Abstract
MicroRNAs are often aberrantly expressed in human neoplasms and are postulated to play a role in neoplastic initiation and progression. miR-221 and miR-222 negatively regulate expression of CDKN1B (p27)and CDKN1C (p57), two cell cycle regulators expressed in ovarian surface epithelium and down-regulated in ovarian carcinomas. We characterized miR-221 and miR-222 expression in 49 sporadic high grade ovarian carcinomas and determined whether somatic mutation or epigenetic alterations explained the differences in expression of these miRNAs. We correlated these findings with protein expression of CDKN1B and CDKN1C as assessed by immunohistochemistry. Expression of miR-221 and miR-222 were closely correlated with each other (P=0.0001). Interestingly, a lower ratio of miR-221 to miR-222 expression was significantly correlated with worse overall survival (P=0.01) and remained a significant predictor of overall survival in multivariate analysis using the co-variate adequacy of surgical cytoreduction (P=0.03). Higher miR-222 and miR-221 expression were significantly associated with decreased CDKN1C expression (P=0.009 and 0.01). In contrast, CDKN1B expression was not associated with miR-221 or miR-222 expression. Neither somatic mutations nor methylation of the studied region explained the alterations in miR-221 and miR-222 expression in most carcinomas.
INTRODUCTION
MicroRNAs (miRNAs) are small non-coding RNAs that function as negative gene regulators at the post-transcriptional level by destabilizing messenger RNA or by directly repressing translation (Bartel, 2004; 2009). MiRNAs may exist as discrete transcriptional units or reside in introns or exons and share a primary transcript and regulatory elements with the host gene (Bartel, 2004). These primary transcripts are termed pri-miRNAs and are cleaved in the nucleus by the enzyme Drosha to form a 60–70 nucleotide stem loop structure known as the pre-miRNA. The pre-miRNA is actively exported out of the nucleus into the cytoplasm, where another key enzyme Dicer further cleaves the pre-miRNA into the mature miRNA, approximately 22 nucleotides in length. One strand is released by Dicer and incorporated into the RNA-induced silencing complex (RISC). MiRNAs have been implicated in the regulation of nearly all cellular functions and are critical in normal development and differentiation.
MiRNAs are often aberrantly expressed in human neoplasms, and may play a key role in the formation and progression of neoplasms (Garzon et al., 2006). Recent findings indicate that numerous miRNAs are up or down-regulated in ovarian carcinoma tissues and cell lines, and expression levels of miRNAs can distinguish malignant and non malignant ovarian epithelium (Iorio et al., 2007; Dahiya et al., 2008; Zhang et al., 2008). The mechanisms of miRNA down-regulation in cancers are not fully understood, however both copy number alteration and epigenetic silencing play some role (Iorio et al., 2007; Zhang et al., 2008). Copy number alterations in key enzymes that regulate miRNA production are also prevalent in human cancers (Zhang et al., 2006). Low Dicer expression in ovarian carcinomas has recently been shown to be an independent predictor of worse survival (Merritt et al., 2008).
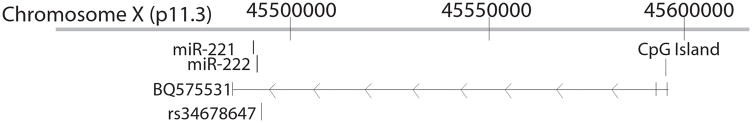
MiR-221 and miR-222 are two miRNAs encoded in tandem on the X chromosome, separated by only 227 nucleotides. A single expressed sequence tag (EST) BQ575531 overlaps with miR-221 and miR-222 and is likely to represent the pri-miRNA (Fig. 1). At least 40% of human miRNAs are encoded in clusters of two or more miRNAs that are processed from a common pri-miRNA (Altuvia et al., 2005). Such polycistronic miRNAs are often related to each other, but even when they do not share sequence homology they may have functional relationships (Bartel, 2004). MiR-221 and MiR-222 both function in cell cycle regulation and recently, miR-221 and miR-222 have been shown to target the negative cell cycle regulators CDKN1B (p27) and CDKN1C (p57) (Larson et al., 2008; Medina et al., 2008). CDKN1B and CDKN1C may function as tumor suppressor genes in a variety of cancer types.
Figure 1.
Schematic depicting the genomic organization of miR-221 and miR-222. On this diagram miR-221 and miR-222 would be transcribed from right to left. The pri-RNA for miR-221 and miR-222 is not defined, but could be the EST BQ575531. We assessed the CpG island in the promoter region of BQ575531 for methylation. Three cases had germline heterozygosity at the single nucleotide polymorphism rs34678647.
