Abstract
Rationale
Voltage-gated sodium channels initiate action potentials in excitable tissues. Mice in which Scn5A (the predominant sodium channel gene in heart) has been knocked out die early in development with cardiac malformations by mechanisms which have yet to be determined.
Objective
Here we addressed this question by investigating the role of cardiac sodium channels in zebrafish heart development.
Methods and Results
Transcripts of the functionally-conserved Scn5a homologs scn5Laa and scn5Lab were detected in the gastrulating zebrafish embryo and subsequently in the embryonic myocardium. Antisense knockdown of either channel resulted in marked cardiac chamber dysmorphogenesis and perturbed looping. These abnormalities were associated with decreased expression of the myocardial precursor genes nkx2.5, gata4, and hand2 in anterior lateral mesoderm and significant deficits in the production of cardiomyocyte progenitors. These early defects did not appear to result from altered membrane electrophysiology, as prolonged pharmacological blockade of sodium current failed to phenocopy channel knockdown. Moreover, embryos grown in calcium channel blocker-containing medium had hearts that did not beat but developed normally.
Conclusions
These findings identify a novel, and possibly non-electrogenic, role for cardiac sodium channels in heart development.
Keywords: Ion Channels, Heart Development, Zebrafish, scn5La
Introduction
The Nav1 family of voltage-gated sodium channels are multi-protein complexes that account for the initial upstroke (phase 0) of the action potential in neurons, myocytes, and other excitable cells by permitting a rapid influx of Na+ ions.1 Ten distinct pore-forming (α) subunit genes (SCNxA) have been cloned,1–3 and SCN5A encodes Nav1.5, the predominant sodium channel isoform in myocardium.4
Perturbed expression or function of Nav1.5 in patients can cause a range of phenotypes including the long QT syndrome, Brugada syndrome, progressive cardiac conduction system disease, and atrial arrhythmias.5, 6 Mice heterozygous for Scn5a deletion display slow conduction and susceptibility to ventricular tachycardia.7 By contrast, Scn5a−/− homozygotes die between embryonic day 10 (E10) and E11 with abnormalities of ventricular morphogenesis, indicating that Scn5a is also required for normal development.7 The mechanisms underlying these defects have not been determined.
Here we used zebrafish to examine the role of sodium channels in the developing heart. Zebrafish embryos are optically transparent and externally fertilized, facilitating the study of early organ formation. Genetic manipulation is readily achieved using antisense morpholinos, and embryos are also permeable to small molecule drugs placed in their medium. The stages of zebrafish cardiac development have been well-delineated: cardiac precursors are located at the blastula margin at 5 hours post fertilization (hpf).8 These bilateral precursors undergo a complex series of movements that result in the formation of a cardiac cone and subsequently a beating heart tube by 22–24hpf.9, 10 The tube then loops to form a two-chambered heart and the ventricular wall begins to thicken concentrically between 48–72hpf.9, 10 Our findings identify a previously-unappreciated role for voltage-gated sodium channels in early cardiac development.
Materials and methods
The following zebrafish strains were used in this study: AB, TuAB, Tg(cmlc2:GFP), Tg(cmlc2:DsRed2-nuc). Whole-mount in situ hybridization was performed using digoxigenin-labeled probes. Gene knockdown was achieved by morpholino antisense oligonucleotides (Gene Tools) designed to inhibit translation of mRNAs or disrupt splicing of pre-mRNAs. Drugs and toxins were delivered to embryos by bath exposure or by pressure injection directly into the yoke sac, trunk circulation, sinus venosus, or pericardial sac, depending on the experiment and developmental stage. Transcript levels were quantified using Power SYBR Green (Applied Biosystems). Confocal images of live embryos were captured using a Zeiss LSM 510 Confocal Microscope System equipped with either 20x or 40x objective lenses. Image analysis and cell counts were performed using Image J software. Photographs of embryos were adjusted for brightness and contrast in Adobe Photoshop and/or Microsoft Powerpoint and identical adjustments were made for active treatment groups and controls. Statistical analyses were performed as indicated in the text and figure legends.
An expanded Materials and methods section is available in the Online Data Supplement at http://circres.ahajournals.org.
Results
Cardiac sodium channels in zebrafish
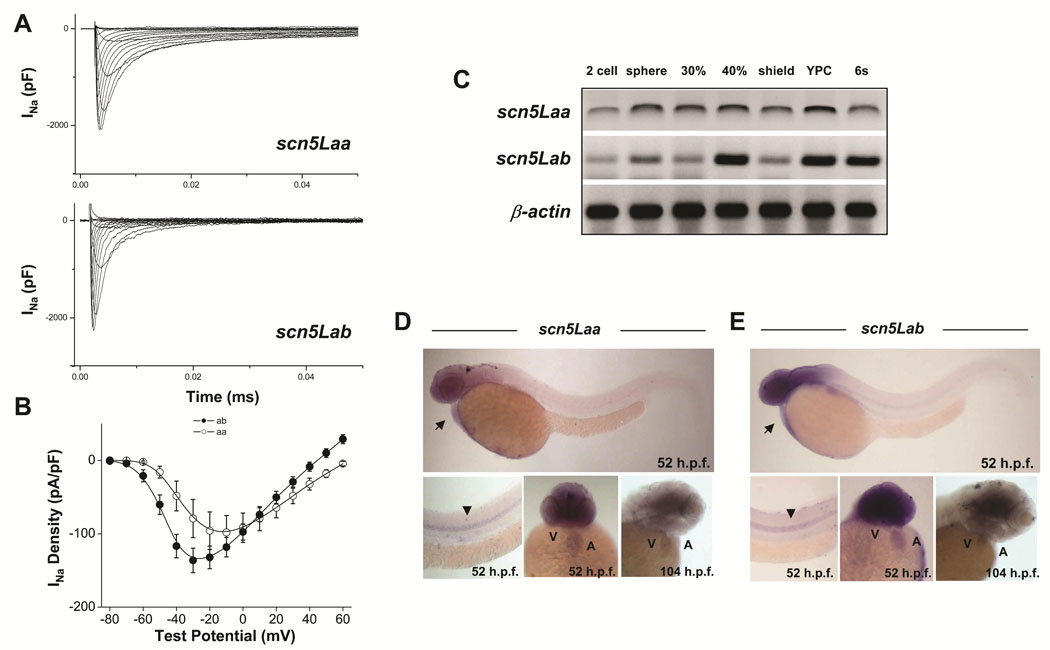
Two SCN5A orthologs have been cloned in zebrafish11 (see also Online Figure I) and are termed scn5Laa and scn5Lab. These encode typical voltage-gated sodium channels sharing 60–65% amino acid identity with SCN5A (Online Figure II, Online Figure III), with even greater conservation in important functional domains including the transmembrane segments, voltage-sensors, pore loops, inactivation gate, and C-terminus (Online Figure III). Expression of full-length scn5Laa and scn5Lab in CHO cells produced typical voltage-gated sodium currents (Figure 1A, B). Transcripts of both genes were detected in gastrulating embryos prior to the expression of early cardiogenic transcription factors and well in advance of myocardial differentiation (Figure 1C). Both sodium channel genes were expressed in the heart, brain, and spinal cord at 52 −104hpf by in situ hybridization (Figure 1D, E).
