Summary
Sequestration of Plasmodium falciparum-infected erythrocytes in the placenta is responsible for many of the harmful effects of malaria during pregnancy. Sequestration occurs as a result of parasite adhesion molecules expressed on the surface of infected erythrocytes binding to host receptors in the placenta such as chondroitin sulphate A (CSA). Identification of the parasite ligand(s) responsible for placental adhesion could lead to the development of a vaccine to induce antibodies to prevent placental sequestration. Such a vaccine would reduce the maternal anaemia and infant deaths that are associated with malaria in pregnancy. Current research indicates that the parasite ligands mediating placental adhesion may be members of the P. falciparum variant surface antigen family PfEMP1, encoded by var genes. Two relatively well-conserved subfamilies of var genes have been implicated in placental adhesion, however, their role remains controversial. This review examines the evidence for and against the involvement of var genes in placental adhesion, and considers whether the most appropriate vaccine candidates have yet been identified.
Introduction
In the late 1990s it was discovered that pregnant women in malaria-endemic countries develop antibodies against conserved epitopes of malaria parasites causing placental infection (Fried et al., 1998; Ricke et al., 2000). This finding caused great excitement among researchers as it suggested that developing a vaccine to mimic this naturally acquired immunity might be relatively straightforward. Such a vaccine could prevent the 75 000–200 000 infant deaths that are estimated to occur annually because of malaria in pregnancy (Steketee et al., 2001). The race was on to identify the parasite molecules that facilitate Plasmodium falciparum infection of the placenta and are the targets of protective immunity. This review summarizes the progress that has been made over the past 5 years towards identifying vaccine candidates to prevent malaria in pregnancy, and focuses on the controversial findings implicating some members of the P. falciparum var gene family in placental infection.
Mechanisms of placental infection
Adults in malaria-endemic countries are usually immune to clinical malaria, having acquired immunity during repeated infections in childhood. During pregnancy, however, women become particularly susceptible to P. falciparum infection and this has adverse effects on both mother and unborn child, causing maternal anaemia and low birthweight babies (Brabin, 1983; McGregor et al., 1983). The increased incidence of low birthweight associated with malaria in pregnancy is a leading cause of death, because low birthweight infants are vulnerable to many life-threatening ailments (McCormick, 1985). The reasons why women are particularly susceptible to malaria during pregnancy have been debated for many years, and it was initially suggested that pregnancy-related immunosuppression could be responsible (Menendez, 1995). However, current thinking has shifted towards explanations based on the placenta providing a unique environment for a subpopulation of malaria parasites that adhere to placental receptors such as chondroitin sulphate A (CSA) (Fried and Duffy, 1996) and hyaluronic acid (Beeson et al., 2000). This adhesion causes the sequestration of mature infected erythrocytes in the placental blood spaces, which allows these parasites to grow and multiply while avoiding the host's spleen-mediated killing mechanisms. For the host, sequestration of infected erythrocytes in the placenta is damaging as it leads to inflammatory responses (Suguitan et al., 2003) and deposition of fibrinoid material (Walter et al., 1982). This may reduce placental blood flow (Dorman et al., 2002) causing impaired fetal growth with subsequent low birthweight and prematurity (Menendez et al., 2000). The binding of infected erythrocytes to CSA and other placental receptors is therefore the basic pathological mechanism primarily responsible for malaria-induced low birthweight.
The importance of placental sequestration was emphasized by the discovery that during pregnancy, malaria-infected women develop antibodies that inhibit binding of infected erythrocytes to CSA, and such antibodies are associated with protection against placental infection and low birthweight in subsequent pregnancies (Fried et al., 1998; Ricke et al., 2000; O'Neil-Dunne et al., 2001; Staalsoe et al., 2001; Duffy and Fried, 2003a). These protective antibodies are thought to recognize conserved parasite antigens because studies from malaria-endemic areas showed that sera from Asian women who had experienced multiple pregnancies prevented African placentally derived parasites from binding to CSA and vice versa (Fried et al., 1998). The implications of this finding were that identification of the parasite ligand(s) that mediates CSA-binding and is the target of adhesion-blocking antibodies could lead to the development of a globally useful vaccine to prevent malaria in pregnancy. It must be stressed that placental sequestration may be mediated via other host receptors, therefore other (non- CSA-binding) parasite ligands may be additional targets of this protective antibody response. This review will mainly discuss parasite interactions with host CSA, as this has been the focus of most of the recent research on placental sequestration.
The P. falciparum var gene family
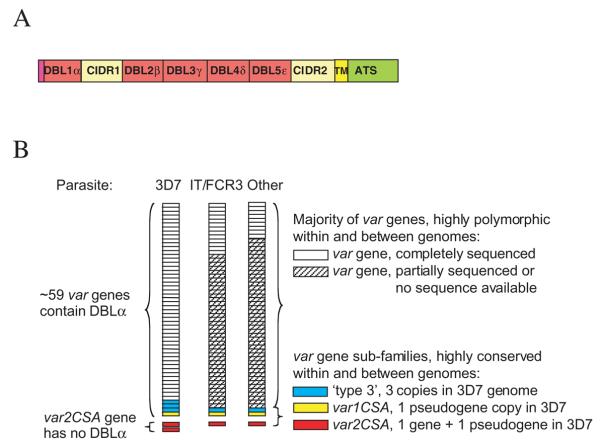
During malaria infections of non-pregnant humans, adhesion of infected erythrocytes to host cells such as endothelium and uninfected erythrocytes is mediated by members of the variant surface antigen family P. falciparum erythrocyte membrane protein one (PfEMP1), encoded by var genes (Baruch et al., 1997; Rowe et al., 1997). PfEMP1 molecules are composed of tandemly arranged cysteine-rich regions (Fig. 1A) known as DBL domains (Duffy Binding-Like; so called because the first such domain was described in the P. vivax Duffy Binding Protein). Each DBL domain is approximately 300 amino acids long, contains 12–18 cysteines and a number of other conserved hydrophobic residues (Su et al., 1995). DBL domains also contain several short conserved motifs (10–20 amino acids) interspersed by regions that are highly variable in sequence. The DBL domains are numbered consecutively from the N-terminus, and have been classified into five types (alpha to epsilon) on the basis of their amino acid sequence (Smith et al., 2000b), plus a sixth heterogeneous group, DBLX (Gardner et al., 2002). Most PfEMP1 variants have a DBL1α domain at the N-terminus, followed by the Cysteinerich interdomain region (CIDR), a degenerate form of DBL domain (Su et al., 1995; Gardner et al., 2002). The remaining DBL domains in different PfEMP1 variants vary in total number and in the order of the domain types. All variants have one trans-membrane region (TM) and a well-conserved intracellular domain (the acidic terminal segment, ATS, Fig. 1A). DBL1α is the most well-conserved extracellular domain (Smith et al., 2000b; Taylor et al., 2000a), although alignment of DBL1α from 19 different PfEMP1 variants from GenBank shows only 36–52% amino acid identity (Rowe et al., 2002).