Deregulation of miR-221 and miR-222 promotes the growth of human glioblastoma, thyroid cancer, prostate cancer, and melanoma cells by inhibiting expression of CDKN1B (le Sage et al., 2007). In human hepatocellular carcinoma, miR-221 has oncogenic function by targeting CDKN1C (Fornari et al., 2008). Dahiya and colleagues identified miR-221 as the most up-regulated miRNA in human ovarian cancer tissues and cell lines compared to an immortalized ovarian surface epithelial culture (Dahiya et al., 2008), though we recently reported miR-221 as under-expressed in ovarian carcinomas compared to non-immmortalized primary ovarian surface epithelial cultures (Wyman et al., 2009). These data suggest that the choice of the normal comparison tissue may markedly affect interpretation of miRNA expression, similar to previous mRNA expression analyses in ovarian carcinomas (Zorn et al., 2003). MiR-222 has also been shown to be up-regulated in ovarian carcinoma cell lines compared to immortalized ovarian surface epithelium (Zhang et al., 2008).
Both CDKN1B and CDKN1C protein expression are frequently decreased in ovarian carcinomas, but the mechanisms explaining alterations of CDKN1B and CDKN1C in ovarian carcinomas are not identified (Masciullo et al., 1999; Sui et al., 1999; Rosenberg et al., 2001; Sui et al., 2002; Khouja et al., 2007). Because miR-221 and miR-222 target the 3′ untranslated regions of the CDKN1B and CDKN1C genes, we hypothesized that miR-221 and miR-222 expression might be important determinants of CDKN1B or CDKN1C expression in ovarian carcinomas. We characterized miR-221 and miR-222 expression and determined if somatic mutation or epigenetic alterations via methylation explain alterations in these miRNAs in ovarian carcinomas. We evaluated CDKN1B and CDKN1C protein expression in the same set of carcinomas and correlated expression with alterations in miR-221 and miR-222.
MATERIALS AND METHODS
Clinical Specimens
Tissue and clinical information was collected from women undergoing surgery for ovarian carcinoma by the University of Washington Gynecologic Oncology Tissue Bank as approved by the Human Subjects Committee of the Institutional Review Board. All patients were treated with combination platinum and taxane chemotherapy as initial therapy. Tumors were surgically staged according to the International Federation of Obstetrics and Gynecology (FIGO) criteria. DNA and RNA were extracted from lymphocytes and from ovarian carcinomas that were microdissected if necessary to attain a neoplastic cellularity of at least 70%. Overall survival was calculated from the date of diagnosis to the date of last follow-up or death. Progression-free interval was calculated from the date of diagnosis to the date of progression. Forty-nine sporadic high grade ovarian carcinomas were chosen. Sporadic carcinomas were defined as those occurring in women without a family history of ovarian or pre-menopausal breast carcinoma.
Taqman MicroRNA Quantitative Reverse-Transcription PCR (qRT-PCR)
Expression of mature miRNAs was detected using the Taqman miRNA qRT-PCR Assay (Applied Biosystems, Carlsbad, CA). Reverse transcription and PCR reactions were run in triplicate in an Applied Biosystems 7300 Fast Real-Time PCR System. Data were analyzed using Sequence Detection Software (V 1.2.3, Applied Biosystems). The comparative CT method was used to calculate relative expression with comparison to the small RNA U47 for each specimen. The ratio of miR-221/miR-222 was calculated by comparing the ratio of the relative quantification for each miRNA for each case.
Methylation-Specific PCR
Bisulfite modification of the tumor DNA was performed using the CpGenome DNA Modification Kit (Chemicon International, Temecula, CA). The CpG island assessed is at ChrX:45,595,214-45,595,923. This CpG island is a putative regulator of miR-221 and miR-222 as it resides in the promoter region of the EST BQ575531 which overlaps with miR-221 and miR-222 (Fig. 1). The BQ575531 CpG island was analyzed for methylation using primers designed for either the methylated or unmethylated DNA sequence. Primer sequences for the unmethylated reaction were 5′-GCGGTCCCAAAAGGGTCAGTGAAGGGAAATGAGGATTATTGTTTT-3′ and 5′-GCGGTCCCAAAAGGGTCAGTCTCAAAAATACCCACCTACA-3′. Primer sequences for the methylated reaction were 5′-GCGGTCCCAAAAGGGTCAGTAGGGAAATGAGGATTATCGTTTC-3′ and 5′-GCGGTCCCAAAAGGGTCAGTCTCGAAAATACCCACCTACG-3′.