FIGURE 1. Function and developmental expression of zebrafish cardiac-type sodium channels.
A, B. Whole cell sodium currents (A), and current-voltage relationships in CHO cells transfected with full-length scn5Laa and scn5Lab (B). C. Temporal expression of scn5Laa and scn5Lab in early embryos (0–12hpf) by RT-PCR. YPC = yoke plug closure stage, s = somites. D, E. By whole-mount in situ hybridization, sodium channel gene expression was detected diffusely in both heart chambers before displaying predominately ventricular expression by 104hpf. Transcripts were also detected in the brain and spinal cord. Developmental stages are as labeled. Arrow, heart. Arrowhead, spinal cord. A, atrium. V, ventricle.
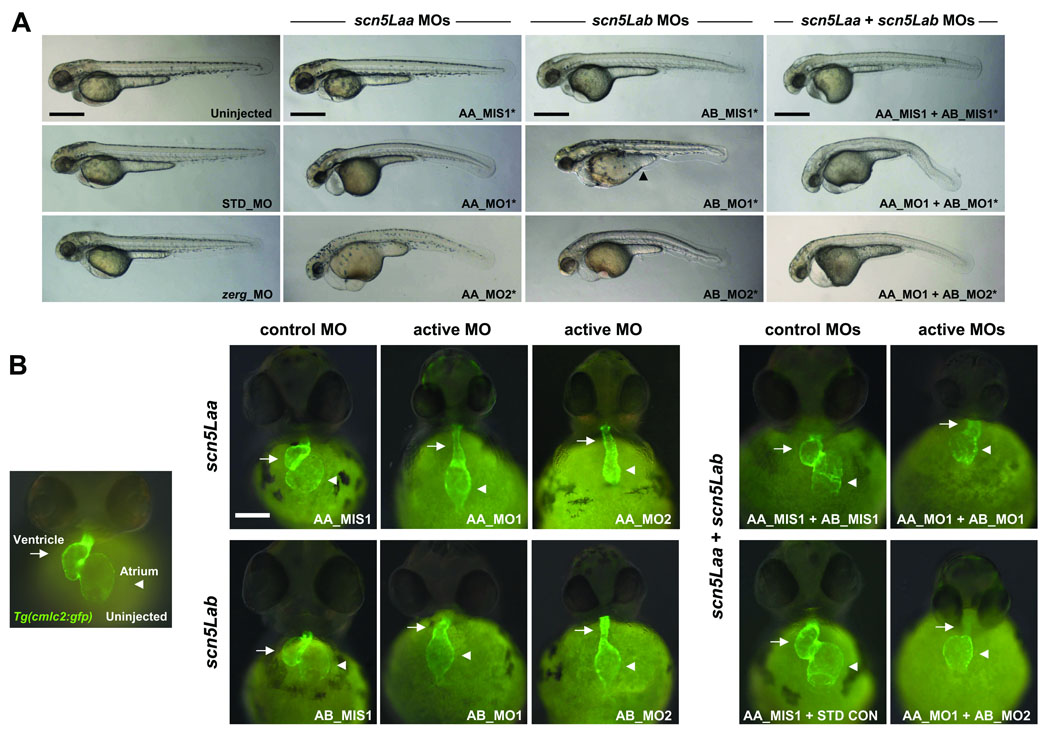
Knockdown of zebrafish cardiac sodium channels results in defects of cardiac morphogenesis
To investigate the role of scn5Laa and scn5Lab in development, we used multiple translation- and splice-blocking morpholino-antisense oligonucleotides (morpholinos) that specifically target each gene as well as 5-nucleotide mismatch control morpholinos (Online Figure IV–Online Figure V, Online Table I). Knockdown of either scn5Laa or scn5Lab in Tg(cmlc2:GFP) zebrafish, a transgenic line expressing green fluorescent protein in the myocardium,12 resulted in embryos with marked defects in cardiac development and function compared to embryos injected with equivalent doses of control morpholinos (Figure 2, Online Table II). Dual knockdown of scn5Laa and scn5Lab produced embryos with cardiac defects that appeared more severe than those resulting from knockdown of either gene alone, primarily with respect to ventricular development (Figure 2B). As an additional control, we knocked-down the cardiac potassium channel zerg which caused arrhythmias by 48hpf but did not perturb development (Figure 2, Online Table II).
Table 1.
Developmental effects of pharmacological modulators of ion channel function.
| target | agent | type | delivery | [drug] | n | phenotypes |
|---|---|---|---|---|---|---|
| VGSC | tetrodotoxin | inhibitor | bath solution | 500µm | 74 |
|
| bath solution | 500µm | 26 |
|
|||
| microinjection | 1mM | 99 |
|
|||
| VGSC | lidocaine | inhibitor | bath solution | 500µm | 200 |
|
| VGSC | anemone toxin II |
activator | bath solution | 10µM | 25 |
|
| microinjection | 100µM | 50 |
|
|||
| VGSC | veratridine | activator | bath solution | 100µM | 34 |
|
| microinjection | 1mM | 50 |
|
|||
| VGSC; ERG | flecainide | inhibitor | bath solution | 400µm | 46 |
|
| bath solution | 400µm | 29 |
|
|||
| LTCC | nisoldipine | inhibitor | bath solution | 10µm | 100 |
|
| LTCC/ TTCC |
mibefradil | inhibitor | bath solution | 250µm | 77 |
|
| ERG | E-4031 | inhibitor | bath solution | 250µm | 93 |
|
VGSC = voltage-gated sodium channel, LTCC = L-type calcium channel, TTCC = T-type calcium channel, ERG = ether-a-go-go related gene potassium channel. For bath immersion, embryos were dechorionated and placed in drug solutions by the 64 cell stage. For microinjection, 3 nanoliters of drug solution were injected in 2 different locations of the yoke sac of 1–4 cell stage embryos. Cardiac lineage specification assessed by in situ hybridization for nkx2.5 at 8 somites. Differentiation assessed by heart tube formation and heart beating in Tg(cmlc2:GFP) embryos at 28–30 h.p.f. See methods for additional details.
FIGURE 2. Zebrafish scn5Laa and scn5Lab are each required for normal cardiac development.
