Abstract
Background: Alterations in plasma fatty acid distribution are linked to metabolic abnormalities related to type 2 diabetes and cardiovascular disease.
Objective: The aim of this study was to investigate genetic factors influencing plasma fatty acid distribution in Alaskan Eskimos from the Genetics of Coronary Artery Disease in Alaska Natives (GOCADAN) study.
Design: Fatty acids in plasma were measured by gas chromatography in 761 related individuals (>35 y of age).
Results: Quantitative genetic analyses showed that fatty acid distribution is significantly heritable (P < 0.001), with heritabilities ranging from 0.33 to 0.55. A genome-wide scan for plasma fatty acids identified a 20-cM region on chromosome 8 (p12–p21) with a quantitative trait locus for monounsaturated fatty acids (logarithm of odds score = 3.8). The same region had a quantitative trait locus for polyunsaturated fatty acids (logarithm of odds score = 2.6). We genotyped single nucleotide polymorphisms (SNPs) in candidate genes in 8p12–p21 and found a significant association between fatty acids and SNPs in apolipoprotein J (APOJ), lipoprotein lipase (LPL), macrophage scavenger receptor 1 (MSR1), and tumor necrosis factor receptor superfamily member 10b (TNFRSF10B). A Bayesian quantitative trait nucleotide analysis based on a measured genotype model showed that SNPs in LPL, TNFRSF10B, and APOJ had strong statistical evidence of a functional effect (posterior probability ≥75%) on plasma fatty acid distribution.
Conclusions: The results indicate that there is strong genetic influence on plasma fatty acid distribution and that genetic variation in APOJ, LPL, and TNFRSF10B may play a role. The GOCADAN study was registered at www.clinicaltrials.gov as NCT00006192.
INTRODUCTION
Plasma fatty acid (FA) concentration and distribution have been closely linked to type 2 diabetes mellitus (T2DM) and cardiovascular disease (CVD) risk. Changes in plasma FA concentrations and distribution depend on the utilization and release of FAs by tissues such as the liver, adipose tissue, and muscle (1). Increases in plasma FAs are associated with the activation of the sympathetic nervous system, which results in an increased risk of CVD (2). Elevated circulating FAs have been reported to affect several tissues (1). Although there is less information available on FA distribution, plasma long-chain polyunsaturated FAs (PUFAs) have been associated with the risk of atherosclerosis, with one of its family (ie, n−3) being linked to reduced risk, whereas another member of its family (ie, n−6) is related to an increased risk of atherosclerosis (3). In a case-control study of men, with and without incident coronary artery disease, the plasma saturated FA (SFA) palmitate was associated with an increased risk of coronary heart disease, whereas n−3 PUFAs were inversely correlated with coronary heart disease (4). n−3 FAs were also associated with a reduced risk of diabetes (5). Thus, it is likely that the concentration and distribution of FAs in plasma may play a key role in influencing the development and progression of T2DM and CVD. Both genetic and environmental factors are likely to affect the distribution of FAs. A recently published genome-wide association study (GWAS) of related individuals found the FA desaturase 1 (FADS1) gene to be a major contributor to the variation observed in plasma PUFAs (6). Although, to date, this is the only study to have investigated the genetic influence on plasma FAs in related individuals, its primary focus was only plasma PUFAs and not all FAs. The population of the current study is Alaska Natives from the Norton Sound region of Alaska who are participating in the Genetics of Coronary Artery Disease in Alaska Natives (GOCADAN) study. The GOCADAN study is focused on investigating the relative contribution of genetic and environmental factors to CVD in Alaska Natives. Traditionally, Alaska Natives have had a low CVD mortality (7). This was assumed to be due to their traditional diet, which was rich in n−3 FAs (8). However, increased Western acculturation has not only changed their dietary habits but has also led to decreased activity levels. Between 1994 and 1998, death rates due to CVD were 30–40% higher in Alaska Natives (aged 25–54 y) than in whites in the United States (9). Similarly, the death rate due to stroke is also higher among Alaska Natives than their US white counterparts (9). Given the high CVD risk in this population along with the changing dietary habits (particularly with regard to the type of fat consumed) and the multiple metabolic consequences of increased plasma FAs, we investigated whether there was any evidence of genetic influence on the variation in plasma FAs in this population.
SUBJECTS AND METHODS
Study design
In the GOCADAN study, 1214 individuals (all 18 y of age and older) were recruited from villages in the Norton Sound region on the northwestern coast of Alaska (see Supplemental Figure 1 under ldquoSupplemental datardquo in the online issue). FA data were available for 761 individuals. All individuals had a baseline examination. Diet, physical activity, and medical history were recorded by using a set of standardized interview instruments. Participants visited the clinics for a fasting blood draw. Blood was drawn by venipuncture, and samples were stored in aliquots at −80 C for phenotypic analysis and DNA extraction. Physical examinations were performed along with electrocardiograms and carotid artery scans. Details of the study recruitment, designs, and methods have been reported by Howard et al (10) and Ebbesson et al (11). The Institutional Review Boards from all participating institutions approved this study, and informed consent was obtained from all participants.