Universal Methylated DNA was used as a positive control for methylation (Chemicon International). In addition to the primary carcinomas, methylation was evaluated in 5 different lymphocyte DNAs, two immortalized ovarian surface epithelial cell lines (IOSE-29, IOSE-80), 8 primary ovarian surface epithelial cultures, one benign ovarian mucinous cystadenoma, and three ovarian cancer cell lines (UWB1.289, HeyA8, and EFO-21).
Sequencing of miR-221 and miR-222
DNA was polymerase chain reaction (PCR) amplified and sequenced for miR-221 and miR-222 that included the coding sequences, the intervening sequence, 52 basepairs upstream, and 87 basepairs downstream of the two miRNAs. PCR primer sequences were 5′-CAAGGAATCATGTATGCTGTAG -3′ and 5′-AGGATGACATTACACCTTATCTC -3′. PCR products of genomic DNA were purified and sequenced with BigDye Terminator chemistry (Applied Biosystems) using ABI 3100 DNA sequencer (Applied Biosystems). Sequencing primer sequences were 5′-ATGGCATTTTCAACATGATGTC -3′ and 5′-GTGTGTGTAATTCAAGGTAAAG -3′. Sequencing data were analyzed using Sequencher Software (Gene Codes Corporation, Ann Arbor, MI). All alterations identified in tumor sequences were reviewed and compared with those from corresponding normal DNA and confirmed in a separate amplification and sequencing reaction.
Immunohistochemical Studies
Briefly, paraffin sections were deparaffinized, re-hydrated, subjected to antigen retrieval and treated with 3% H2O2. Sections were washed with PBS and blocked with 2% bovine serum albumin in PBS. Primary antibodies were the mouse monoclonals 57/kip1/p27 at 1:1000 dilution (BD Transduction Laboratories, San Jose, CA) for CDKN1B and Kip2/57P06 at 1:400 dilution (Lab Vision Corporation Neomarkers, Fremont, CA) for CDKN1C and were applied for 30 min at room temperature. Secondary antibody and streptavidin biotin-peroxidase were from Universal Large Volume LSAB+, Peroxidase kit (DAKO) and were each applied for 30 minutes at room temperature. DAB (3,3′ diaminobenzidine) chromogen was used to visualize antibody complexes and sections were counterstained with hematoxylin. Negative controls were sections incubated with normal mouse serum instead of primary antibody. Stain was read as a continuous variable as a percent of moderate to strongly stained neoplastic cells by an investigator blinded to miRNA expression results.
RESULTS
The majority of cases studied were high grade, advanced stage serous ovarian carcinomas. All patients received surgery by a board certified gynecologic oncologist and approximately half were optimally debulked at initial surgery with a maximum tumor diameter less than one centimeter. Clinical characteristics of these cases are outlined in Table 1.
TABLE 1.
Clincopathological Characteristics of Ovarian Carcinomas.
| Age | |
| Range | 37–88 years |
| Median | 57 years |
| Stage | |
| I | 2 (4%) |
| II | 1 (2%) |
| III | 38 (78%) |
| IV | 8 (16%) |
| Histology | |
| Serous | 34 (69%) |
| Endometrioid | 1 (2%) |
| Carcinoma | 2 (4%) |
| MMMT | 1 (2%) |
| Clear cell | 2 (4%) |
| Adenocarcinoma | 9 (18%) |
| Grade | |
| Grade 1 | 1 (2%) |
| Grade 2 | 1 (2%) |
| Grade 3 | 47 (96%) |
| Cytoreduction | |
| Optimal (<1cm) | 25 (51%) |
| Suboptimal | 24 (49%) |
| Total | 49 |
MMMT: malignant mixed mullerian tumor (carcinosarcoma)
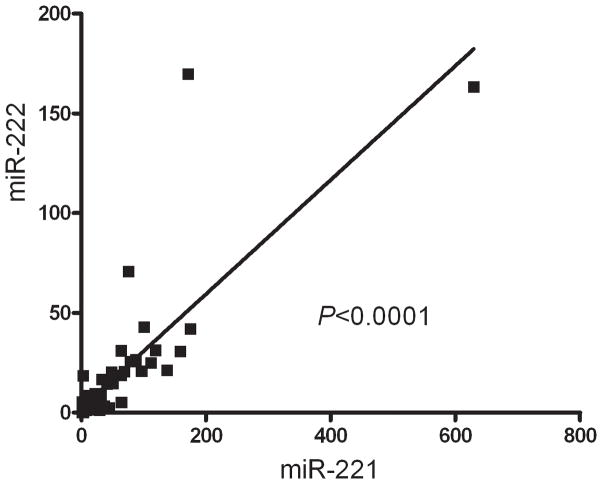
mRNA expression of miR-221 and miR-222 was evaluated by a TaqMan miRNA quantitative R-PCR assay and normalized to the small RNA U47 for each sample. This expression analysis revealed a tight correlation between miR-221 and miR-222 within neoplasms (correlation coefficient r2=0.65, P<0.0001, linear regression) consistent with their tandem expression in a common primary transcript or pri-miRNA (Fig. 2).