Fig. 1.
Plasmodium falciparum var gene structure and repertoires.
A. Schematic diagram of a typical P. falciparum var gene. DBL, Duffy Binding-Like domain; CIDR, cysteine-rich interdomain region; TM, transmembrane region; ATS, acidic terminal segment.
B. Different P. falciparum lines have distinct var gene repertoires with little overlap, except for a small number of well-conserved var gene sub-families. The var gene repertoire is represented as a pile of stacked boxes. The full sequence of all var genes is only known for the 3D7 parasite clone (and its parental line NF54).
Every P. falciparum isolate/line/clone1 has a repertoire of approximately 50–60 var genes (Fig. 1B). Only one PfEMP1 variant is thought to be expressed at the surface of an infected erythrocyte (Chen et al., 1998), and switching of expression from one variant to another brings about antigenic variation in malaria (Smith et al., 1995). It was originally thought that there was little overlap between the var gene repertoires of different parasite lines (Su et al., 1995; Freitas-Junior et al., 2000; Taylor et al., 2000a), although recombinatorial shuffling may have generated short blocks of homologous sequence in var genes from different isolates (Ward et al., 1999). Recently, however, a small number of var genes that are well-conserved throughout their entire length have been identified in diverse parasite isolates (Fig. 1B, Rowe et al., 2002; Salanti et al., 2002; Winter et al., 2003). These well-conserved var gene subfamilies will be described in more detail below.
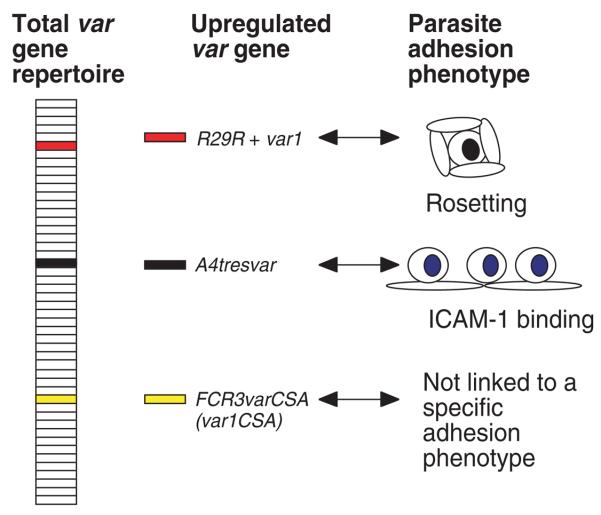
Research into the role of PfEMP1 in malaria pathogenesis in non-placental infections has identified some of the var gene family members and PfEMP1 domains responsible for adhesion to host receptors such as CD36 (Baruch et al., 1997), erythrocyte CR1 (rosetting) (Rowe et al., 1997) and ICAM-1 (Smith et al., 2000a). In these examples, the var/PfEMP1 responsible for adhesion was identified from the total var gene repertoire because transcription of the variant responsible for adhesion was upregulated in parasites selected for the ability to bind to the receptor under study (Fig. 2). In addition, heterologous expression studies were used to show specific binding of PfEMP1 domains to the host receptor (Rowe et al., 1997; Smith et al., 2000a). For a more detailed review of var gene/PfEMP1 structure and function in non-placental malaria see Smith et al., 2001.
Fig. 2.
Transcription of a specific var gene is upregulated in a P. falciparum clone selected for binding to a particular host receptor. The entire var gene repertoire of approximately 50–60 var genes from the IT/FCR3 parasite line is represented as stacked boxes. Unselected parasites tend to express a variety of different var genes. In a parasite clone derived from IT/FCR3 selected for high levels of rosetting (R29), the transcription of the R29R + var1 gene is upregulated in comparison to isogenic non-rosetting parasites (Rowe et al., 1997). Similarly, a parasite clone selected for high levels of ICAM-1 binding upregulates a different var gene, A4tresvar(Smith et al., 2000a). Heterologous expression studies showed that the PfEMP1 variant encoded by R29R + var1 binds RBC, while that encoded by A4tresvar binds ICAM-1 (Rowe et al., 1997; Smith et al., 2000a). This pattern of upregulation of a single specific var gene in a parasite clone selected for adhesion to a particular receptor has been widely demonstrated, although the transcriptional control mechanisms responsible for regulation of var gene expression are not well understood. In contrast, at least one var gene, FCR3varCSA, has many unusual features. It is well-conserved and widely transcribed among different parasite isolates/lines and its transcription is not associated with a particular adhesion phenotype (Kyes et al., 2003).