A. Embryos at 58–62hpf following treatment with active or control antisense morpholinos. All embryos were also treated with the p53 morpholino. Injection of p53 morpholino alone did not cause any identifiable phenotype (Online Figure VII). Note that head size is reduced in all sodium channel morphants and that many embryos injected with AB_MO1 display a yoke sac extension abnormality (arrowhead). All scale bars = 500µM. B. Cardiac developmental defects in morphant Tg(cmlc2:GFP) embryos. AA_MO1 and AB_MO2 caused the most severe cardiac phenotypes at the lowest doses. Knockdown of both scn5Laa and scn5Lab resulted in cardiac defects that were more severe than knockdown of either sodium channel alone, particularly with respect to ventricular morphogenesis. Arrows = ventricle. Arrowheads = atrium. Scale bar = 150µM. Treatments are as labeled: STD_MO = standard control morpholino, zerg_MO = cardiac potassium channel morpholino, AA_MIS1 = scn5Laa 5-mismatch control morpholino, AA_MO1 = scn5Laa translation-blocking morpholino (ATG initiation site), AA_MO2 = second scn5Laa translation-blocking morpholino (5’UTR), AB_MIS1 = scn5Lab 5-mismatch control morpholino, AB_MO1 = scn5Lab splice-blocking morpholino (E6I6), AB_MO2 = second scn5Lab splice-blocking morpholino (E25I25).
Non-specific morpholino toxicity or cell death can be minimized by concomitant knockdown of the zebrafish p53 gene.13 Co-injection of sodium channel and p53 morpholinos resulted in embryos with increased head size and overall body length but equally severe defects in cardiac development compared to embryos injected with sodium channel morpholinos alone (Figure 2, Online Figure VI). Acridine orange staining of morphant embryos identified no apoptotic cells in the dysmorphic, hypoplastic sodium channel morphant hearts (Online Figure VII).
Sodium channels are required for the production of physiological numbers of embryonic cardiomyocytes
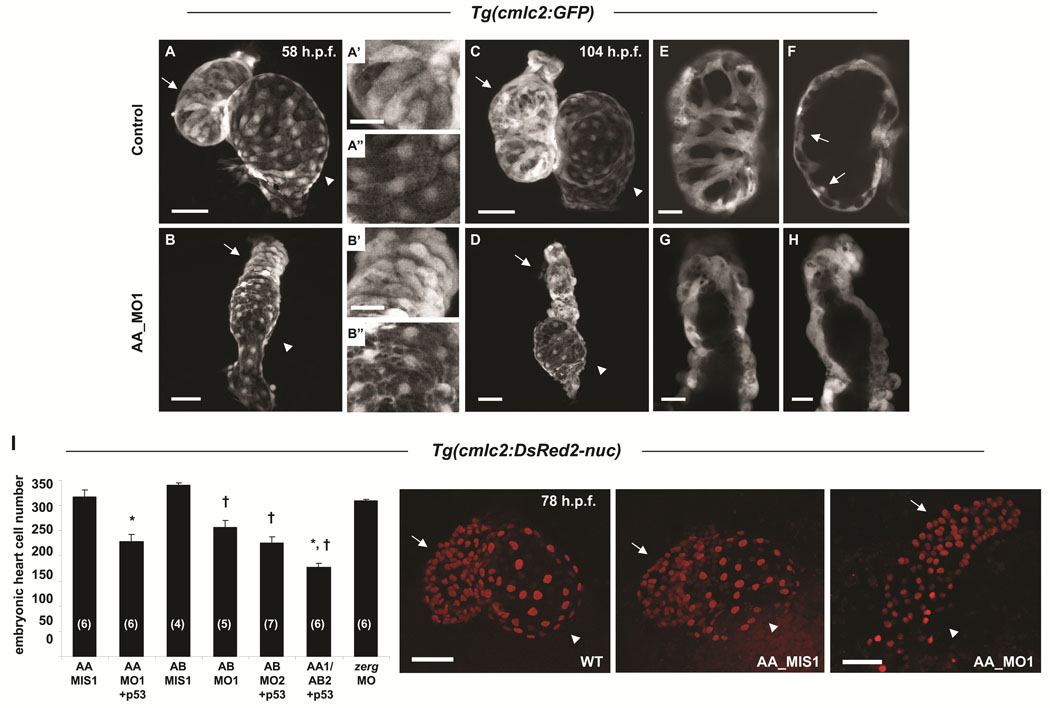
Scn5Laa morphant hearts displayed marked abnormalities of both atrial and ventricular chamber morphogenesis and looping at 58hpf (Figure 3A, B). At 104hpf, when growth of the zebrafish heart is achieved primarily by cardiomyocyte proliferation, scn5Laa morphant cardiac chambers remained small (Figure 3C, D). While control embryo hearts added cardiomyocytes to the interior of the ventricular wall to form trabeculae, scn5Laa morphant ventricles remained a single cell layer lacking trabeculated myocardium (Figure 3E–H). These findings indicate that sodium channel knockdown compromises both early chamber formation and normal patterned growth of the ventricle. Quantification of cardiomyocyte number in Tg(cmlc2:DsRed2-nuc) zebrafish revealed significant deficits in scn5Laa and scn5Lab morphants at 60–62hpf (Figure 3I). Dual scn5Laa/scn5Lab morphants had the fewest number of cardiomyocytes (40% less than controls) (Figure 3I).
FIGURE 3. Sodium channel morphant embryos have reduced numbers of embryonic cardiomyocytes.
A–H. Confocal reconstructions of hearts of control Tg(cmlc2:GFP) embryos (A, C) or clutchmates injected with the scn5Laa translation inhibitor morpholino (AA_MO1) (B, D) illustrate that sodium channels are required for normal numbers of embryonic cardiomyocytes. Arrows, ventricle and arrowheads, atrium at 58hpf (A, B) and 104hpf (C, D), respectively. Higher magnification of ventricular and atrial chambers presented in A’, B’ and A”, B”, respectively, illustrate similar chamber-specific cellular morphology in both control (A’, A”) and morphant (B’, B”) embryos. Scale bars = 50µm (A, B, C, D) and 20µm (A’, A”, B’, B”). E, F. Serial confocal sections through the ventricular chamber of control embryos revealed trabeculation by 104hpf (arrows in F). Scale bar = 20µm. G, H. At the same timepoint, scn5Laa morphant ventricles remained a single layer (shown are ventricles from two different morphant embryos). Scale bars = 20µm. I. Confocal reconstructions of the embryonic heart in Tg(cmlc2:DsRed2-nuc) embryos permitted quantification of deficits of embryonic cardiomyocytes in morphant embryos at 60–62hpf. Scn5Laa and scn5Lab morphant hearts have significantly fewer embryonic cardiomyocytes than those of mismatch control morpholino-injected clutchmates and embryos injected with a morpholino targeting the zerg cardiac potassium channel. Results are mean ± s.e.m. (N) in bar graph = number of hearts analyzed. *, P<0.01 versus AA_MIS1 and †, P<0.01 versus AB_MIS1, ANOVA. Embryos injected with both AA_MO1 and AB_MO2 morpholinos (scn5Laa, scn5Lab double knockdown) had fewer cardiomyocytes than embryos injected with either AA_MO1 or AB_MO2 alone (177±8 versus 228±14 and 226±12, respectively) but the results were not statistically significant following ANOVA of all 7 groups. Where indicated, p53 morpholino was co-injected with active morpholinos as a control for non-specific morpholino toxicity. Photos are representative wild type, scn5Laa control, and scn5Laa morphant embryo hearts at 78hpf. Arrows, ventricle. Arrowheads, atrium. Scale bars = 50µm.