Figure 2.
Correlation Between miR-221 and miR-222 expression. Expression analysis revealed a tight correlation between miR-221 and miR-222, consistent with their transcription as a common primary transcript before processing into individual miRNAs (correlation coefficient r2=0.65, P<0.0001, linear regression). Both miR-221 and miR-222 expression is plotted as the relative expression compared to the small RNA U47 for each specimen.
DNA from all carcinomas was evaluated for somatic mutation in the miR-221 and miR-222 region. DNA sequencing revealed one case (Carcinoma 121) with a somatic deletion of 1,049 basepairs at chrX:45,490,682-45,491,731 that deleted miR-222 while leaving miR-221 intact. The median ratio of miR-221 to miR-222 for all carcinomas was 3.7 (range 0.15–23.4). Carcinoma 121 had a similar ratio of 4.7, despite the monoallelic deletion of miR-222. In addition to this somatic deletion, we identified three cases with germline heterozygosity (G/T) at the known SNP rs34678647 (Fig. 1). There was no obvious difference in expression level of either miRNA in cases with or without this SNP. Likewise, there was no clear difference in the ratio of expression of the two miRNAs in cases with the SNP, but power was limited to detect such a difference given the small number of cases with the SNP.
Next, we evaluated methylation of the putative regulatory region for miR-221 and miR-222 using methylation sensitive PCR to determine if alterations in methylation of this area impacted miRNA expression in ovarian carcinomas. This region was methylated in 83% of ovarian carcinomas. However, methylation of this region did not correlate with miR-221 or miR-222 expression nor with alterations in the ratio of the two miRNAs. A similar 80% frequency of methylation was identified in normal tissues tested which included: 4 of 5 lymphocytes, one mucinous ovarian cystadenoma, all eight ovarian surface epithelium primary cultures, one of two immortalized ovarian surface epithelial cell lines, and all three ovarian carcinoma cell lines.
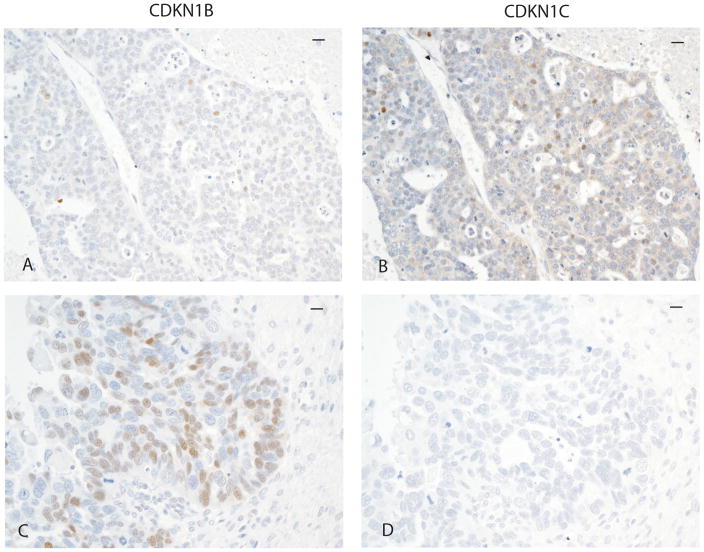
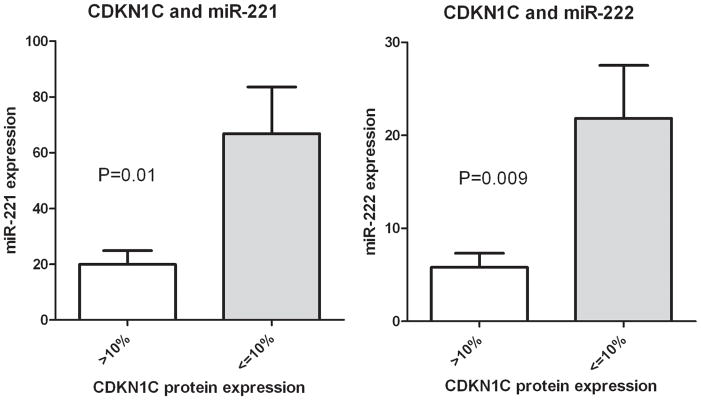
We next evaluated CDKN1C protein expression using immunohistochemistry to determine the relationship of expression of this cell cycle checkpoint protein to expression of miR-221 and miR-222, since CDKN1C is a validated target of these miRNAs. CDKN1C protein expression varied between carcinomas and was present in 0–60% of neoplastic cells. CDKN1C was present in greater than 10% of neoplastic cells in 8/47 (17%) of carcinomas. Representative immunostaining is demonstrated in Fig. 3. Cases with ≥10% of neoplastic cells with CDKN1C immunostaining had significantly lower miR-222 expression and miR-221 expression than carcinomas without CDKN1C expression (P=0.009 and P=0.01, respectively, unpaired T test with Welch’s correction, two-tailed, Fig. 4). Therefore, CDKN1C protein expression was related to mRNA expression levels of both miR-221 and miR-222.