Identification of the parasite CSA-binding ligand(s)
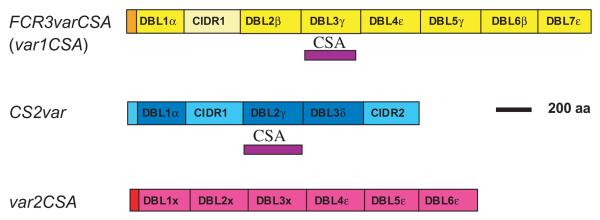
Given the known role of PfEMP1 in adhesion to host receptors, it seemed possible that adhesion of placentally derived parasites to CSA would also be attributed to PfEMP1. In 1999, two groups identified PfEMP1 variants implicated in CSA-binding, encoded by the genes FCR3varCSA (Buffet et al., 1999) and CS2var (Reeder et al., 1999) (Fig. 3). These PfEMP1 variants were identified using degenerate primers to examine the var genes transcribed in the parasite lines FCR3CSA and CS2, which were selected for high CSA-binding in vitro by panning. The FCR3varCSA and CS2var genes appeared to be the predominantly transcribed var genes in each CSA-selected parasite line, although subsequent work has cast doubt upon this (see below). In both cases the domain of PfEMP1 shown to mediate specific binding to CSA in vitro was a DBLγ type domain (Buffet et al., 1999; Reeder et al., 1999) (Fig. 3). However, it was surprising, given the serological evidence for conserved epitopes in the parasite CSA-binding ligand, that there was no obvious sequence homology between the two CSA-binding DBLγ domains. This cast doubt in the minds of some investigators, who questioned whether PfEMP1 could be the true CSA-binding ligand. In particular, given the known extreme diversity of the PfEMP1 family, how could identification of PfEMP1 as the CSA ligand explain the conserved epitopes indicated by sero-epidemiological data? Could other, more conserved parasite antigens be responsible for the CSA-binding phenotype? Research over the past few years has provided partial answers to these questions, however, definitive proof of the identity of the placental parasite CSA-binding ligand remains elusive. Table 1 indicates criteria that we would expect to be fulfilled by any putative ligand, and the relative merits of the current candidate CSA-binding ligands are shown in Table 1 and discussed below. It is of note that the ability of a heterologously expressed parasite protein to bind CSA in in vitro assays has not proved to be sufficient to unequivocally identify the CSA-binding ligand that is functional in vivo in the infected placenta.
Fig. 3.
Schematic diagram showing the extra-cellular domains of three P. falciparum var genes that have been implicated in CSA-binding. The CSA-binding regions demonstrated by heterologous expression and in vitro adhesion assays (Buffet et al., 1999; Reeder et al., 2000) are shown as purple blocks.
Table 1.
Criteria for identification of the P. falciparum CSA-binding ligand.
| Candidates |
|||
|---|---|---|---|
| Criterion |
Var1CSA (e.g. FCR3varCSA) |
CS2var | Var2CSA |
| 1. The gene should be transcribed and/or upregulated in CSA-binding parasites compared to non-CSA-binding parasites. | No | No | Yes |
| 2. The gene should be transcribed and/or upregulated in placental isolates compared to peripheral blood isolates from children. | No | No | Yes |
| 3. The protein should be present on the surface of CSA-binding infected erythrocytes and placentally derived parasites. | ND | ND | ND |
| 4. The protein should bind to CSA. | Yes | Yes | ND |
| 5. Antibodies raised to the protein should block CSA-binding. | Yes | Yes | ND |
| 6. Parasite CSA-binding should be lost when the gene is knocked out.a | No | ND | ND |
| 7. The protein should be recognized by sera from multigravid women from malarious countries with natural immunity to malaria in pregnancy and not recognized by adult males and children (i.e. gender-specific recognition). | No | ND | ND |
| 8. Sera from multigravid women from malarious countries should show enhanced recognition of the protein compared to sera from primigravid women (i.e. gravidity-dependent recognition). | No | ND | ND |
| 9. The protein, or epitopes within it, should be conserved among parasite isolates from different parts of the world. | Yes | No | Yes |
This would not apply if there is more than one CSA-binding protein per parasite (see text).
ND, not determined.
Candidate CSA-binding ligands – the var1CSA subfamily
Evidence for
The first var gene implicated in placental adhesion, FCR3varCSA (Buffet et al., 1999), has been the most intensively studied. The DBL3γ domain of FCR3varCSA, which binds to CSA (Fig. 3), is considered by many to be the front-runner as a vaccine candidate. This view was supported by the finding that almost all genetically distinct parasite isolates from around the world have a well-conserved FCR3varCSA-like gene (Rowe et al., 2002; Salanti et al., 2002; Winter et al., 2003). This subfamily of FCR3varCSA-like genes has now been called var1CSA. The occurrence of a well-conserved var gene subfamily was unexpected, as up until this time, var genes were thought to be extremely diverse both within and between different parasite isolates (Su et al., 1995; Ward et al., 1999; Taylor et al., 2000b). It therefore seemed that the conundrum of how the conserved CSA-binding ligand could be a member of the highly variable PfEMP1 family was resolved – var genes/PfEMP1 variants are not always as variable as first thought.
Further crucial support for var1CSA as a vaccine candidate came from work suggesting that antibodies raised to the DBL3γ domain of var1CSA from the IT/FCR3 parasite line are ‘pan-reactive’ and recognize the surface of infected erythrocytes of a wide range of different CSA-binding parasite lines (Lekana Douki et al., 2002; Costa et al., 2003). It therefore appeared that the PfEMP1 variant encoded by var1CSA has one of the crucial attributes of a potential vaccine, the ability to elicit antibodies that can recognize many (all?) CSA-binding P. falciparum isolates. It has not yet been shown whether the antibodies induced by var1CSA immunization actually block CSA-binding in multiple parasite isolates. It is also unclear whether adhesion-blocking activity is necessary for functional immunity, as it is possible that the binding of antibodies to the infected erythrocyte surface could lead to parasite clearance by mechanisms such as phagocytosis or complement activation.