Knockdown of sodium channels disrupts the expression of cardiogenic transcription factors in anterior lateral mesoderm
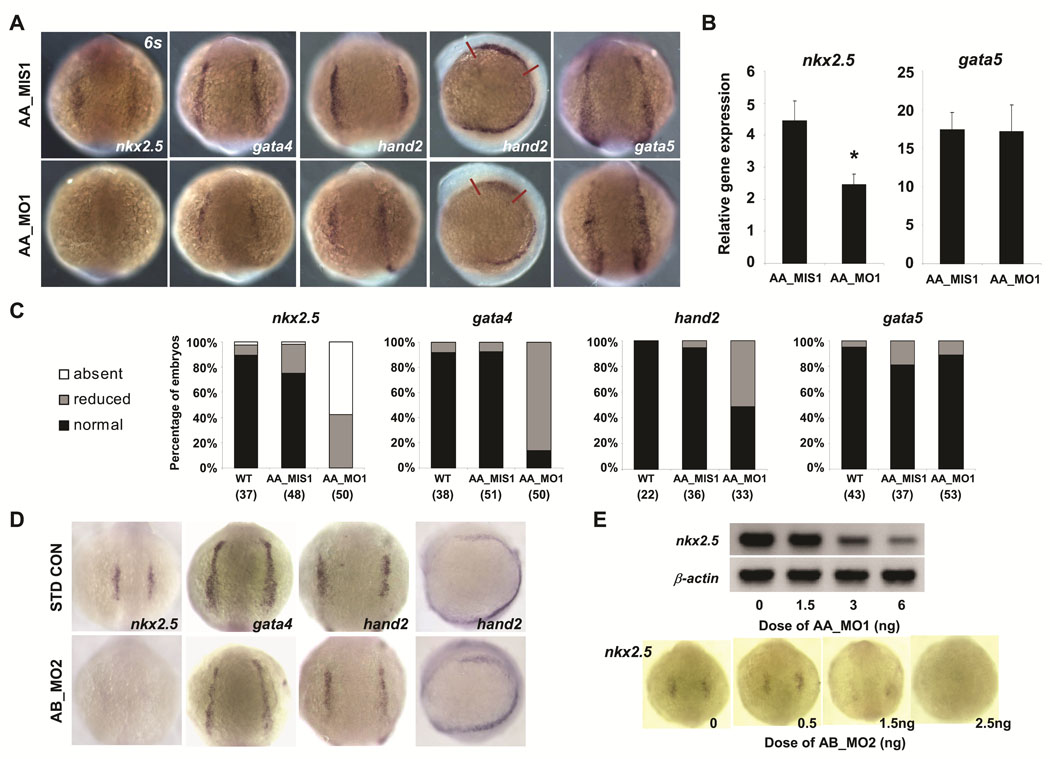
The expression of scn5Laa and scn5Lab during gastrulation and the diminished production of cardiomyocytes in morphant embryos together suggested that sodium channels may be required for the specification of normal numbers of cardiac progenitor cells. To test this hypothesis, we used in situ hybridization to examine gene expression in the heart-forming region of anterior lateral plate mesoderm (ALPM), a population of undifferentiated cells with cardiac potential demarcated by the expression of nkx2.5 and the overlapping expression domains of gata4 and hand2.14 Compared to control embryos, scn5Laa morphants displayed decreased expression of all three transcription factors in ALPM at the 6-somite stage (Figure 4A, C). Somite staging was used to confirm that the observed differences were not the result of developmental delay. Although gata5 is a potent inducer of nkx2.5 expression in zebrafish,15 gata5 expression levels were indistinguishable in scn5Laa morphant and control embryos (Figure 4A). These findings were independently corroborated by real-time RT-PCR (Figure 4B). Similar to scn5Laa, knockdown of scn5Lab also resulted in mispatterning of anterior lateral mesoderm with decreased expression of both nkx2.5 and gata4 (Figure 4D). The effect of scn5Laa and scn5Lab morpholinos on nkx2.5 expression at 6–8 somites was dose-dependent (Figure 4E). These results indicate that zebrafish cardiac sodium channels are required for the normal expression of cardiac fate-determining genes in anterior lateral mesoderm and for specification of appropriate numbers cardiac progenitor cells during development.
FIGURE 4. Sodium channel knockdown perturbs the expression of cardiac precursor genes in anterior lateral mesoderm.
A. Analysis of gene expression in the early heart-forming region of anterior lateral plate mesoderm at the 6-somite stage by in situ hybridization. Embryos injected with scn5Laa translation inhibitor morpholino (AA_MO1) had reduced expression of nkx2.5 gata4, and hand2 but no change in gata5 expression compared to embryos injected with 5-mismatch control morpholino (AA_MIS1). All embryos shown are from the same injected clutch. Scale bar = 200µm. B. Real-time quantitative RT-PCR performed using independent clutches of embryos also revealed significantly reduced nkx2.5 expression but no change in gata5 expression following injection of AA_MO1 compared to injection of AA_MIS1 control morpholino. Results are mean ± s.e.m for three independent experiments, each performed in triplicate. P<0.05 for nkx2.5, T-test. C. Quantification of numbers (n) of scn5Laa morphant embryos displaying absent, reduced or normal (comparable to wild type) expression levels of cardiac precursor genes as assessed by in situ hybridization. D. Knockdown of scn5Lab also resulted in markedly-reduced expression of nkx2.5 and gata4 at 6 somites. However, no significant change in the expression of hand2 was observed. E. Sodium channel morpholinos reduce nkx2.5 expression in a dose-dependent manner as shown by RT-PCR (AA_MO1) and in situ hybridization at 6 somites (AB_MO2).
Sodium channel knockdown results in reduced numbers of differentiating cardiomyocytes
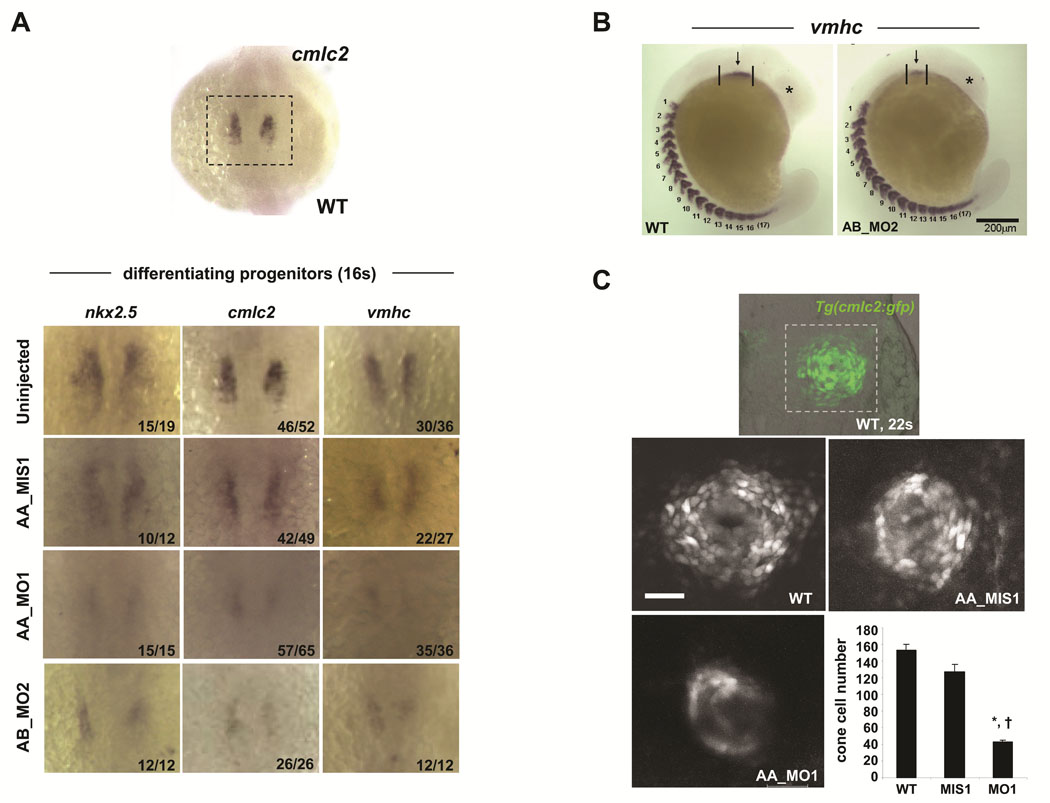
At the 16-somite stage, scn5Laa and scn5Lab morphants displayed reduced expression not only of nkx2.5 but of the sarcomeric genes cmlc2 and vmhc (Figure 5A). Moreover, examination of the number of somites (marked by vmhc expression) revealed that reductions in the size of the field of differentiating cardiomyocytes were not due to a global developmental delay. To assess the magnitude of the reduction in differentiating cardiomyocytes, we quantified total cardiac cell number in the cardiac cone of Tg(cmlc2:GFP) zebrafish at the 22-somite stage, the earliest timepoint when we could detect fluorescence in this line of transgenic zebrafish. At this stage, we found a significant reduction in the total number of newly differentiated myocytes in scn5Laa morphants compared to control embryos (Figure 5C).
FIGURE 5. Sodium channel knockdown results in reduced numbers of differentiating cardiomyocytes.
A. By in situ hybridization, scn5Laa and scn5Lab morphant embryos at the 16 somite stage display markedly reduced expression of nkx2.5 and of the myocardial sarcomeric genes cmlc2 and vmhc compared to control embryos. Treatment groups are as labeled. The number of embryos displaying the phenotype/the total number of embryos assessed is indicated in the lower right corner of each panel. B. Deficiencies in cardiac differentiation are not due to developmental delay. Shown is an embryo injected with the AB_MO2 morpholino compared to an uninjected clutchmate. Despite dramatic differences in the size of the heart field and head, both embryos have equivalent numbers of somites. C. Confocal reconstructions of the developing heart cone in Tg(cmlc2:GFP) embryos at the 22-somite stage (embryo in top panel is shown for orientation). Scale bar = 50µm. Scn5Laa morphant heart cones (AA_MO1, n=4) have significantly fewer differentiating cardiomyocytes than the heart cones of wild type (WT, n=6) and control-injected (AA_MIS1, n=3) clutchmates. Results are mean ± s.e.m. *, P<0.01 versus wild-type and †, P<0.01 versus AA_MIS1-injected, ANOVA.
Voltage-gated sodium channels may regulate early cardiac development independent of membrane electrophysiology
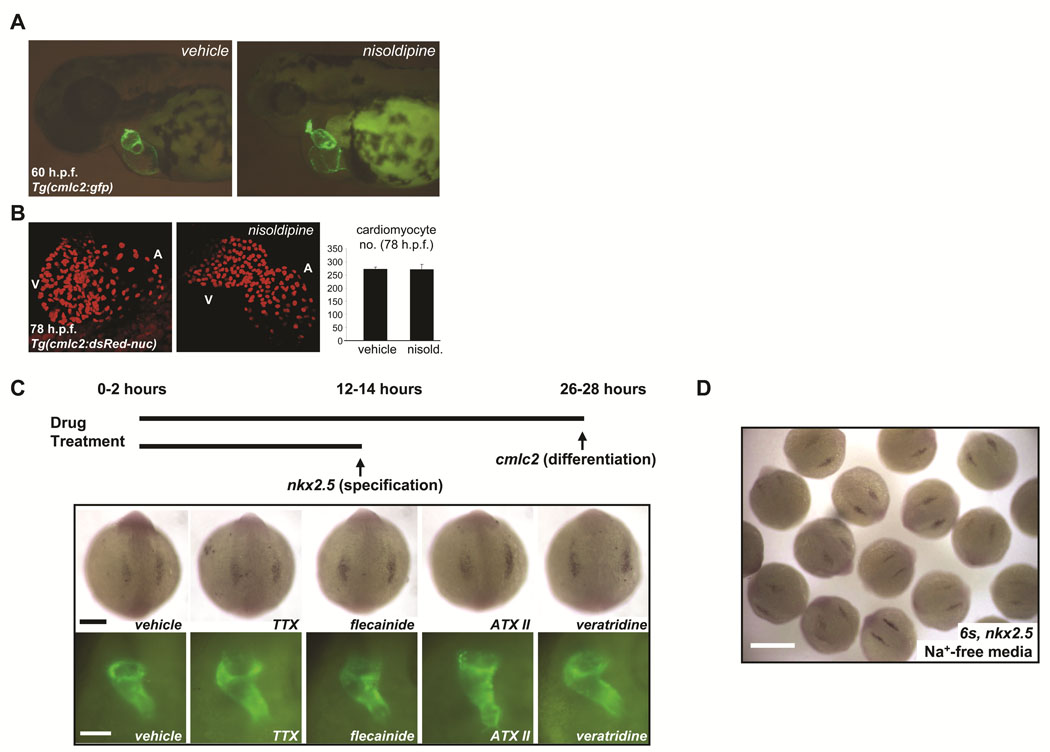
Prior studies in mice and chicks suggest that coordinated electrical activity in the early heart does not require fast sodium current (INa).16–18 Consistent with these prior observations, delivery of sodium channel blockers (e.g. TTX) and activators (e.g. ATX) directly into the pericardial space of zebrafish embryos failed to perturb heart-beating at early stages but caused conduction abnormalities at later stages (Online Figure VIII, Online Table III, Online Movie I–Online Movie IV). By contrast, the L-type calcium channel blocker nisoldipine silenced the heart at every stage examined. Therefore, we reared embryos in a high concentration of nisoldipine (10µM) from cleavage-stages through 78hpf. Nisoldipine-treated embryos failed to initiate heart beating at any stage examined but displayed normal cardiac morphogenesis through 60hpf and normal cardiomyocyte number compared to vehicle-treated embryos (Figures 6A, B, Table 1). This result is similar to that previously observed in zebrafish harboring mutations in cardiac troponin T (silent heart) in which heart formation is unaffected despite an absence of both contractility and circulation.19 These findings demonstrate that neither coordinated electrical activity nor beating is required for the earliest steps of cardiac development.
FIGURE 6. Voltage-gated sodium channels may regulate early cardiac development independent of membrane electrophysiology.
A. Rearing of embryos in 10µM solution of the L-type calcium blocker nisoldipine inhibited heart-beating at all stages examined through 78hpf but did not perturb early chamber formation or the initiation of looping. B. Analysis of vehicle-treated and nisoldipine-treated Tg(cmlc2:DsRed2-nuc) embryos indicated that L-type calcium channel-blockade did not affect embryonic cardiomyocyte number through 78hpf. Results are mean ± s.e.m for vehicle-treated (n = 2) and nisoldipine-treated (n = 3) embryos, respectively. C. Prolonged exposure of developing embryos to pharmacological modulators of sodium channel function failed to disrupt either cardiac specification or differentiation. In situ hybridization for nkx2.5 at the 6–8 somite stage (top) and fluorescent microscopy of the heart tubes of Tg(cmlc2:GFP) embryos at 26–28hpf (bottom), after the indicated treatments (see Table 2). TTX = tetrodotoxin, ATX II = anemone toxin II. Scale bars = 200µm (top) and 100µm (bottom). D. In situ hybridization for nkx2.5 at the 6 somite stage after incubation from the 1 cell stage in sodium-free media, to inhibit inward sodium current (INa). Early heart development proceeded normally. Heart tubes also form normally (not shown). Scale bar = 500µm.
We next hypothesized that sodium current (INa) may have an important role in early cardiac development prior to the onset of heart-beating. However, high concentrations of a range of drugs that activate or block INa had no effect on cardiac specification or differentiation when delivered to embryos in the bath solution or by microinjection (Figure 6C, Table 1). Importantly, TTX-treated embryos displayed complete neuromuscular paralysis at the end of the treatment period (∼28hpf) (Table 1, Online Video V), and treatment with channel activators (e.g. ATX II, veratridine) resulted in neuromuscular hyperexcitability (Table 1, Online Video VI). These findings indicate adequate compound penetration and confirmed that these toxins interact with other voltage-gated sodium channels in vivo, even at early stages. Early cardiogenesis also proceeded normally in embryos reared from the 1 cell stage in sodium-free media, another approach that inhibits inward INa (Figure 6D). Taken together, these results indicate that sodium channel expression but not sodium current is required for early cardiac development in zebrafish.
Discussion
Nav1 voltage-gated sodium channels underlie the upstroke of the action potential and are thus the principal determinants of membrane excitability in the nervous system, skeletal muscle, heart and other tissues.1 In the present study, we identified a previously-unappreciated role for cardiac-type sodium channels in the developing heart. We detected expression of scn5Laa and scn5Lab in the gastrulating embryo, prior to the differentiation of excitable tissues. Using both translation- and splice-blocking morpholino antisense oligonucleotides, we found that reduced expression of either isoform disrupted early cardiogenic gene expression in anterior lateral mesoderm and compromised the generation of sufficient numbers of progenitor cells for normal cardiac morphogenesis. We and others demonstrated that hearts that do not beat still display normal early development.19 Moreover, pharmacological manipulation of sodium current (INa) in vivo generated neuromuscular phenotypes but did not affect cardiac gene expression or morphogenesis, suggesting that sodium channel expression but not INa is required for cardiac development. These findings indicate that voltage-gated sodium channels have important roles in vertebrate heart development and support the hypothesis that these roles are mediated by non-electrogenic functions of the channel complex.
Voltage-gated sodium channels are required for early cardiac gene expression
A key defect we observed in sodium channel morphant embryos was a deficiency in the expression of multiple transcription factors that contribute to the specification of cardiac progenitor cells from anterior lateral mesoderm. Although reduced in number, these progenitors appear to normally differentiate into nascent cardiomyocytes. Early cardiogenic mesoderm is distinguished by the overlapping expression of a group of highly conserved regulatory genes belonging to the NK2, GATA, MEF2, T-box, and basic helix loop helix (bHLH) families of transcription factors.20, 21 The markedly reduced expression of nkx2.5 following knockdown of either scn5Laa or scn5lab suggests a disruption of one or more of the molecular mechanisms that normally act to specify the cardiac cell fate during gastrulation.
Prior studies in zebrafish have demonstrated that gata5 is a potent positive regulator of nkx2.5 expression, potentially analogous to the role played by gata4 or gata6 in mammals.15, 22 Notably, we found that scn5Laa regulates nkx2.5 expression without affecting levels of gata5, analogous to fgf8 signaling.23 This suggests that scn5Laa may act downstream of gata5 or fgf8 or in a parallel, previously-undefined pathway to regulate cardiac development. Knockdown of scn5Laa and scn5Lab also resulted in changes in the expression of gata4 and hand2 in the anterior lateral mesoderm. In zebrafish, both gata4 and hand2 are expressed in larger domains of anterior lateral mesoderm than nkx2.5 and are important for normal cardiogenesis.14, 24, 25 Reduced gata4 expression was observed to cause defects in cardiac chamber growth and looping, while loss of hand2 resulted in embryos with significantly fewer embryonic cardiomyocytes.24, 25 Moreover, the expression domain of hand2 appears to more accurately mark the population of early myocardial progenitors in zebrafish than nkx2.5.14 Rather than being required for the patterning or boundaries of the heart-forming region in mesoderm, hand2 appears to have a direct, permissive role in promoting cardiac differentiation within this cell population.14, 25 Taken together, the diminished number of cardiac progenitor cells observed in zebrafish cardiac sodium channel morphants is likely to be a consequence of the reduced expression level and/or domain of nkx2.5, gata4, and hand2, leading to a reduction in the size of the heart-forming region in anterior lateral mesoderm and limiting overall myocardial potential.
Voltage-gated sodium channels may contribute to early cardiogenesis independent of membrane electrophysiology
Supratherapeutic doses of TTX or other sodium channel active drugs did not perturb cardiac development. Cardiogenesis also proceeded normally when sodium was withdrawn from the external media in which embryos developed, another method for blocking inward INa. These findings thus suggest that zebrafish cardiac sodium channels may act in development via mechanisms that are independent of membrane depolarization.
Mounting evidence indicates that voltage-gated sodium channels subserve multiple functions whose disruption may underlie the phenotypes we describe. For example, sodium channel complexes play a role in cell adhesion.26 Pore-forming Nav1 sodium channel α subunits are known to interact with one or more ancillary β subunits (encoded by SCN1B-4B) that modulate α subunit expression and function.1, 2 Through their extracellular immunoglobulin (IG) domains, β subunits also mediate interactions between the sodium channel complex and other cell adhesion molecules and extracellular matrix proteins.26 Sodium channels are also known to interact with many other functionally-diverse proteins including 14-3-3, Nedd-4 type ubiquitinases, calmodulin, ankyrin G, syntrophins, plakophilin, and fibroblast growth factor homologous factors,27, 28 so it is possible that the sodium channel α subunit itself could act as a scaffold to facilitate intracellular signaling by these associated proteins within the channel complex.
Alternatively, sodium channel α subunits may generate non-electrogenic intracellular signals via mechanisms that are intrinsic to the channel protein itself. The C-terminal tail of L-type calcium channels, for example, is cleaved from the pore-forming α subunit and translocates from the membrane to the nucleus where it regulates gene expression.29 In this context, it is notable that zebrafish harboring a nonsense mutation in the cardiac L-type calcium channel α1c gene (island beat) display atrial arrhythmias and a small, non-contractile, hypoplastic ventricle by 72hpf.30 By contrast, we found that pharmacological blockade of calcium entry through L-type calcium channels over a similar time period did not significantly affect ventricular development. Future studies are required for additional insight into the possible non-electrogenic functions of cardiac voltage-gated sodium and calcium channels.
Limitations
In these experiments, we cannot exclude the possibility that intracellular sodium conductance (e.g. channels acting on the membrane of an organelle rather than at the cell surface) is required for normal development. Second, rescue of morphant phenotypes with full length scn5Laa or scn5Lab mRNA has not been possible to date. This is likely due to inadequate translation of the 6kb transcripts, as we have been unable to detect an epitope tag engineered into the rescue construct. Finally, knockdown of either scn5Laa or scn5Lab resulted in a severe cardiac phenotype. It is possible that the two genes, which have significantly diverged in amino acid sequence, may subserve different functions during heart development that were not uncovered by our assays. Alternatively, it is possible that manipulating the expression of one gene altered the expression or function of the other.
In summary, the results presented here argue that sodium channels are required for heart development in addition to their canonical role as regulators of heart rhythm. Moreover, the data suggest that this developmental role is mediated by a non-electrogenic function of the channel.
Novelty and Significance
What is known?
Activation of the Voltage-gated sodium channels leads to the depolarization of excitable membranes in mature myocardium, skeletal muscle and nerve.
Mutations in Scn5a, the predominant cardiac sodium channel gene, are associated with multiple heritable disorders of heart rhythm.
While Scn5a +/− mice develop myocardial fibrosis in senescence, Scn5a −/− mice develop marked abnormalities of ventricular morphogenesis by embryonic day 10.5, and die by day 11.5.
What new information does this article contribute?
In zebrafish, scn5a orthologs are expressed in the gastrulating embryo, prior to the differentiation of excitable tissues.
Antisense knockdown of either zebrafish cardiac sodium channel resulted in reduced expression of early markers of cardiomyocyte fate and marked chamber dysmorphogenesis.
These findings suggest that the zebrafish cardiac sodium channels affect heart development via a non-electrogenic mechanism
Voltage-gated sodium channels are the principal determinants of membrane excitability. In the heart, mutations in the cardiac sodium channel gene Scn5a are well-described in heritable arrhythmia syndromes. While sodium channels play a critical role in the mature heart, the function of Scn5a during cardiac development is less well understood. Here we used zebrafish embryos to investigate the role of sodium channels in cardiogenesis. We observed that zebrafish cardiac sodium channel genes are expressed before myocardial differentiation. Embryos treated with sodium channel antisense displayed reduced expression of early cardiogenic transcription factors including nkx2.5, gata4, and hand2, leading to diminished numbers of cardiac progenitor cells, cardiac chamber dysmorphogenesis, and perturbed looping. Prolonged pharmacological blockade of sodium current failed to phenocopy channel knockdown, suggesting that sodium channels act in early development via mechanisms that are independent of membrane electrophysiology. These studies may contribute to an improved understanding of the spectrum of clinical phenotypes linked to mutations in Scn5a.
Supplementary Material
Acknowledgements
We thank A. George, B. Appel, J. Barnett, R. Emeson and L. Solnica-Krezel for their advice and suggestions; Q. Guan, H. Jia, J. Denton, P. Yang, K. Liu, J. Bennett, and C. Campbell for their technical assistance; and J. Stassun for her administrative support. We also thank members of the imaging core for their invaluable advice and suggestions. Scn5Laa and scn5Lab probes for in situ hybridization were kindly provided by the Ribera lab.
Sources of Funding
This work was supported by Public Health Service award T32 GM07347 from the National Institute of General Medical Studies for the Vanderbilt Medical-Scientist Training Program, funding from the Vanderbilt University Functional Genomics Initiative, and HL49989.