Figure 3.
CDKN1B and CDKN1C immunostaining in ovarian carcinomas.
Representative immunostaining for CDKN1C and CDKN1B protein in two carcinomas. The bars in the upper right carner of each micrograph represents 10 μm. The first carcinoma demonstrates rare positive neoplastic cells (brown nuclear staining) for CDKN1B (A) and approximately 20% of neoplastic cells positive for CDKN1C (B). The second carcinoma demonstrates about 60% of neoplastic cells positive for CDKN1B (brown nuclear staining, C) and negative CDKN1C staining (D).
Figure 4.
Association between miR221, miR-222 expression and CDKN1C expression. Cases with ≥10% of neoplastic cells with CDKN1C protein have significantly lower miR-221 and miR-222 expression than carcinomas without CDKN1C expression (P=0.009 and P=0.01 respectively, unpaired T test with Welch’s correction, two-tailed). MiR-221 and miR-222 expression was calculated as the relative expression with comparison to the small RNA U47 for each case.
We also evaluated the relationship of the cell cycle checkpoint CDKN1B, another validated target of miR-221 and miR-222, to message expression of these miRNAs. CDKN1B protein expression also varied among carcinomas and was identified in 0–80% of neoplastic cells. Representative immunostaining is demonstrated in Fig. 3. CDKN1B was present in greater than 20% of cells in 64% of carcinomas. Unlike CDKN1C protein expression, there was no correlation between CDKN1B protein expression and miR-221 or miR-222 expression within ovarian carcinomas (data not shown).
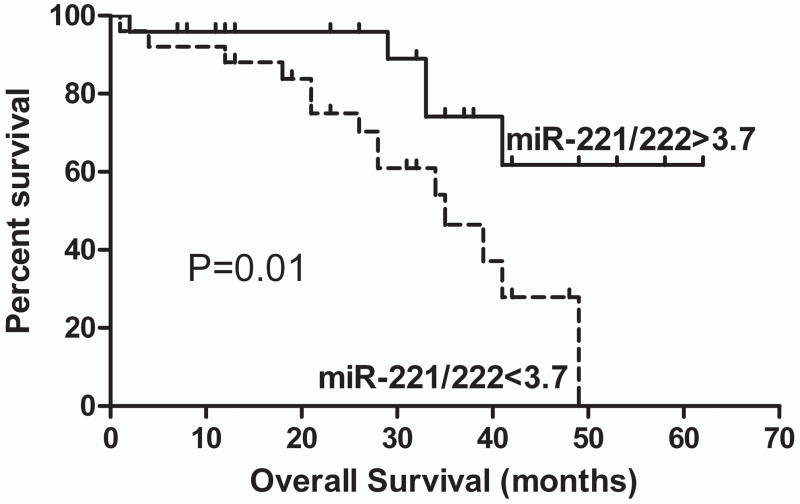
We tested whether miRNA expression at the mRNA level correlated with prognosis in our cases. Neither miR-221 nor miR-222 expression levels were related to overall survival or progression-free interval. However, overall survival was significantly associated with the ratio of the relative expressions of miR-221 to that of miR-222. Women whose cancers had a miR-221/miR-222 ratio less than the median ratio of 3.7 had a median overall survival of 35 months, while those with a ratio greater than 3.7 had a longer overall survival for which the median value has not yet been reached (Fig. 5, P=0.01, Log-Rank Test, Hazard Ratio (HR) 0.32, 95% confidence interval of HR 0.13–0.82). The miR-221/miR-222 ratio was not associated with other clinical risk factors including stage, histology or likelihood of optimal cytoreduction. There was no relationship between either CDKN1B or CDKN1C protein expression and overall survival. In this small series, stage was not significantly associated with survival, but 46/49 cases were advanced stage (stage III or IV, Table 1) minimizing the impact of survival on stage. The only other predictor of survival in this sample set was the adequacy of surgical cytoreduction. In the advanced stage cases, optimal cytoreduction to a maximum tumor diameter <1 cm was associated with better overall survival (P=0.049). A Cox regression model was created using the co-variates relative miR-221/miR-222 ratio > or < 3.7 and optimal cytoreduction. In this model, cytoreduction was no longer significant (P=0.28), but the miRNA ratio remained significantly related to overall survival (P=0.03).