Demographic and phenotypic data
Demographic and genealogic data collected during the interviews included name, sex, date, place of birth, current home of the participant and his or her spouse, and first-degree relatives of all household members. Anthropometric measurements, height, weight, and waist and hip circumferences were measured by standardized procedures. Body mass index (BMI) was calculated by dividing weight (in kg) by height squared (in m). Body fat was measured by using a Quantum II bioelectrical body-composition analyzer (RJL Systems, Clinton Township, MI). Total, HDL, and LDL cholesterol and triglycerides were measured by using an auto analyzer (model 717; Hitachi, Amposta, Spain).
Total plasma FA concentrations were estimated in 761 individuals following the method previously described by Ebbesson et al (8). In short, analysis of the composition of total plasma FAs was performed on a Hewlett-Packard 5890 gas chromatograph equipped with a 30 nm × 0.32 mm SP2330 capillary column. Each FA was computed by using a regression equation in which the ratio of the area of each FA peak to the internal standard was plotted against the weight ratio of the FA and the internal standard. All FAs are expressed as percentages (mol/100 mL total FAs).
Genotypic data
DNA was isolated from buffy coats by using organic solvents. For each of the participants, 400 short tandem repeat markers (spaced at an average interval of 10 cM throughout the genome) were amplified in separate polymerase chain reactions (PCRs) by using fluorescently labeled primer pairs (ABIPRISM Linkage Mapping Set MD 10 version 2; Applied Biosystems, Foster City, CA). Pedigree and Mendelian errors were detected and corrected by using the software PREST (pedigree relation statistical tests) and SIMWALK2 (12). Multipoint identity-by-descent matrices for genome-wide linkage analyses were calculated by using the linkage analysis package (LOKI) (13). The chromosomal map used in these computations was based on marker locations reported by DeCode genetics (14).
Single nucleotide polymorphism genotyping
Single nucleotide polymorphisms (SNPs) were typed by using the multiplex VeraCode technology from Illumina according to the manufacturer's protocol (Illumina, San Diego, CA). Briefly, the technology is based on allele-specific primer extension. Genomic DNA (250 ng) was activated chemically with biotin and then hybridized to a pool of locus-specific oligos (OPA, Oligo Pool All; Illumina). After removal of nonspecific unbound oligos, a PCR reaction was performed by using fluorescent-labeled primers (Cy3 and Cy5). All steps through the PCR were performed on a Tecan Freedom EVO 150cm liquid handler (Tecan, Männedorf, Switzerland) with Illumina GTS Robot Control software (Illumina). PCR products were cleaned and denatured, and single-stranded fluorescent-labeled DNAs were hybridized to VeraCode beads, which were scanned on a BeadXpress reader by using VeraScan software (Illumina). Raw data, consisting of intensities of fluorescence, were then imported into the analysis software BeadStudio (Illumina). Cluster calls were checked for accuracy, and genotypes were exported as text files for further use in association analysis. Replica samples were included as controls for genotyping and allele calling consistencies. Furthermore, Illumina has included sample-dependent and -independent controls to test for the accuracy of the procedure. We typed 239 SNPs from 5 candidate genes: apolipoprotein J (APOJ), β-3-adrenergic receptor (ADRB3), lipoprotein lipase (LPL), tumor necrosis factor superfamily, member 10B (TNFRSF10B), and macrophage scavenger receptor 1 (MSR1). These SNPs were selected on the basis of being previously reported to be polymorphic in both Asian and European populations.
Statistical analyses
Univariate genetic analysis
A variance components decomposition method was used to estimate heritability and linkage to chromosomal locations affecting variation in plasma FA distribution via genome-wide linkage analysis. This method is implemented in the software program Sequential Oligogenic Linkage Analysis Routines (SOLAR; Southwest Foundation for Biomedical Research, San Antonio, TX; available from http://www.sfbr.org/departments/genetics_detail.aspx?p=37) and has been described in detail elsewhere (15). Before conducting genetic analyses, distributional properties of all traits were evaluated. All values beyond 4 SDs were removed, and the remaining traits were transformed by inverse normalization before analysis, to meet assumptions of normality. For the quantitative genetic analysis (linkage and SNP association) we used SOLAR version 4.0 to analyze the data (15).
Bivariate genetic analysis
Phenotypic, genetic, and environmental correlations were calculated between plasma FAs and other adiposity-related traits as summarized by the following model:
where h12 and h22 are heritabilities of the 2 phenotypes being studied, and rhoG and rhoE are the additive genetic and environmental correlations between the traits, respectively (16).
A model in which all parameters are estimated was compared with a model in which the genetic correlation is constrained to zero. To test for complete pleiotropy between the 2 traits, a model in which the genetic correlation was constrained to one was compared with a model in which all parameters are estimated. Twice the difference of logarithm likelihood of the 2 models asymptotically yields a distribution of chi square with 1 df (17). Evidence of pleiotropy (a common set of genes influencing more than one trait) was indicated by a genetic correlation significantly different from 0.
Measured genotype analysis
Genotype frequencies for each SNP were calculated by using all individuals (18) and were tested for departures from Hardy-Weinberg equilibrium. