylation was found be to positively associated with breast cancer 5-year mortality [9]. The adenomatous polyposis coli (APC) gene is a tumor suppressor gene associated with both familial and sporadic cancer [10]. Its best understood function is destabilization of β̣-catenin and aberrant nuclear and cytoplasmic localization of β-catenin in human breast cancer [11]. Promoter methylation in APC was observed in breast cancer [12] and was associated with poor prognosis [13]. p16INK4A is a predominant tumor suppressor gene in a wide variety of human cancers and p16 promoter methylation has been identified as a major mechanism for p16 inactivation in a number of primary cancers [14]. Methylation of the 5’ promoter has been observed in both human breast cancer cell lines and 20–30% of breast cancers [15]. However, its role in breast cancer progression is less known. Despite a high rate of loss of heterozygosity at these three loci in breast cancer, somatic mutations of the remaining allele are rare in sporadic cancer [5]. Therefore, other mechanisms for loss of function must exist. DNA methylation has been proposed as an alternative mechanism to inactivate these genes [16].
We previously reported that intakes of B vitamins as well as common polymorphisms in one-carbon metabolizing genes were associated with breast cancer survival in the population-based Long Island Breast Cancer Study Project [17]. Herein, we investigated promoter methylation status of a panel of breast cancer genes in relation to clinical/pathological factors and survival in the context of breast cancer.
MATERIALS AND METHODS
Study population
We utilized the resources of the parent case-control as well as the follow-up study of the Long Island Breast Cancer Study Project, a population-based study. Briefly, eligible cases were women residing in Nassau or Suffolk counties, who spoke English, were 20 years of age or older, and were newly diagnosed with in situ or invasive breast cancer between August 1, 1996, and July 31, 1997. A total of 1,508 women with breast cancer participated in the baseline, case-control study interview. As previously reported [18], at the time of the first primary breast cancer diagnosis, the mean age was 58.8 years (range: 25.1–98.1); 94% were white, 4% were African American, and 2% were other; 235 (15.6%) had carcinoma in situ and 1273 (84.4%) had invasive tumors. Case women, or their next of kin, were interviewed again approximately five years after diagnosis. Medical records were abstracted during the case-control study and again as part of the follow-up study. Details of the study design have been described previously [18–20]. The study protocol was approved by the Institutional Review Boards of the collaborating institutions. REMARK criteria were used through this report [21].
Exposure Assessment
Participants information used in this study were obtained as part of the 1) case-control (baseline) in-home interview; 2) follow-up telephone interview; and 3) medical record abstraction. The questionnaires were administrated to assess the demographic characteristics, breast cancer-related factors, and treatment information. Treatment information provided by the subject at the time of the follow-up interview was found to be in excellent agreement with the data abstracted from the medical record [radiation therapy (Kappa = 0.97), chemotherapy (Kappa = 0.96), and hormone therapy (Kappa = 0.92)], thus we used the self-reported data. Characteristics of the tumor, including estrogen and progesterone receptor (ER/PR) status, were extracted from medical records. Among cases with this information available, 583 (58.9%) were ER+/PR+; 143 (14.4%) were ER+/PR−; 52 (5.3%) were ER−/PR+; and 212 (21.4%) were ER−/PR−. For a smaller subset of women, we were able to obtain medical record information on tumor size (n=321) and node involvement (n=327).
Tumor block retrieval and DNA extraction
Archived pathology blocks were requested from the 33 hospitals in the Long Island study area and successfully retrieved for 962 (66.2%) women. After the review by trained pathologists, the tumor tissues from 859 subjects (89.3%) were available for laboratory analyses. We compared the demographic and clinico/pathological features between cases with or without tumor block available for methylation analysis in our study. Although most characteristics are similar between these two groups, some factors were different. Case women who had tumor samples available for methylation analysis tended to be older (mean age 59.6 vs. 57.9 years; p=0.005); to have an invasive tumor (87.8% vs. 80.1%; p<0.001); and to be post-menopausal (70.7% vs. 64.6%; p=0.01).
The paraffin blocks from each case participant were used to generate 2x 10 micron thick slides. Tumor tissues were isolated from paraffin sections by microdissection. Tumor DNA was isolated by adding 30 µl of proteinase K-digestion buffer (50mM Tris, pH 8.1, 1 mM EDTA, 0.5% Tween 20, 10 µg/ml proteinase K) to the tube and by incubating overnight at 37°C. Proteinase K was inactivated incubating the samples at 95°C for 10 min and centrifugation.
Analysis of gene promoter methylation
Tumor DNA first underwent bisulfite modification to convert unmethylated cytosine residues to uracil using the CpGnome DNA Modification Kit (Chemicon International, Purchase, NY) following the protocol from the manufacturer. BRCA1 promoter methylation was determined by methylation-specific PCR (MSP) as described previously [9]. The MethyLight assay was used for determining the methylation status of APC and p16 as described previously [22, 23]. The percentage of methylation was calculated by the 2−ΔΔCT method, where ΔΔCT = (CT,Target-CT,Actin)sample - (CT,Target-CT,Actin) fully methylated DNA[24] and multiplying by 100. Samples containing ≥ 4% fully methylated molecules were designated as methylated, whereas samples containing ≤ 4% were designated as unmethylated. We obtained methylation data on 851 tumor tissues for BRCA1 and on 800 for APC or p16, respectively. The main reason for missing methylation data was insufficient DNA that was obtained from the tumor blocks.
Study outcome
The National Death Index was used to ascertain all-cause and breast cancer-specific mortality. Information on vital status of our study subjects was requested from the National Death Index (NDI), using social security numbers, birth date and other information provided to us by the study subject at the time of the baseline interview. The NDI provided us with potential death matches, including date and cause of death. These data were manually inspected by our research study staff to determine whether the information provided was a likely match to individual subjects within our study cohort. Among the entire cohort of 1508 women diagnosed with breast cancer in 1996–1997, 308 (20.2%) deaths occurred by December 31, 2005, of which 164 (53.2%) were due to breast cancer. Among the subset of women with methylation data available (n = up to 851), a total of 122 (14.3%) deaths (all-cause mortality) were observed, of which 79 (64.8%) were due to breast cancer. The mean follow up time was 8.0 years (range: 0.3–9.4).
Statistical Analysis
The association between gene promoter methylation status of the tumor tissue and the clinical/pathological factors was examined using the chi-square statistic or fisher exact test for categorical variables and by the two-sample t-test for continuous variables [25]. Percentage methylation for APC and p16 was dichotomized to methylated or unmethylated using a cut-off of 4%. The Cox proportional hazard regression [26] was used to estimate the hazard ratio (HR) and 95% confidence interval (CI) for the association between gene promoter methylation status and breast cancer-specific as well as all-cause mortality. Potential confounding by other factors might influence survival among breast cancer patients was evaluated by adjustment in the Cox model. These factors include age at diagnosis, ER/PR status [positive group = ER and PR both positive (ER+/PR+) vs. negative group = any negative receptor (ER+/PR−, ER−/PR+, ER−/PR−)], cancer stage (in situ vs. invasive), menopausal status (pre- vs. post-), race (white vs. other), family history of breast cancer (a mother, sister or daughter) and history of benign breast disease diagnosed by biopsy. If eliminating a covariate from the full Cox regression model changed the effect estimate by 10% or more, the covariate was considered a confounder and kept in the model [27]. Otherwise that covariate was dropped from the multivariate model. None of the covariates tested met this criterion, and thus, only results from the crude model are presented. The HRs and 95% CIs for the association between promoter methylation and mortality stratified by ER/PR status of the case were also estimated by Cox model. All statistical analyses were performed using SAS statistical software version 9.1(SAS Institute, Cary, NC).
RESULTS
The prevalence of the promoter methylation status of BRCA1, APC and p16, as assessed in the archived breast tumor samples of this population-based cohort of up to 851 women (including 104 in situ and 747 invasive cases), was 59.0%, 48.4% and 3.6% for BRCA1, APC and p16, respectively.
Table 1 summarizes the relationship between gene promoter methylation and clinical/pathological factors. BRCA1 promoter methylation was more frequent in cancers that were classified as invasive (p=0.02) and among premenopausal women (p=0.05). BRCA1 promoter methylation was more frequent in cancers with at least one node involved (p=0.003) and with tumor size greater than 2 cm (p=0.001). Promoter methylation status of APC and p16 were not associated with factors examined in Table 1.
Table 1.
Associations between gene promoter methylation assessed in archived tumor tissue and clinical/pathological factors assessed at the baseline interview, among a population-based cohort of case women diagnosed with a first primary breast cancer in 1996–1997 on Long Island, NY.
| Gene | BRCA1 | APC | p16 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Factors | U | M | P | U | M | P | U | M | P |
| 347(40.8) | 504(59.2) | 413(51.6) | 387(48.4) | 771(96.4) | 29(3.6) | ||||
| Age at diagnosis (y) | |||||||||
| ≤60 | 175(40.3) | 259(59.7) | 0.78 | 217(52.4) | 197(47.6) | 0.64 | 395(95.4) | 19(4.6) | 0.13 |
| >60 | 172(41.2) | 245(58.8) | 196(50.8) | 190(49.2) | 376(97.4) | 10(2.6) | |||
| mean age (t-test) | 60.4 | 59.0 | 0.12 | 58.7 | 60.2 | 0.11 | 59.4 | 59 | 0.88 |
| Race | |||||||||
| White | 329(41.5) | 463(58.5) | 0.21 | 380(51.0) | 365(49.0) | 0.30 | 718(96.4) | 27(3.6) | 0.79 |
| Black | 13(31.0) | 29(69.0) | 21(53.9) | 18(46.2) | 37(94.9) | 2(5.1) | |||
| Other | 4(26.7) | 11(73.3) | 10(71.4) | 4(28.6) | 14(100.0) | 0(0.0) | |||
| Cancer type | |||||||||
| in situ | 53(51.0) | 51(49.0) | 0.02 | 41(43.2) | 54(56.8) | 0.08 | 90(94.7) | 5(5.3) | 0.36 |
| invasive | 294(39.4) | 453(60.6) | 372(52.8) | 333(47.2) | 681(96.6) | 24(3.4) | |||
| Menopausal status | |||||||||
| pre- | 87(35.7) | 157(64.3) | 0.05 | 131(56.0) | 103(44.0) | 0.09 | 228(97.4) | 6(2.6) | 0.32 |
| post- | 253(43.0) | 336(57.0) | 271(49.4) | 278(50.6) | 527(96.0) | 22(4.0) | |||
| FHBC | |||||||||
| no | 267(40.1) | 399(59.9) | 0.57 | 326(52.1) | 300(47.9) | 0.88 | 601(96.0) | 25(4.0) | 0.14 |
| yes | 66(42.6) | 89(57.4) | 75(51.4) | 71(48.6) | 144(98.6) | 2(1.4) | |||
| BBD | |||||||||
| no | 276(39.8) | 417(60.2) | 0.21 | 340(52.2) | 312(47.9) | 0.59 | 627(96.2) | 25(3.8) | 0.63 |
| yes | 71(45.2) | 86(54.8) | 73(49.7) | 74(50.3) | 143(97.3) | 4(2.7) | |||
| ER status | |||||||||
| negative | 60(40.3) | 89(59.7) | 0.95 | 82(57.3) | 61(42.7) | 0.13 | 135(94.4) | 8(5.6) | 0.10 |
| positive | 193(40.5) | 283(59.5) | 223(50.1) | 222(49.9) | 433(97.3) | 12(2.7) | |||
| PR status | |||||||||
| negative | 93(40.8) | 135(59.2) | 0.90 | 110(51.2) | 105(48.8) | 0.79 | 205(95.4) | 10(4.7) | 0.20 |
| positive | 160(40.3) | 237(59.7) | 195(52.3) | 178(47.7) | 363(97.3) | 10(2.7) | |||
| Node | |||||||||
| 0 | 106(42.7) | 142(57.3) | 0.003 | 128(55.4) | 103(44.6) | 0.61 | 205(95.4) | 8(3.5) | 0.46 |
| 1 | 19(24.0) | 60(76.0) | 39(52.0) | 36(48.0) | 363(97.3) | 1(1.3) | |||
| Tumor zise | |||||||||
| <2cm | 141(57.1) | 106(42.9) | 0.001 | 127(55.7) | 101(44.3) | 0.66 | 224(98.3) | 4(1.8) | 0.08 |
| >2cm | 58(78.4) | 16(21.6) | 38(52.7) | 34(47.3) | 68(94.4) | 4(5.6) |
Methylation status: U-unmethylated; M-methylated; percentages shown were methylated or unmethylated cases among all; FHBC: family history of breast cancer; BBD: history of benign breast disease; P: p value for chi-square or fisher exact test.
As shown in Table 2, promoter methylation status of three genes, BRCA1, APC and p16, was associated with breast cancer-specific mortality in this study population. Compared to cases with an unmethylated promoter, those who had a methylated promoter had an 81% increased risk of dying from breast cancer at the end of follow up for BRCA1 (HR: 1.81; 95% CI: 1.18–2.78) and a 46% increased risk for APC (HR: 1.46; 95% CI: 0.98–2.17). The strongest effect was observed for p16, with a more than 3-fold increase of risk of dying from breast cancer at the end of follow up (HR: 3.53; 95% CI: 1.83–6.78). Similar associations were observed for all-cause mortality (Table 2). These associations were not attenuated after adjusting for other well-established prognostic factors such as age at diagnosis and ER/PR status (data not shown). We also explored whether there was heterogeneity by cancer type (in situ vs. invasive), but results did not vary substantially (data not shown).
Table 2.
Hazard ratios (HRs) and 95% confidence intervals (CIs) for the associations of gene promoter methylation status and mortality among a population-based cohort of case women diagnosed with a first primary breast cancer in 1996–1997 on Long Island, NY and followed for vital status through the end of 2005.
| Breast cancer-specific mortality | All-cause mortality | |||||||
|---|---|---|---|---|---|---|---|---|
| Gene | Methylation | Case no. | No. of death | P for log- rank test |
HR(95% CI) | No. of death | P for log- rank test |
HR(95% CI) |
| BRCA1 | U | 347 | 29 | 0.006 | 1.00(ref.) | 68 | 0.105 | 1.00(ref.) |
| M | 504 | 74 | 1.81(1.18–2.78) | 122 | 1.28(0.95–1.72) | |||
| APC | U | 413 | 43 | 0.061 | 1.00(ref.) | 81 | 0.037 | 1.00(ref.) |
| M | 387 | 56 | 1.46(0.98–2.17) | 98 | 1.37(1.02–1.84) | |||
| p16 | U | 771 | 89 | <0.0001 | 1.00(ref.) | 168 | 0.016 | 1.00(ref.) |
| M | 29 | 10 | 3.53(1.83–6.78) | 11 | 2.09(1.14–3.84) | |||
Methylation status: U-unmethylated; M-methylated.
When considering promoter methylation status of the three genes simultaneously, a dose-response association emerged; as the number of methylated genes increased, the magnitude of the association with breast cancer-specific mortality also increased (p, trend=0.011) (Table 3). Cases with all three genes methylated had the highest risk of dying from breast cancer at the end of follow up (HR: 3.58; 95% CI: 1.52–8.42). Similar but weaker associations were observed for all-cause mortality (p, trend=0.056).
Table 3.
Number of methylated genes in relation to breast cancer-specific and all-cause mortality among a population-based cohort of women diagnosed with a first primary breast cancer in 1996–1997 on Long Island, NY and followed for vital status through the end of 2005.
| No. of genes methylated |
Case no. | No. of death |
P for log- rank test |
HR(95%CI) |
|---|---|---|---|---|
|
BC-specific mortality |
0.011 | |||
| 0 | 137 | 11 | 1.00(ref.) | |
| 1 | 356 | 43 | 1.52(0.78–2.94) | |
| 2 | 268 | 35 | 1.70(0.86–3.35) | |
| 3 | 38 | 10 | 3.58(1.52–8.42) | |
|
All-cause mortality |
0.294 | |||
| 0 | 137 | 25 | 1.00(ref.) | |
| 1 | 356 | 77 | 1.20(0.77–1.89) | |
| 2 | 268 | 66 | 1.42(0.90–2.25) | |
| 3 | 38 | 11 | 1.76(0.87–3.58) | |
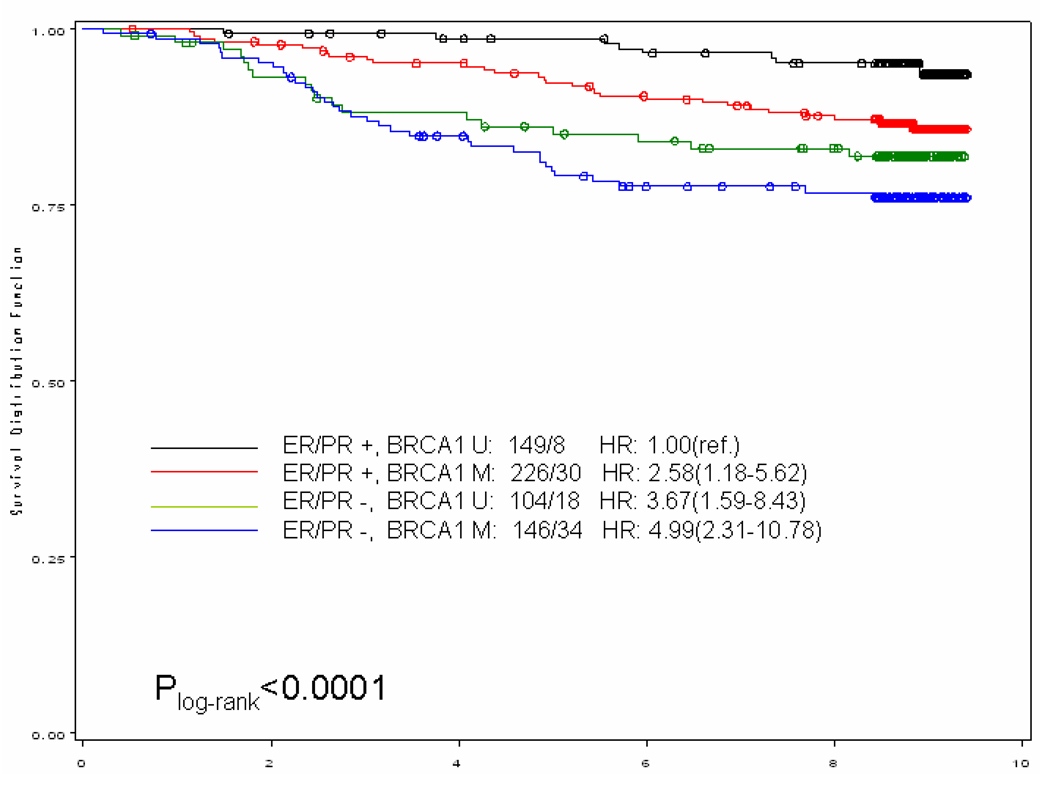
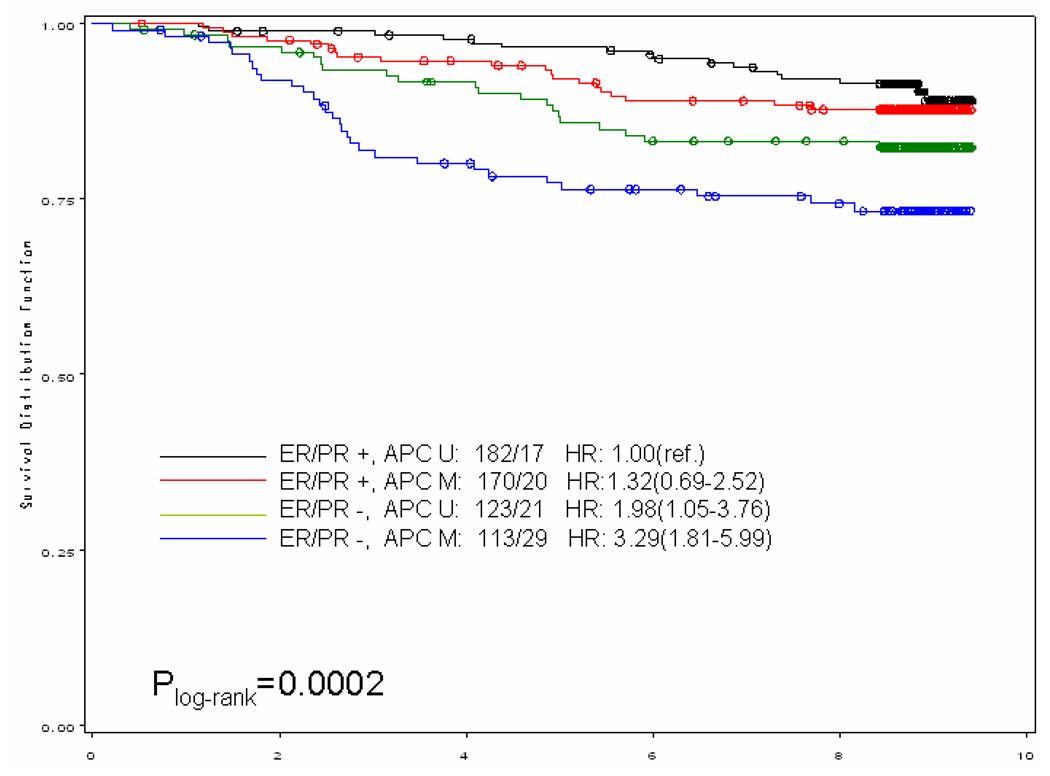
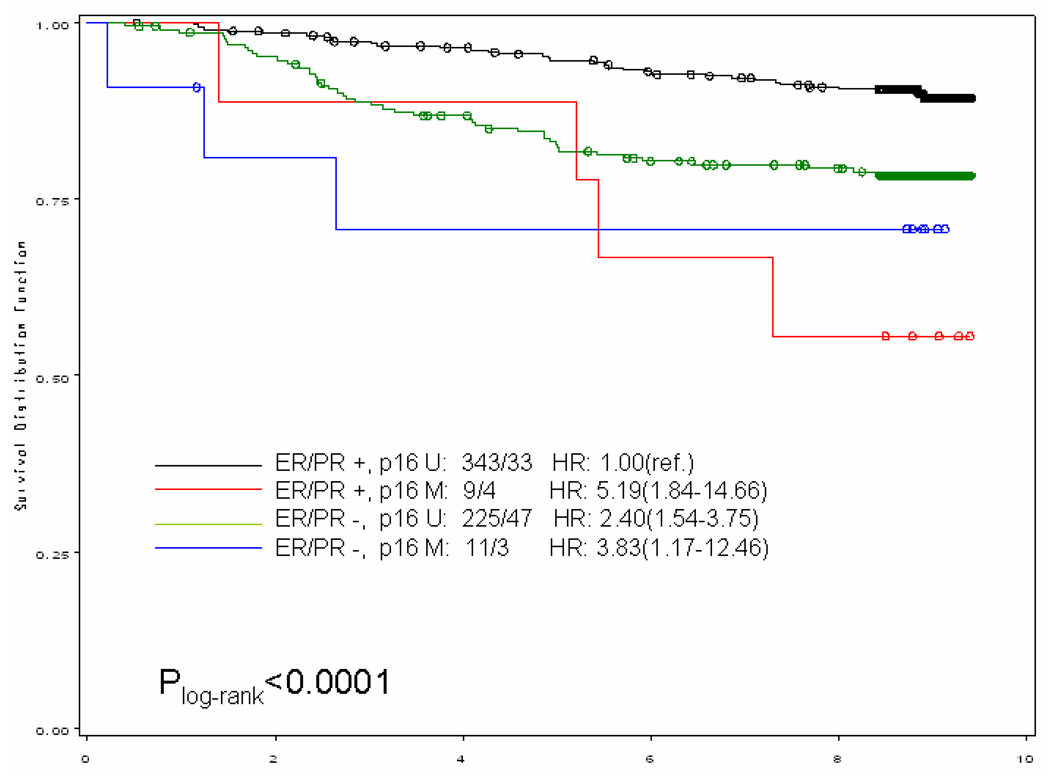
When we stratified our analysis by ER/PR status (both receptors positive vs. any negative receptor) (Figure 1), breast cancer-specific mortality was higher among women with methylated gene promoter regardless of the hormone receptor status of the tumor. In general, it appeared that women with the worse prognosis were those with ER/PR negative breast tumor with methylated gene promoter; for example, among the ER/PR negative subgroup those who also had a methylated BRCA1 promoter, the hazards ratio was more than doubled (HR and 95% CI: 2.58;1.18–5.62) compared to those with unmethylated BRCA1 promoter. An exception to this general pattern were women with ER/PR positive tumors who also had methylated p16 promoter; their risk of dying from breast cancer was more than 5-fold (HR and 95% CI: 5.19; 1.84–14.66) compared to those with unmethylated p16 promoter.
Figure 1.
Kaplan–Meier curve for breast cancer-specific survival of gene promoter methylation by ER/PR status among a population-based cohort of women diagnosed with a first primary breast cancer in 1996–1997 on Long Island, NY and followed for vital status through the end of 2005. Numbers showed before the hazards ratio (HR) are numbers of case / death. (U: unmethylated; M, methylated)
When survival analysis was performed according to treatment groups, the relationship between gene promoter methylation status and survival did not differ between those who received the treatments (radiation, chemotherapy, and/or hormonal therapy) and those who did not (data not shown).
DISCUSSION
Only part of the variation in the risk of mortality among women with breast cancer can be explained by known pathologic and clinical parameters [28]. Thus, to effectively reduce the disease burden of breast cancer, identification of genetic, environmental and behavioral factors that influence survival among breast cancer survivors is needed. We chose to examine promoter methylation status of three genes that were selected based on their biologic importance in breast carcinogenesis [29]. Our study is based on data drawn from a large population-based sample, and is the first to report on the prognostic value of gene promoter methylation status in breast cancer in an epidemiologic study. Although our analysis of methylation-survival association was based on a sub-sample of a population-based parent study, we can quantify any potential biases due to tumor tissue being unavailable for analyses, which is different from the clinic based studies that are unable to quantify to which segment of the population that is generalizable. The population-based study design, in which cases encompassed a broad range of ages and were drawn from a defined geographic area, yields results that are more generalizable than a series of cases from a narrow age range or from a single institution. In addition, the relatively large sample size allows multiple factors to be taken into consideration in studying associations, with the ability to conduct stratified analyses and adjustment in multivariate models.
We found promoter methylation status of three tumor suppressor genes, BRCA1, APC and p16, was associated with breast cancer-specific mortality in our study. The potential prognostic value of mutation in these genes has been explored. A recent review [30] summarized differences in survival outcome in relation to BRCA1 germline mutations. Carriers of a gene mutation had functionally defective proteins, usually resulting in some loss of function. Promoter methylation represented an alternative mechanism for loss of function of these tumor-suppressors. The observed decreased survival associated with methylated gene promoters corroborates the previous findings on mutations. More interestingly, when the number of dysregulated genes (presumably by aberrant promoter methylation) increased, the survival outcome deteriorated, as illustrated by the dose-dependent relationship between the number of methylated genes and mortality (Table 3). These findings lend support to the notion that promoter methylation could silence expression of these of tumor-suppressor genes, affecting disease progression.
We found that the gene promoter methylation status of BRCA1, APC and p16 could further identify cases with statistically significantly different survival profiles among patients with the same ER/PR status (Figure 1). The estrogen receptor (ER) and progesterone receptor (PR) play an important role in regulating growth and differentiation of normal breast epithelium and are well established prognostic factors of breast cancer [31]. Positive receptor status correlates with favorable prognostic features including a lower rate of cell proliferation and histologic evidence of tumor differentiation [32]. Our findings that promoter methylation of BRCA1, APC and p16 can further categorize cases with the same ER/PR status with respect to survival profile may have clinical significance; such information may potentially aid in tailoring treatment strategies based on these epigenetic markers. However, some subgroup analyses are based on small numbers of women (i.e. low frequency of p16 hypermethylation), as reflected in the wide confidence intervals; these results should be interpreted with caution.
The relationship between clinical/pathological factors and gene promoter methylation status has not been well-examined. One study found a higher frequency of APC methylation in inflammatory breast cancer [33] while another reported that APC methylation was less frequent in ER-negative and HER2/neu-negative breast cancers [34]. One report showed that p16 hypermethylation was more frequent among ER-negative cases than ER-positive cases [35]. In our analysis, only BRCA1 methylation status showed some correlations with clinical/pathological factors of breast cancer. BRCA1 promoter methylation was more frequent in invasive than in in situ carcinomas. Different methylation profiles observed in subtypes of breast cancer tissues may imply different etiologic roles of methylation in these disease phenotypes.
Limitations in our study include the following concerns. First, some known, validated, independent breast cancer prognostic factors were not completed on the medical records for all cases. We adjusted for tumor size and node involvement (individually and jointly) on the subset of the population with available information and found the associations were not attenuated. Second, gene expression was not measured in the tumor tissues, thus we could not explore the functional consequence of DNA methylation. Studying gene expression in archived samples (i.e. formaldehyde fixed paraffin-embedded tissues) has been problematic [36] and conflicting results are found in the literature [37]. Nevertheless, the fact that our results corroborate those from gene mutation studies suggests that promoter methylation indeed results in gene silencing or loss of gene function. Third, Long Island Breast Cancer Study Project was conducted in multiple hospitals, so the treatment information was not as detailed as data from a single institutional study. This limited our ability to conduct a more detailed investigation on the predictive effect of gene methylation status. For those cases who had treatment information available, we performed separate analyses according to treatment groups within that subset of population; no substantial differences were observed. Lastly, one potential limitation is that we can not rule out the possibility that for some subjects, cause of death may have been misclassified. The accuracy of cause of death on death certificates has been questioned [38], and is an ongoing issue in most cohort studies of cause-specific mortality. However, the National Death Index completeness has been found to be reasonably validated [39], and thus it remains a standard source of mortality data for epidemiologic research.
In summary, we examined promoter methylation status of three breast cancer genes and explored their relationship with clinical/pathological factors as well as breast cancer survival. Our results indicate that promoter methylation of BRCA1, APC and p16 could offer additional value in breast cancer prognosis beyond established prognostic factors such as ER/PR status.
ACKNOWLEDGEMENTS
This work was supported by grants from the National Institutes of Health (CA109753) and in part by grants from Department of Defense (BC031746), National Institutes of Health (UO1CA/ES66572, UO1CA66572, P30CA013696, P30ES009089 and P30ES10126). Xu, X. is a recipient of the Predoctoral Traineeship Award (W81XWH-06-1-0298) of Department of Defense Breast Cancer Research Program.
REFERENCES
- 1.Landis SH, Murray T, Bolden S, et al. Cancer statistics, 1999. CA Cancer J Clin. 1999;49:8–31. doi: 10.3322/canjclin.49.1.8. [DOI] [PubMed] [Google Scholar]
- 2.Gralow J, Ozols RF, Bajorin DF, et al. Clinical Cancer Advances 2007: Major Research Advances in Cancer Treatment, Prevention, and Screening A Report From the American Society of Clinical Oncology. J Clin Oncol. 2008;26:313–325. doi: 10.1200/JCO.2007.15.4088. [DOI] [PubMed] [Google Scholar]
- 3.Richiardi L, Fiano V, Vizzini L, et al. Promoter Methylation in APC, RUNX3, and GSTP1 and Mortality in Prostate Cancer Patients. J Clin Oncol. 2009;27:3161–3168. doi: 10.1200/JCO.2008.18.2485. [DOI] [PubMed] [Google Scholar]
- 4.Herman JG. Hypermethylation of tumor suppressor genes in cancer. Semin Cancer Biol. 1999;9:359–367. doi: 10.1006/scbi.1999.0138. [DOI] [PubMed] [Google Scholar]
- 5.Widschwendter M, Jones PA. DNA methylation and breast carcinogenesis. Oncogene. 2002;21:5462–5482. doi: 10.1038/sj.onc.1205606. [DOI] [PubMed] [Google Scholar]
- 6.Ralhan R, Kaur J, Kreienberg R, et al. Links between DNA double strand break repair and breast cancer: accumulating evidence from both familial and nonfamilial cases. Cancer Lett. 2007;248:1–17. doi: 10.1016/j.canlet.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 7.Hopper JL. Genetic epidemiology of female breast cancer. Semin Cancer Biol. 2001;11:367–374. doi: 10.1006/scbi.2001.0392. [DOI] [PubMed] [Google Scholar]
- 8.Moller P, Evans DG, Reis MM, et al. Surveillance for familial breast cancer: Differences in outcome according to BRCA mutation status. Int J Cancer. 2007;121:1017–1020. doi: 10.1002/ijc.22789. [DOI] [PubMed] [Google Scholar]
- 9.Xu X, Gammon MD, Zhang Y, et al. BRCA1 promoter methylation is associated with increased mortality among women with breast cancer. Breast Cancer Res Treat. 2009;115:397–404. doi: 10.1007/s10549-008-0075-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Polakis P. The adenomatous polyposis coli (APC) tumor suppressor. Biochimica et Biophysica Acta (BBA) - Reviews on Cancer. 1997;1332:F127–F147. doi: 10.1016/s0304-419x(97)00008-5. [DOI] [PubMed] [Google Scholar]
- 11.Lin S, Xia W, Wang JC, et al. β-Catenin, a novel prognostic marker for breast cancer: Its roles in cyclin D1 expression and cancer progression. Proc Natl Acad Sci U S A. 2000;97:4262–4266. doi: 10.1073/pnas.060025397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klarmann GJ, Decker A, Farrar WL. Epigenetic gene silencing in the Wnt pathway in breast cancer. Epigenetics. 2008;3:59–63. doi: 10.4161/epi.3.2.5899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Muller HM, Widschwendter A, Fiegl H, et al. DNA Methylation in Serum of Breast Cancer Patients: An Independent Prognostic Marker. Cancer Res. 2003;63:7641–7645. [PubMed] [Google Scholar]
- 14.Rocco JW, Sidransky D. p16(MTS-1/CDKN2/INK4a) in Cancer Progression. Experimental Cell Research. 2001;264:42–55. doi: 10.1006/excr.2000.5149. [DOI] [PubMed] [Google Scholar]
- 15.Yang X, Yan L, Davidson NE. DNA methylation in breast cancer. Endocr Relat Cancer. 2001;8:115–127. doi: 10.1677/erc.0.0080115. [DOI] [PubMed] [Google Scholar]
- 16.Birgisdottir V, Stefansson OA, Bodvarsdottir SK, et al. Epigenetic silencing and deletion of the BRCA1 gene in sporadic breast cancer. Breast Cancer Res. 2006;8:R38. doi: 10.1186/bcr1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu X, Gammon MD, Wetmur JG, et al. B-Vitamin Intake, One-Carbon Metabolism, and Survival in a Population-Based Study of Women with Breast Cancer. Cancer Epidemiol Biomarkers Prev. 2008;17:2109–2116. doi: 10.1158/1055-9965.EPI-07-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gammon MD, Neugut AI, Santella RM, et al. The Long Island Breast Cancer Study Project: description of a multi-institutional collaboration to identify environmental risk factors for breast cancer. Breast Cancer Res Treat. 2002;74:235–254. doi: 10.1023/a:1016387020854. [DOI] [PubMed] [Google Scholar]
- 19.Gammon MD, Santella RM, Neugut AI, et al. Environmental toxins and breast cancer on Long Island. I. Polycyclic aromatic hydrocarbon DNA adducts. Cancer Epidemiol Biomarkers Prev. 2002;11:677–685. [PubMed] [Google Scholar]
- 20.Cleveland RJ, Eng SM, Abrahamson PE, et al. Weight gain prior to diagnosis and survival from breast cancer. Cancer Epidemiol Biomarkers Prev. 2007;16:1803–1811. doi: 10.1158/1055-9965.EPI-06-0889. [DOI] [PubMed] [Google Scholar]
- 21.McShane LM, Altman DG, Sauerbrei W, et al. REporting recommendations for tumor MARKer prognostic studies (REMARK) Breast Cancer Res Treat. 2006;100:229–235. doi: 10.1007/s10549-006-9242-8. [DOI] [PubMed] [Google Scholar]
- 22.Eads CA, Danenberg KD, Kawakami K, et al. CpG Island Hypermethylation in Human Colorectal Tumors Is Not Associated with DNA Methyltransferase Overexpression. Cancer Res ; Cancer Res. 1999;59:2302–2306. [PubMed] [Google Scholar]
- 23.Eads CA, Lord RV, Kurumboor SK, et al. Fields of Aberrant CpG Island Hypermethylation in Barrett's Esophagus and Associated Adenocarcinoma. Cancer Res ; Cancer Res. 2000;60:5021–5026. [PubMed] [Google Scholar]
- 24.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 25.Rosner BA. Fundamentals of Biostatistics. Duxbury, Australia ; Pacific Grove, CA: 2000. [Google Scholar]
- 26.Hosmer DW. Applied survival analysis : regression modeling of time to event data. New York: Wiley; 1999. [Google Scholar]
- 27.Rothman KJ, Greenland S. Modern Epidemiology. Philadelphia, Pa: Lippincott-Raven Publishers; 1998. [Google Scholar]
- 28.Anonymous. Cancer Medicine. Hamilton, London: BC Decker Inc; 2003. [Google Scholar]
- 29.Rosen EM, Fan S, Pestell RG, et al. BRCA1 gene in breast cancer. J Cell Physiol. 2003;196:19–41. doi: 10.1002/jcp.10257. [DOI] [PubMed] [Google Scholar]
- 30.Liebens FP, Carly B, Pastijn A, et al. Management of BRCA1/2 associated breast cancer: a systematic qualitative review of the state of knowledge in 2006. Eur J Cancer. 2007;43:238–257. doi: 10.1016/j.ejca.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 31.Silvestrini R, Daidone MG, Luisi A, et al. Biologic and clinicopathologic factors as indicators of specific relapse types in node-negative breast cancer. J Clin Oncol. 1995;13:697–704. doi: 10.1200/JCO.1995.13.3.697. [DOI] [PubMed] [Google Scholar]
- 32.Osborne CK. Steroid hormone receptors in breast cancer management. Breast Cancer Res Treat. 1998;51:227–238. doi: 10.1023/a:1006132427948. [DOI] [PubMed] [Google Scholar]
- 33.Van der Auwera I, Van Laere SJ, Van den Bosch SM, et al. Aberrant methylation of the Adenomatous Polyposis Coli (APC) gene promoter is associated with the inflammatory breast cancer phenotype. Br J Cancer. 2008;99:1735–1742. doi: 10.1038/sj.bjc.6604705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sunami E, Shinozaki M, Sim MS, et al. Estrogen receptor and HER2/neu status affect epigenetic differences of tumor-related genes in primary breast tumors. Breast Cancer Res. 2008;10:R46. doi: 10.1186/bcr2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tao MH, Shields PG, Nie J, et al. DNA hypermethylation and clinicopathological features in breast cancer: the Western New York Exposures and Breast Cancer (WEB) Study. Breast Cancer Res Treat. 2009;114:559–568. doi: 10.1007/s10549-008-0028-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wilson CA, Ramos L, Villasenor MR, et al. Localization of human BRCA1 and its loss in high-grade, non-inherited breast carcinomas. Nat Genet. 1999;21:236–240. doi: 10.1038/6029. [DOI] [PubMed] [Google Scholar]
- 37.Al-Mulla F, Abdulrahman M, Varadharaj G, et al. BRCA1 gene expression in breast cancer: a correlative study between real-time RT-PCR and immunohistochemistry. J Histochem Cytochem. 2005;53:621–629. doi: 10.1369/jhc.4A6544.2005. [DOI] [PubMed] [Google Scholar]
- 38.Sington JD, Cottrell BJ. Analysis of the sensitivity of death certificates in 440 hospital deaths: a comparison with necropsy findings. J Clin Pathol. 2002;55:499–502. doi: 10.1136/jcp.55.7.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rich-Edwards JW, Corsano KA, Stampfer MJ. Test of the National Death Index and Equifax Nationwide Death Search. Am J Epidemiol. 1994;140:1016–1019. doi: 10.1093/oxfordjournals.aje.a117191. [DOI] [PubMed] [Google Scholar]