Figure 5.
Ratio of miR-221/miR-222 and overall survival. Relative expression of miR-222 (normalized to U47 expression for each specimen) was divided by the relative expression of miR-221 (also normalized to U47 for each specimen). The cases were divided into two near equal groups by the median miR-221/miR-222 ratio of 3.7. Overall survival was significantly associated with the ratio of miR-221/miR-222. A miR-221/miR-222 ratio less than 3.7 was associated with a median survival of 35 months. A miR-221/miR-222 ratio greater than 3.7 was associated with significantly longer survival for which the median value has not yet been reached (P=0.01, Log-Rank Test, Hazard Ratio (HR) 0.32, 95% confidence interval of HR 0.13–0.82).
DISCUSSION
Expression of miR-221 and miR-222 are closely correlated in ovarian carcinomas, consistent with their transcription as a common pri-miRNA before processing into individual miRNAs. Interestingly, a lower ratio of miR-221 to miR-222 was significantly correlated with worse overall survival. This relationship remained significant after accounting for optimal versus suboptimal surgical cytoreduction in a multivariate model. The ratio of miR-221 to miR-222 was the only independently significant predictor of overall survival in our small set of predominantly high grade, advanced stage sporadic ovarian carcinomas. To our knowledge this is the first report showing that alterations in the ratio of co-expressed miRNAs may have prognostic importance in cancer.
Co-expressed miRNAs may share functional relationships (Bartel, 2004), such as the cell cycle regulatory function of miR-221 and miR-222. The relative expression of such miRNAs are likely to be important in fine tuning gene and protein expression. The mechanisms that control the relative expression of co-expressed miRNAs are not known but could involve variation in miRNA turnover. Alterations in the ratio of miR-221 and miR-222 could directly impact tumor behavior by regulating target genes such as CDKN1B and CDKN1C. Alternatively, alterations in the relative ratio of miR-221 and miR-222 could impact prognosis by signaling a more global disruption of miRNA balance at the post-transcriptional level.
There are few experimentally validated targets for most miRNAs and large numbers of potential targets predicted by various algorithms. TargetScan (http://www.targetscan.org/) predicts 317 targets for miR-221, while miRBase (http://microrna.sanger.ac.uk/targets/v5/) predicts 742 tragets for miR-221 and 744 for miR-222. While miRNA-221 and miRNA-222 are likely to have multiple targets in ovarian carcinomas, we chose to focus on validated as opposed to putative targets, especially since the currently validated targets for these miRNAs (CDKN1B and CDKN1C) have been previously implicated as tumor suppressor genes. Recently, CDKN1C has been identified as a regulatory target of miR-221 in hepatocellular carcinomas (Fornari et al., 2008). We found that ovarian carcinomas with CDKN1C expression in less than 10% of cells had statistically higher miR-221 and miR-222 expression than those with greater CDKN1C expression (Fig. 4). These data suggest that the upregulation of these miRNAs in ovarian carcinoma may promote proliferation by decreasing expression of the cell cycle inhibitor CDKN1C. To date, the role of CDKN1C in ovarian carcinomas has not been studied extensively. One previous study also demonstrated loss of CDKN1C protein was frequent, but as in our study, CDKN1C expression was not correlated with overall survival (Khouja et al., 2007).
miR-221 and miR-222 have also been verified as regulators of CDKN1B. Higher levels of these miRNAs correlate with lower levels of CDKN1B protein in glioblastomas and human hepatocellular carcinoma (Gillies and Lorimer, 2007; Fornari et al., 2008). CDKN1B is an important cell cycle inhibitor and tumor suppressor, and deregulated expression of miR-221 and miR-222 may promote malignant proliferation (le Sage et al., 2007). CDKN1B expression is frequently decreased in ovarian carcinoma and loss of CDKN1B has been shown to be a negative prognostic factor in some studies (Baekelandt et al., 1999; Masciullo et al., 1999; Sui et al., 1999). We found CDKN1B protein was frequently decreased in ovarian carcinomas, consistent with previous studies. However, unlike CDKN1C, CDKN1B protein expression was not correlated to miR-221 or miR-222 expression and was not associated with overall survival in our sample set.