Evidence against
Unfortunately, not all studies have reached encouraging conclusions regarding the identity of var1CSA as the parasite ligand mediating placental adhesion. It quickly became apparent that although var1CSA occurs in almost all parasite isolates, the gene is either only rarely transcribed in parasites derived from placental infections (Rowe et al., 2002), or is transcribed equally in parasites from placental infections and infections of children (Fried and Duffy, 2002; Winter et al., 2003). Closer examination of var1CSA in laboratory parasite lines showed that transcription of the gene does not correlate with CSA-binding phenotype, a finding demonstrated by three independent groups using a range of different parasite lines (Kyes et al., 2003; Salanti et al., 2003; M. Duffy, T.J. Byrne, S.J. Rogerson, J.G. Beeson, and G.V. Brown, submitted). In other words, many non-CSA binding parasites transcribe the var1CSA gene, and transcription is not increased when parasites are selected for CSA-binding (Fig. 2). In fact, CSA-selected parasites were found to transcribe a different var gene that could be responsible for their adhesive properties (see below). The var1CSA genes are transcribed at the mature trophozoite stage of parasite development (approximately 20–36 h after erythrocyte invasion) (Kyes et al., 2003), unlike other var genes in which the full-length mRNA is transcribed predominantly at ring-stage (3–18 h post invasion) (Kyes et al., 2000). The transcription of var1CSA by mature trophozoites may explain why this gene was identified by Buffet et al. (1999), who used mature trophozoites to study the var genes transcribed in a CSA-selected parasite clone. Why the var1CSA genes are widely transcribed and show different timing of transcription compared to other var genes are important questions that remain to be answered.
Doubts have also been raised about the finding that immunization with recombinant DBLγ from var1CSA induces ‘pan-reactive’ antibodies that recognize the surface of infected erythrocytes from a wide range of CSA-binding parasite isolates (Lekana Douki et al., 2002; Costa et al., 2003). Flick et al. showed that placental parasites bind non-immune IgG and IgM (natural antibodies) from normal human serum (Flick et al., 2001), and it was later shown that CSA-selected parasites bind both mouse and human IgM non-specifically (Creasey et al., 2003). Many of the pan-reactive reagents raised to var1CSA were mouse monoclonal antibodies (mAbs) of the IgM class, therefore it is possible that the pan-reactive epitopes described by Lekana Douki et al. (2002) are merely the result of mouse mAbs binding non-specifically to CSA-selected parasites. More recent work suggests that some mouse IgG mAbs and polyclonal sera raised in monkeys to the var1CSA DBLγ domain also recognize the surface of many different CSA-selected parasite lines (Costa et al., 2003). It is difficult to interpret these experiments given the suggestion that placental parasites may bind IgG non-specifically (Flick et al., 2001), although this finding was not confirmed by another group (Creasey et al., 2003). The extent to which CSA-selected parasites can bind different antibody classes from different species requires further clarification. Most importantly, any non-specific interactions must be considered when interpreting the results of experiments using specific antibodies raised to individual CSA-binding ligands. This would be made easier if studies always reported the results of appropriate negative controls, such as preimmune sera, isotype controls and secondary antibody-only controls.
A further piece of evidence that does not support a role for var1CSA in placental adhesion is that recombinant proteins encoded by var1CSA are not specifically recognized by sera from multigravid women from malarious regions (Jensen et al., 2003). Multigravid women who have been exposed to malaria during previous pregnancies are immune to the most severe effects of placental infection and their sera recognize antigens on the surface of CSA-binding parasite lines (Ricke et al., 2000; Staalsoe et al., 2001). These antigens are not recognized by sera from men or children from the same region, and it has been suggested that this ‘gender-specific recognition’ should be the gold standard in identification of the CSA-binding ligand (Staalsoe et al., 2002). The lack of gender-specific recognition of var1CSA argues against its involvement in placental malaria, although only the N-terminal domains and not the CSA-binding DBLγ domain of var1CSA was tested (Jensen et al., 2003). An alternative explanation for the lack of gender-specific recognition of the var1CSA recombinant proteins could be that the bacterially expressed proteins did not have the correct conformation. Given the large number of cysteine residues in DBL domains this explanation is certainly plausible.
Knock-out studies have the potential to provide important information on gene function, and a knock-out of the var1CSA gene from the IT/FCR3 parasite has been performed (Andrews et al., 2003). It was found that var1CSA knock-out parasites could still be selected for CSA-binding, implying either that var1CSA is not the CSA-binding ligand, or that IT/FCR3 parasites have more than one CSA-binding ligand. The latter explanation is certainly possible, as in other parasite adhesion phenotypes, such as rosetting, there can be at least four distinct rosette-mediating variants within the var gene repertoire of a single parasite genotype (J.A. Rowe, unpubl. data). The knock-out experiment therefore neither supports nor refutes a role for var1CSA in parasite CSA-binding. Obviously it would be of interest to know which var genes are transcribed by the CSA-binding var1CSA knock-out parasites, but this has not yet been reported.
One important piece of data that is missing from all studies to date on var1CSA is confirmation that the PfEMP1 variant encoded by var1CSA is on the surface of infected erythrocytes. It remains possible that var1CSA is the parasite CSA-binding ligand, and that although both CSA-binding and non-CSA-binding parasites transcribe the gene, they differ in surface expression of the mature protein. Resolving the question of where the PfEMP1 variant encoded by var1CSA is localized is important in evaluating the role of this molecule as a vaccine candidate.
In summary, although the DBLγ domain of var1CSA binds to CSA in in vitro assays, and antibodies raised to it inhibit CSA-binding, the transcription patterns of the gene do not fit with what would be expected of a placental parasite adhesion ligand. In addition, there is considerable uncertainty in interpretation of existing data obtained using antibodies raised to var1CSA because of possible non-specific effects. The role of the var1CSA subfamily in CSA-binding and placental malaria therefore remains open to question, and it would seem premature to conclude that var1CSA should be developed as a vaccine to prevent malaria in pregnancy. Future studies to clarify the specificity of antibodies raised to var1CSA and to determine the localization of the var1CSA PfEMP1 protein would help to resolve some of the current uncertainty.
Candidate CSA-binding ligands – CS2var
Evidence for
The second putative CSA-binding ligand is the PfEMP1 variant encoded by the CS2var gene (Reeder et al., 1999) (Fig. 3). This gene was first identified as being transcribed in a parasite clone selected for CSA-binding (CS2) (Reeder et al., 1999), and has received relatively little attention compared to var1CSA. Both the CIDR1 and DBL2γ domains of the PfEMP1 variant encoded by CS2var bind to CSA (and other glycosaminoglycans)(Reeder et al., 2000), while antibodies raised to the DBL2γ domain showed specific inhibition of CS2 parasite binding to CSA (Reeder et al., 1999).
Evidence against
The CS2var gene does not appear to be well-conserved among different isolates (Rowe et al., 2002), and its transcription is not upregulated in CSA-selected parasite lines (M. Duffy, T.J. Byrne, S.J. Rogerson, J.G. Beeson, and G.V. Brown, submitted). Indeed, the parasite clone in which CS2var was identified, has now been shown to upregulate a different var gene following CSA-selection (see below, M. Duffy, T.J. Byrne, S.J. Rogerson, J.G. Beeson, and G.V. Brown, submitted), and it seems possible that the adhesion blocking activity of antibodies raised against the CS2var was caused by fortuitous cross-reactivity. It therefore seems unlikely that the CS2var is directly involved in placental adhesion and unlikely that this variant will be a component of a vaccine to prevent malaria in pregnancy.
Candidate CSA-binding ligands – the var2CSA subfamily
Evidence for
A second well-conserved var gene subfamily that fulfils some of the predicted criteria for the CSA-binding ligand has been identified, known as var2CSA (Salanti et al., 2003) (Fig. 3; Table 1). This gene was missed in initial screens for the var genes transcribed by CSA-binding parasites because it does not contain a DBLα domain used to design ‘universal’ primers to detect var gene expression (Taylor et al., 2000b). Instead, members of the var2CSA family consist of three DBL domains that do not fit into any of the currently recognized domain types (DBLX) and three DBLε domains (Fig. 3). The var2CSA gene is well-conserved among different parasite isolates and its transcription is upregulated following CSA-selection (Salanti et al., 2003). This was shown using quantitative real-time polymerase chain reaction (PCR) with primer pairs to all of the var genes in the genome of the parasite clone NF54 and the CSA-selected line NF54CSA (Salanti et al., 2003). A second group has independently confirmed this finding, using other non-selected and CSA-selected parasite lines (M. Duffy, T.J. Byrne, S.J. Rogerson, J.G. Beeson, and G.V. Brown, submitted). It has also been shown that transcription of var2CSA is upregulated in placental isolates compared to peripheral blood isolates from children, although only five isolates were studied (Salanti et al., 2003). Therefore unlike var1CSA and CS2var, the transcription of var2CSA is consistent with the pattern that would be expected for a placental parasite CSA-binding ligand.
Evidence against
To date there is no convincing experimental evidence against the role of var2CSA in placental adhesion, however, many crucial experiments remain to be performed (Table 1). It has not yet been demonstrated that the var2CSA encodes a protein that is present on the surface of infected cells, nor that the var2CSA PfEMP1 mediates CSA-binding. It has not yet been shown that antibodies to var2CSA inhibit CSA-binding nor that the var2CSA protein is recognized in a gender-specific, parity-dependent manner. Clearly more work needs to be done before it is known whether the PfEMP1 variant encoded by var2CSA is the parasite CSA-binding ligand responsible for placental adhesion.
Identifying proteins on the surface of infected erythrocytes
For both var1CSA and var2CSA, demonstrating the presence of the PfEMP1 variants that they encode on the surface of infected erythrocytes derived from infected placentas and CSA-binding laboratory parasites would be a major step in promoting further development towards a vaccine. How can such experimental evidence be obtained? Antibodies are not useful because of the potential for confusing cross-reactions described above. A recent study has attempted to carry out mass spectrometric analysis of the PfEMP1 proteins on the surface of placental infected erythrocytes (Fried et al., 2004). It was found that the PfEMP1 variants encoded by var1CSA and var2CSA were not detected on the surface of placental infected erythrocytes, but four other PfEMP1 variants were found. Further work is needed to characterize the four novel PfEMP1 variants and determine whether they play a role in placental sequestration. Proteomics techniques clearly show great promise, however, the detection of low abundance surface proteins remains a challenge, and the possibility that some PfEMP1 variants were missed because of sensitivity limitations cannot be excluded.
How many different CSA-binding ligands are there?
It is possible that other PfEMP1 variants will be implicated in CSA-binding or that other parasite molecules that bind to CSA will be identified. Even if CSA-binding is entirely attributed to PfEMP1, on the basis of current data it is uncertain how many different PfEMP1 variants need to be recognized by the host's immune system before an immune response that prevents placental infection develops. Epidemiological evidence suggests that women in low to moderate malaria transmission areas may need to experience at least three or four pregnancies before developing immunity that prevents malaria-associated low birthweight (Nosten et al., 1991; Shulman et al., 2001). This may suggest that several infections and exposure to several different PfEMP1 variants and/or other parasite molecules are required before functional immunity develops.
The role of other receptor–ligand interactions in placental sequestration?
Although CSA and the parasite CSA-binding ligand(s) are of major importance in placental sequestration, a role for other placental receptors and parasite ligands cannot be excluded. Indeed, it has been shown that although many parasite isolates derived from placental infections bind to CSA (Fried and Duffy, 1996), some do not (Beeson et al., 1999). Another glycosaminoglycan molecule, hyaluronic acid (HA) has been implicated in placental binding (Beeson et al., 2000; Beeson and Brown, 2004). The parasite ligand mediating binding to HA has not yet been identified. Placental sequestration via Fcγ receptors has also been suggested (Flick et al., 2001), although Fcγ receptors are not thought to be accessible to infected erythrocytes in the placental blood spaces (Duffy and Fried, 2003b). IgM natural antibodies bind to CSA-selected parasite clones, therefore it is possible that interaction between infected erythrocytes and Fcμ receptors, if expressed in the placenta, could enhance placental sequestration (Creasey et al., 2003). Further receptors may remain to be identified. This is an important area of research that should not be neglected, because if other receptors are important in bringing about placental sequestration, using a vaccine to raise antibodies to block CSA-binding may be insufficient to prevent placental malaria infection.