Non-standard Abbreviations and Acronyms
- ALPM
Anterior lateral plate mesoderm
- ATX II
Anemone toxin
- CHO
Chinese hamster ovary
- DsRed
Discosoma sp. red fluorescent protein
- E
embryonic day
- GFP
green fluorescent protein
- Hpf
hours post fertilization
- Nav1.5
sodium channel alpha subunit 1.5
- nuc
nuclear
- TTX
Tetrodotoxin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures
None
References
- 1.Yu FH, Catterall WA. Overview of the voltage-gated sodium channel family. Genome Biol. 2003;4:207. doi: 10.1186/gb-2003-4-3-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goldin AL. Resurgence of sodium channel research. Annu Rev Physiol. 2001;63:871–894. doi: 10.1146/annurev.physiol.63.1.871. [DOI] [PubMed] [Google Scholar]
- 3.Catterall WA, Goldin AL, Waxman SG. International Union of Pharmacology. XLVII. Nomenclature and structure-function relationships of voltage-gated sodium channels. Pharmacol Rev. 2005;57:397–409. doi: 10.1124/pr.57.4.4. [DOI] [PubMed] [Google Scholar]
- 4.Roden DM, Balser JR, George AL, Jr, Anderson ME. Cardiac ion channels. Annu Rev Physiol. 2002;64:431–475. doi: 10.1146/annurev.physiol.64.083101.145105. 431–75. [DOI] [PubMed] [Google Scholar]
- 5.George AL., Jr Inherited disorders of voltage-gated sodium channels. J Clin Invest. 2005;115:1990–1999. doi: 10.1172/JCI25505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koopmann TT, Bezzina CR, Wilde AA. Voltage-gated sodium channels: action players with many faces. Ann Med. 2006;38:472–482. doi: 10.1080/07853890600969072. [DOI] [PubMed] [Google Scholar]
- 7.Papadatos GA, Wallerstein PM, Head CE, Ratcliff R, Brady PA, Benndorf K, Saumarez RC, Trezise AE, Huang CL, Vandenberg JI, Colledge WH, Grace AA. Slowed conduction and ventricular tachycardia after targeted disruption of the cardiac sodium channel gene Scn5a. Proc Natl Acad Sci U S A. 2002;99:6210–6215. doi: 10.1073/pnas.082121299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Keegan BR, Meyer D, Yelon D. Organization of cardiac chamber progenitors in the zebrafish blastula. Development. 2004;131:3081–3091. doi: 10.1242/dev.01185. [DOI] [PubMed] [Google Scholar]
- 9.Glickman NS, Yelon D. Cardiac development in zebrafish: coordination of form and function. Semin Cell Dev Biol. 2002;13:507–513. doi: 10.1016/s1084952102001040. [DOI] [PubMed] [Google Scholar]
- 10.Stainier DY. Zebrafish genetics and vertebrate heart formation. Nat Rev Genet. 2001;2:39–48. doi: 10.1038/35047564. [DOI] [PubMed] [Google Scholar]
- 11.Novak AE, Jost MC, Lu Y, Taylor AD, Zakon HH, Ribera AB. Gene duplications and evolution of vertebrate voltage-gated sodium channels. J Mol Evol. 2006;63:208–221. doi: 10.1007/s00239-005-0287-9. [DOI] [PubMed] [Google Scholar]
- 12.Burns CG, Milan DJ, Grande EJ, Rottbauer W, Macrae CA, Fishman MC. High-throughput assay for small molecules that modulate zebrafish embryonic heart rate. Nat Chem Biol. 2005;1:263–264. doi: 10.1038/nchembio732. [DOI] [PubMed] [Google Scholar]
- 13.Robu ME, Larson JD, Nasevicius A, Beiraghi S, Brenner C, Farber SA, Ekker SC. p53 activation by knockdown technologies. PLoS Genet. 2007;3:e78. doi: 10.1371/journal.pgen.0030078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schoenebeck JJ, Keegan BR, Yelon D. Vessel and blood specification override cardiac potential in anterior mesoderm. Dev Cell. 2007;13:254–267. doi: 10.1016/j.devcel.2007.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reiter JF, Alexander J, Rodaway A, Yelon D, Patient R, Holder N, Stainier DY. Gata5 is required for the development of the heart and endoderm in zebrafish. Genes Dev. 1999;13:2983–2995. doi: 10.1101/gad.13.22.2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sperelakis N, Pappano AJ. Physiology and pharmacology of developing heart cells. Pharmacol Ther. 1983;22:1–39. doi: 10.1016/0163-7258(83)90050-5. [DOI] [PubMed] [Google Scholar]
- 17.Davies MP, An RH, Doevendans P, Kubalak S, Chien KR, Kass RS. Developmental changes in ionic channel activity in the embryonic murine heart. Circ Res. 1996;78:15–25. doi: 10.1161/01.res.78.1.15. [DOI] [PubMed] [Google Scholar]
- 18.Renaud JF, Romey G, Lombet A, Lazdunski M. Differentiation of the fast Na+ channel in embryonic heart cells: interaction of the channel with neurotoxins. Proc Natl Acad Sci U S A. 1981;78:5348–5352. doi: 10.1073/pnas.78.9.5348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sehnert AJ, Huq A, Weinstein BM, Walker C, Fishman M, Stainier DY. Cardiac troponin T is essential in sarcomere assembly and cardiac contractility. Nat Genet. 2002;31:106–110. doi: 10.1038/ng875. [DOI] [PubMed] [Google Scholar]
- 20.Olson EN. Gene regulatory networks in the evolution and development of the heart. Science. 2006;313:1922–1927. doi: 10.1126/science.1132292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Srivastava D. Making or breaking the heart: from lineage determination to morphogenesis. Cell. 2006;126:1037–1048. doi: 10.1016/j.cell.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 22.Reiter JF, Verkade H, Stainier DY. Bmp2b and Oep promote early myocardial differentiation through their regulation of gata5. Dev Biol. 2001;234:330–338. doi: 10.1006/dbio.2001.0259. [DOI] [PubMed] [Google Scholar]
- 23.Reifers F, Walsh EC, Leger S, Stainier DY, Brand M. Induction and differentiation of the zebrafish heart requires fibroblast growth factor 8 (fgf8/acerebellar) Development. 2000;127:225–235. doi: 10.1242/dev.127.2.225. [DOI] [PubMed] [Google Scholar]
- 24.Holtzinger A, Evans T. Gata4 regulates the formation of multiple organs. Development. 2005;132:4005–4014. doi: 10.1242/dev.01978. [DOI] [PubMed] [Google Scholar]
- 25.Yelon D, Ticho B, Halpern ME, Ruvinsky I, Ho RK, Silver LM, Stainier DY. The bHLH transcription factor hand2 plays parallel roles in zebrafish heart and pectoral fin development. Development. 2000;127:2573–2582. doi: 10.1242/dev.127.12.2573. [DOI] [PubMed] [Google Scholar]
- 26.Isom LL. The role of sodium channels in cell adhesion. Front Biosci. 2002;7:12–23. doi: 10.2741/isom. [DOI] [PubMed] [Google Scholar]
- 27.Abriel H, Kass RS. Regulation of the voltage-gated cardiac sodium channel Nav1.5 by interacting proteins. Trends Cardiovasc Med. 2005;15:35–40. doi: 10.1016/j.tcm.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 28.Meadows LS, Isom LL. Sodium channels as macromolecular complexes: implications for inherited arrhythmia syndromes. Cardiovasc Res. 2005;67:448–458. doi: 10.1016/j.cardiores.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 29.Gomez-Ospina N, Tsuruta F, Barreto-Chang O, Hu L, Dolmetsch R. The C terminus of the L-type voltage-gated calcium channel Ca(V)1.2 encodes a transcription factor. Cell. 2006;127:591–606. doi: 10.1016/j.cell.2006.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rottbauer W, Baker K, Wo ZG, Mohideen MA, Cantiello HF, Fishman MC. Growth and function of the embryonic heart depend upon the cardiac-specific L-type calcium channel alpha1 subunit. Dev Cell. 2001;1:265–275. doi: 10.1016/s1534-5807(01)00023-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.