Some studies have identified CpG island hypermethylation as a mechanism of miRNA deregulation in cancer, and aberrant hypermethylation of critical regions of miRNAs may play a role in metastasis (Lujambio et al., 2008). We evaluated methylation of the CpG island in the promoter of the EST that overlaps with miR-221 and miR-222. Methylation of this region was common in ovarian carcinomas and in normal lymphocytes, but was not clearly associated with relative expression levels of these miRNAs. Corcoran et al. (2009) recently identified promoter regions of miRNAs using a ChIP-chip approach to identify Pol II binding of regions up to 50 kb upstream of known miRNAs. These investigators identified a putative promoter region for miR-221 and miR-222 about 13 kb upstream. Our target CpG island is greater than 50 kb upstream of miR-221 and miR-222 and was consequently not evaluated in that study (Fig. 1). That CpG island could still have regulatory importance given its location in the promoter region of the one transcript known to overlap with these two miRNAs, and our methylation assay may not have assessed the critical CpG dinucleotides. However, it seems more likely that this CpG island is not relevant to the regulation of miR-221 and miR-222.
Somatic alterations of miR-221 and miR-222 were infrequent in ovarian carcinoma, but we did identify one carcinoma with a focal deletion of miR-222. We also identified three cases with heterozygosity at a germline SNP just upstream of miR-222 and within the pri-miRNA. However, the functional significance of this SNP is not known.
In summary, miRNA-221 and miRNA-222 may be important in regulating CDKN1C in ovarian carcinomas. The ratio of these co-expressed miRNAs is strongly associated with overall survival in women with advanced ovarian carcinoma.
Acknowledgments
We thank William Grady MD, Piri Welsch PhD, and Mary-Claire King PhD for technical advice and assistance.
Supported by a gift from the Yvonne Betson Trust. M. Tewari acknowledges support from the V Foundation (Scholar Grant), the FHCRC Molecular Diagnostics Pilot Project Program, the Pacific Ovarian Cancer Research Consortium (grant number P50 CA83636, PI: Nicole Urban) and from the Listwin Family Foundation and FHCRC (New Development funds).
References
- Altuvia Y, Landgraf P, Lithwick G, Elefant N, Pfeffer S, Aravin A, Brownstein MJ, Tuschl T, Margalit H. Clustering and conservation patterns of human microRNAs. Nucleic Acids Res. 2005;33:2697–2706. doi: 10.1093/nar/gki567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baekelandt M, Holm R, Trope CG, Nesland JM, Kristensen GB. Lack of independent prognostic significance of p21 and p27 expression in advanced ovarian cancer: an immunohistochemical study. Clin Cancer Res. 1999;5:2848–2853. [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahiya N, Sherman-Baust CA, Wang TL, Davidson B, Shih Ie M, Zhang Y, Wood W, 3rd, Becker KG, Morin PJ. MicroRNA expression and identification of putative miRNA targets in ovarian cancer. PLoS ONE. 2008;3:e2436. doi: 10.1371/journal.pone.0002436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornari F, Gramantieri L, Ferracin M, Veronese A, Sabbioni S, Calin GA, Grazi GL, Giovannini C, Croce CM, Bolondi L, Negrini M. MiR-221 controls CDKN1C/p57 and CDKN1B/p27 expression in human hepatocellular carcinoma. Oncogene. 2008;27:5651–5661. doi: 10.1038/onc.2008.178. [DOI] [PubMed] [Google Scholar]
- Garzon R, Fabbri M, Cimmino A, Calin GA, Croce CM. MicroRNA expression and function in cancer. Trends Mol Med. 2006;12:580–587. doi: 10.1016/j.molmed.2006.10.006. [DOI] [PubMed] [Google Scholar]
- Gillies JK, Lorimer IA. Regulation of p27Kip1 by miRNA 221/222 in glioblastoma. Cell Cycle. 2007;6:2005–2009. doi: 10.4161/cc.6.16.4526. [DOI] [PubMed] [Google Scholar]
- Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, Calin GA, Menard S, Croce CM. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- Khouja MH, Baekelandt M, Nesland JM, Holm R. The clinical importance of Ki-67, p16, p14, and p57 expression in patients with advanced ovarian carcinoma. Int J Gynecol Pathol. 2007;26:418–425. doi: 10.1097/pgp.0b013e31804216a0. [DOI] [PubMed] [Google Scholar]
- Larson PS, Schlechter BL, King CL, Yang Q, Glass CN, Mack C, Pistey R, de Las Morenas A, Rosenberg CL. CDKN1C/p57kip2 is a candidate tumor suppressor gene in human breast cancer. BMC Cancer. 2008;8:68. doi: 10.1186/1471-2407-8-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- le Sage C, Nagel R, Egan DA, Schrier M, Mesman E, Mangiola A, Anile C, Maira G, Mercatelli N, Ciafre SA, Farace MG, Agami R. Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. Embo J. 2007;26:3699–3708. doi: 10.1038/sj.emboj.7601790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lujambio A, Calin GA, Villanueva A, Ropero S, Sanchez-Cespedes M, Blanco D, Montuenga LM, Rossi S, Nicoloso MS, Faller WJ, Gallagher WM, Eccles SA, Croce CM, Esteller M. A microRNA DNA methylation signature for human cancer metastasis. Proc Natl Acad Sci U S A. 2008;105:13556–13561. doi: 10.1073/pnas.0803055105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masciullo V, Sgambato A, Pacilio C, Pucci B, Ferrandina G, Palazzo J, Carbone A, Cittadini A, Mancuso S, Scambia G, Giordano A. Frequent loss of expression of the cyclin-dependent kinase inhibitor p27 in epithelial ovarian cancer. Cancer Res. 1999;59:3790–3794. [PubMed] [Google Scholar]
- Medina R, Zaidi SK, Liu CG, Stein JL, van Wijnen AJ, Croce CM, Stein GS. MicroRNAs 221 and 222 bypass quiescence and compromise cell survival. Cancer Res. 2008;68:2773–2780. doi: 10.1158/0008-5472.CAN-07-6754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merritt WM, Lin YG, Han LY, Kamat AA, Spannuth WA, Schmandt R, Urbauer D, Pennacchio LA, Cheng JF, Nick AM, Deavers MT, Mourad-Zeidan A, Wang H, Mueller P, Lenburg ME, Gray JW, Mok S, Birrer MJ, Lopez-Berestein G, Coleman RL, Bar-Eli M, Sood AK. Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med. 2008;359:2641–2650. doi: 10.1056/NEJMoa0803785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg E, Demopoulos RI, Zeleniuch-Jacquotte A, Yee H, Sorich J, Speyer JL, Newcomb EW. Expression of cell cycle regulators p57(KIP2), cyclin D1, and cyclin E in epithelial ovarian tumors and survival. Hum Pathol. 2001;32:808–813. doi: 10.1053/hupa.2001.26462. [DOI] [PubMed] [Google Scholar]
- Sui L, Dong Y, Ohno M, Watanabe Y, Sugimoto K, Tokuda M. Expression of p57kip2 and its clinical relevance in epithelial ovarian tumors. Anticancer Res. 2002;22:3191–3196. [PubMed] [Google Scholar]
- Sui L, Tokuda M, Ohno M, Hatase O, Hando T. The concurrent expression of p27(kip1) and cyclin D1 in epithelial ovarian tumors. Gynecol Oncol. 1999;73:202–209. doi: 10.1006/gyno.1999.5373. [DOI] [PubMed] [Google Scholar]
- Wyman SK, Parkin RK, Mitchell PS, Fritz BR, O’Briant K, Godwin AK, Urban N, Drescher CW, Knudsen BS, Tewari M. Repertoire of microRNAs in epithelial ovarian cancer as determined by next generation sequencing of small RNA cDNA libraries. PLoS ONE. 2009;4:e5311. doi: 10.1371/journal.pone.0005311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Huang J, Yang N, Greshock J, Megraw MS, Giannakakis A, Liang S, Naylor TL, Barchetti A, Ward MR, Yao G, Medina A, O’Brien-Jenkins A, Katsaros D, Hatzigeorgiou A, Gimotty PA, Weber BL, Coukos G. microRNAs exhibit high frequency genomic alterations in human cancer. Proc Natl Acad Sci U S A. 2006;103:9136–9141. doi: 10.1073/pnas.0508889103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Volinia S, Bonome T, Calin GA, Greshock J, Yang N, Liu CG, Giannakakis A, Alexiou P, Hasegawa K, Johnstone CN, Megraw MS, Adams S, Lassus H, Huang J, Kaur S, Liang S, Sethupathy P, Leminen A, Simossis VA, Sandaltzopoulos R, Naomoto Y, Katsaros D, Gimotty PA, DeMichele A, Huang Q, Butzow R, Rustgi AK, Weber BL, Birrer MJ, Hatzigeorgiou AG, Croce CM, Coukos G. Genomic and epigenetic alterations deregulate microRNA expression in human epithelial ovarian cancer. Proc Natl Acad Sci U S A. 2008;105:7004–7009. doi: 10.1073/pnas.0801615105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorn KK, Jazaeri AA, Awtrey CS, Gardner GJ, Mok SC, Boyd J, Birrer MJ. Choice of normal ovarian control influences determination of differentially expressed genes in ovarian cancer expression profiling studies. Clin Cancer Res. 2003;9:4811–4818. [PubMed] [Google Scholar]