Conclusions
Substantial progress has been made in understanding how infected erythrocytes bind to CSA in the placenta, however, the parasite CSA-binding ligand has not yet been identified unequivocally. The identification and pursuit of var1CSA as a vaccine candidate may yet prove to be a false start in the race for a vaccine against malaria in pregnancy. The var2CSA gene is a promising candidate, but much work still needs to be done to clarify its role. Above all, the temptation to base a vaccine on results from a small number of laboratory parasite strains should be avoided, and greater emphasis should be put on examining the expression of adhesion ligands in Plasmodium falciparum isolates derived directly from placental infections.
Acknowledgements
We are grateful to Dave Arnot, Ian Cockburn, Jean-Philippe Semblat and Anne-Marie Deans for comments on the manuscript.
Footnotes
The nomenclature used in this review for P. falciparum parasites is as follows: an isolate is a population of parasites derived from a patient; a line is an isolate that has been adapted to grow in vitro; a clone is derived from a single cell.
References
- Andrews KT, Pirrit LA, Przyborski JM, Sanchez CP, Sterkers Y, Ricken S, et al. Recovery of adhesion to chondroitin-4-sulphate in Plasmodium falciparum varCSA disruption mutants by antigenically similar PfEMP1 variants. Mol Microbiol. 2003;49:655–669. doi: 10.1046/j.1365-2958.2003.03595.x. [DOI] [PubMed] [Google Scholar]
- Baruch DI, Ma XC, Singh HB, Bi X, Pasloske BL, Howard RJ. Identification of a region of PfEMP1 that mediates adherence of Plasmodium falci parum infected erythrocytes to CD36: conserved function with variant sequence. Blood. 1997;90:3766–3775. [PubMed] [Google Scholar]
- Beeson JG, Brown GV. Plasmodium falciparum-infected erythrocytes demonstrate dual specificity for adhesion to hyaluronic acid and chondroitin sulfate A and have distinct adhesive properties. J Infect Dis. 2004;189:169–179. doi: 10.1086/380975. [DOI] [PubMed] [Google Scholar]
- Beeson JG, Brown GV, Molyneux ME, Mhango C, Dzinjalamala F, Rogerson SJ. Plasmodium falciparum isolates from infected pregnant women and children are associated with distinct adhesive and antigenic properties. J Infect Dis. 1999;180:464–472. doi: 10.1086/314899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beeson JG, Rogerson SJ, Cooke BM, Reeder JC, Chai W, Lawson AM, et al. Adhesion of Plasmodium falciparum-infected erythrocytes to hyaluronic acid in placental malaria. Nat Med. 2000;6:86–90. doi: 10.1038/71582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brabin BJ. An analysis of malaria in pregnancy in Africa. Bull WHO. 1983;61:1005–1016. [PMC free article] [PubMed] [Google Scholar]
- Buffet PA, Gamain B, Scheidig C, Baruch D, Smith JD, Hernandez-Rivas R, et al. Plasmodium falciparum domain mediating adhesion to chondroitin sulfate A: a receptor for human placental infection. Proc Natl Acad Sci USA. 1999;96:12743–12748. doi: 10.1073/pnas.96.22.12743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Fernandez V, Sundstrom A, Schlichtherle M, Datta S, Hagblom P, Wahlgren M. Developmental selection of var gene expression in Plasmodium falciparum. Nature. 1998;394:392–395. doi: 10.1038/28660. [DOI] [PubMed] [Google Scholar]
- Costa FT, Fusai T, Parzy D, Sterkers Y, Torrentino M, J.B. D, et al. Immunization with recombinant duffy binding-like-gamma3 induces pan-reactive and adhesion-blocking antibodies against placental chondroitin sulfate A-binding Plasmodium falciparum parasites. J Infect Dis. 2003;188:153–164. doi: 10.1086/375800. [DOI] [PubMed] [Google Scholar]
- Creasey AM, Staalsoe T, Raza A, Arnot DE, Rowe JA. Nonspecific immunoglobulin M binding and chondroitin sulfate A binding are linked phenotypes of Plasmodium falciparum isolates implicated in malaria during pregnancy. Infect Immun. 2003;71:4767–4771. doi: 10.1128/IAI.71.8.4767-4771.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorman EK, Shulman CE, Kingdom J, Bulmer JN, Mwendwa J, Peshu N, Marsh K. Impaired uteroplacental blood flow in pregnancies complicated by falciparum malaria. Ultrasound Obstet Gynecol. 2002;19:165–170. doi: 10.1046/j.0960-7692.2001.00545.x. [DOI] [PubMed] [Google Scholar]
- Duffy PE, Fried M. Antibodies that inhibit Plasmodium falciparum adhesion to chondroitin sulfate A are associated with increased birthweight and the gestational age of newborns. Infect Immun. 2003a;71:6620–6623. doi: 10.1128/IAI.71.11.6620-6623.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffy PE, Fried M. Plasmodium falciparum adhesion in the placenta. Curr Opin Microbiol. 2003b;6:371–376. doi: 10.1016/s1369-5274(03)00090-0. [DOI] [PubMed] [Google Scholar]
- Flick K, Scholander C, Chen Q, Fernandez V, Pouvelle B, Gysin J, Wahlgren M. Role of nonimmune IgG bound to PfEMP1 in placental malaria. Science. 2001;293:2098–2100. doi: 10.1126/science.1062891. [DOI] [PubMed] [Google Scholar]
- Freitas-Junior LH, Bottius E, Pirrit LA, Deitsch KW, Scheidig C, Guinet F, et al. Frequent ectopic recombination of virulence factor genes in telomeric chromosome clusters of P. falciparum. Nature. 2000;407:1018–1022. doi: 10.1038/35039531. [DOI] [PubMed] [Google Scholar]
- Fried M, Duffy PE. Adherence of Plasmodium falciparum to chondroitin sulphate A in the human placenta. Science. 1996;272:1502–1504. doi: 10.1126/science.272.5267.1502. [DOI] [PubMed] [Google Scholar]
- Fried M, Duffy PE. Two DBLgamma subtypes are commonly expressed by placental isolates of Plasmodium falciparum. Mol Biochem Parasitol. 2002;122:201–210. doi: 10.1016/s0166-6851(02)00103-2. [DOI] [PubMed] [Google Scholar]
- Fried M, Nosten F, Brockman A, Brabin BJ, Duffy PE. Maternal antibodies block malaria. Nature. 1998;395:851–852. doi: 10.1038/27570. [DOI] [PubMed] [Google Scholar]
- Fried M, Wendler JP, Mutabingwa TK, Duffy PE. Mass spectrometric analysis of Plasmodium falciparum erythrocyte membrane protein-1 variants expressed by placental malaria parasites. Proteomics. 2004;4:1086–1093. doi: 10.1002/pmic.200300666. [DOI] [PubMed] [Google Scholar]
- Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen AT, Zornig HD, Buhmann C, Salanti A, Koram KA, Riley EM, et al. Lack of gender-specific antibody recognition of products from domains of a var gene implicated in pregnancy-associated Plasmodium falciparum malaria. Infect Immun. 2003;71:4193–4196. doi: 10.1128/IAI.71.7.4193-4196.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyes S, Pinches R, Newbold C. A simple RNA analysis method shows var and rif multigene family expression patterns in Plasmodium falciparum. Mol Biochem Parasitol. 2000;105:311–315. doi: 10.1016/s0166-6851(99)00193-0. [DOI] [PubMed] [Google Scholar]
- Kyes SA, Christodoulou Z, Raza A, Horrocks P, Pinches R, Rowe JA, Newbold CI. A well-conserved Plasmodium falciparum var gene shows an unusual stage-specific transcript pattern. Mol Microbiol. 2003;48:1339–1348. doi: 10.1046/j.1365-2958.2003.03505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lekana Douki JB, Traore B, Costa FT, Fusai T, Pouvelle B, Sterkers Y, et al. Sequestration of Plasmodium falciparum-infected erythrocytes to chondroitin sulfate A, a receptor for maternal malaria: monoclonal antibodies against the native parasite ligand reveal pan-reactive epitopes in placental isolates. Blood. 2002;100:1478–1483. doi: 10.1182/blood-2002-01-0315. [DOI] [PubMed] [Google Scholar]
- McCormick MC. The contribution of low birth weight to infant mortality and childhood morbidity. N Eng J Med. 1985;312:82–90. doi: 10.1056/NEJM198501103120204. [DOI] [PubMed] [Google Scholar]
- McGregor IA, Wilson ME, Billewicz WZ. Malaria infection of the placenta in The Gambia, West Africa; its incidence and relationship to stillbirth, birth-weight and placental weight. Trans R Soc Trop Med Hyg. 1983;77:232–244. doi: 10.1016/0035-9203(83)90081-0. [DOI] [PubMed] [Google Scholar]
- Menendez C. Malaria during pregnancy: a priority area of malaria research and control. Parasitol Today. 1995;11:178–183. doi: 10.1016/0169-4758(95)80151-0. [DOI] [PubMed] [Google Scholar]
- Menendez C, Ordi J, Ismail MR, Ventura PJ, Aponte JJ, Kahigwa E, et al. The impact of placental malaria on gestational age and birth weight. J Infect Dis. 2000;181:1740–1745. doi: 10.1086/315449. [DOI] [PubMed] [Google Scholar]
- Nosten F, ter Kuile F, Maelankirri L, Decludt B, White NJ. Malaria during pregnancy in an area of unstable endemicity. Trans R Soc Trop Med Hyg. 1991;85:424–429. doi: 10.1016/0035-9203(91)90205-d. [DOI] [PubMed] [Google Scholar]
- O'Neil-Dunne I, Achur RN, Agbor-Enoh ST, Valiyaveettil M, Naik RS, Ockenhouse CF, et al. Gravidity-dependent production of antibodies that inhibit binding of Plasmodium falciparum-infected erythrocytes to placental chondroitin sulfate proteoglycan during pregnancy. Infect Immun. 2001;69:7487–7492. doi: 10.1128/IAI.69.12.7487-7492.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder JC, Cowman AF, Davern KM, Beeson JG, Thompson JK, Rogerson SJ, Brown GV. The adhesion of Plasmodium falciparum-infected erythrocytes to chondroitin sulfate A is mediated by P. falciparum erythrocyte membrane protein 1. Proc Natl Acad Sci USA. 1999;96:5198–5202. doi: 10.1073/pnas.96.9.5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeder JC, Hodder AN, Beeson JG, Brown GV. Identification of glycosaminoglycan binding domains in Plasmodium falciparum erythrocyte membrane protein 1 of a chondroitin sulfate A-adherent parasite. Infect Immun. 2000;68:3923–3926. doi: 10.1128/iai.68.7.3923-3926.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricke CH, Staalsoe T, Koram K, Akanmori BD, Riley EM, Theander TG, Hviid L. Plasma antibodies from malaria-exposed pregnant women recognize variant surface antigens on Plasmodium falciparum-infected erythrocytes in a parity-dependent manner and block parasite adhesion to chondroitin sulfate A. J Immunol. 2000;165:3309–3316. doi: 10.4049/jimmunol.165.6.3309. [DOI] [PubMed] [Google Scholar]
- Rowe JA, Kyes SA, Rogerson SJ, Babiker HA, Raza A. Identification of a conserved Plasmodium falciparum var gene implicated in malaria in pregnancy. J Infect Dis. 2002;185:1207–1211. doi: 10.1086/339684. [DOI] [PubMed] [Google Scholar]
- Rowe JA, Moulds JM, Newbold CI, Miller LH. P. falciparum rosetting mediated by a parasite-variant erythrocyte membrane protein and complement-receptor 1. Nature. 1997;388:292–295. doi: 10.1038/40888. [DOI] [PubMed] [Google Scholar]
- Salanti A, Jensen AT, Zornig HD, Staalsoe T, Joergensen L, Nielsen MA, et al. A sub-family of common and highly conserved Plasmodium falciparum var genes. Mol Biochem Parasitol. 2002;122:111–115. doi: 10.1016/s0166-6851(02)00080-4. [DOI] [PubMed] [Google Scholar]
- Salanti A, Staalsoe T, Lavstsen T, Jensen AT, Sowa MP, Arnot DE, et al. Selective upregulation of a single distinctly structured var gene in chondroitin sulphate A-adhering Plasmodium falciparum involved in pregnancy-associated malaria. Mol Microbiol. 2003;49:179–191. doi: 10.1046/j.1365-2958.2003.03570.x. [DOI] [PubMed] [Google Scholar]
- Shulman CE, Marshall T, Dorman EK, Bulmer JN, Cutts F, Peshu N, Marsh K. Malaria in pregnancy: adverse effects on haemoglobin levels and birth-weight in primigravidae and multigravidae. Trop Med Int Health. 2001;6:770–778. doi: 10.1046/j.1365-3156.2001.00786.x. [DOI] [PubMed] [Google Scholar]
- Smith JD, Chitnis CE, Craig AG, Roberts DJ, Hudson-Taylor DE, Peterson DS, et al. Switches in expression of Plasmodium falciparum var genes correlate with changes in antigenic and cytoadherent phenotypes of infected erythrocytes. Cell. 1995;82:101–110. doi: 10.1016/0092-8674(95)90056-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JD, Craig AG, Kriek N, Hudson-Taylor D, Kyes S, Fagen T, et al. Identification of a Plasmodium falciparum intercellular adhesion molecule-1 binding domain: a parasite adhesion trait implicated in cerebral malaria. Proc Natl Acad Sci USA. 2000a;97:1766–1771. doi: 10.1073/pnas.040545897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JD, Gamain B, Baruch DI, Kyes S. Decoding the language of var genes and Plasmodium falciparum sequestration. Trends Parasitol. 2001;17:538–545. doi: 10.1016/s1471-4922(01)02079-7. [DOI] [PubMed] [Google Scholar]
- Smith JD, Subramanian G, Gamain B, Baruch DI, Miller LH. Classification of adhesive domains in the Plasmodium falciparum erythrocyte membrane protein 1 family. Mol Biochem Parasitol. 2000b;110:293–310. doi: 10.1016/s0166-6851(00)00279-6. [DOI] [PubMed] [Google Scholar]
- Staalsoe T, Jensen AT, Theander TG, Hviid L. Novel Plasmodium falciparum malaria vaccines: evidence-based searching for variant surface antigens as candidates for vaccination against pregnancy-associated malaria. Immunol Lett. 2002;84:133–136. doi: 10.1016/s0165-2478(02)00159-1. [DOI] [PubMed] [Google Scholar]
- Staalsoe T, Megnekou R, Fievet N, Ricke CH, Zornig HD, Leke R, et al. Acquisition and decay of antibodies to pregnancy-associated variant antigens on the surface of Plasmodium falciparum-infected erythrocytes that protect against placental parasitemia. J Infect Dis. 2001;184:618–626. doi: 10.1086/322809. [DOI] [PubMed] [Google Scholar]
- Steketee RW, Nahlen BL, Parise ME, Menendez C. The burden of malaria in pregnancy in malaria-endemic areas. Am J Trop Med Hyg. 2001;64(1–2 Suppl.):28–35. doi: 10.4269/ajtmh.2001.64.28. [DOI] [PubMed] [Google Scholar]
- Su X-Z, Heatwole VM, Wertheimer SP, Guinet F, Herrfeldt JA, Peterson DS, et al. A large and diverse family gene family (var) encodes 200–350 kD proteins implicated in the antigenic variation and cytoadherence of Plasmodium falciparum-infected erythocytes. Cell. 1995;82:89–99. doi: 10.1016/0092-8674(95)90055-1. [DOI] [PubMed] [Google Scholar]
- Suguitan ALJ, Leke RG, Fouda G, Zhou A, Thuita L, Metenou S, et al. Changes in the levels of chemokines and cytokines in the placentas of women with Plasmodium falciparum malaria. J Infect Dis. 2003;188:1074–1082. doi: 10.1086/378500. [DOI] [PubMed] [Google Scholar]
- Taylor HM, Kyes SA, Harris D, Kriek N, Newbold CI. A study of var gene transcription in vitro using universal var gene primers. Mol Biochem Parasitol. 2000a;105:13–23. doi: 10.1016/s0166-6851(99)00159-0. [DOI] [PubMed] [Google Scholar]
- Taylor HM, Kyes SA, Newbold CI. Var gene diversity in Plasmodium falciparum is generated by frequent recombination events. Mol Biochem Parasitol. 2000b;110:391–397. doi: 10.1016/s0166-6851(00)00286-3. [DOI] [PubMed] [Google Scholar]
- Walter PR, Garin Y, Blot P. Placental pathologic changes in malaria. A histologic and ultrastructural study. Am J Pathol. 1982;109:330–342. [PMC free article] [PubMed] [Google Scholar]
- Ward CP, Clottey GT, Dorris M, Ji DD, Arnot DE. Analysis of Plasmodium falciparum PfEMP-1/var genes suggests that recombination rearranges constrained sequences. Mol Biochem Parasitol. 1999;102:167–177. doi: 10.1016/s0166-6851(99)00106-1. [DOI] [PubMed] [Google Scholar]
- Winter G, Chen Q, Flick K, Kremsner P, Fernandez V, Wahlgren M. The 3D7var5.2 (var COMMON) type var gene family is commonly expressed in non-placental Plasmodium falciparum malaria. Mol Biochem Parasitol. 2003;127:179–191. doi: 10.1016/s0166-6851(03)00004-5. [DOI] [PubMed] [Google Scholar]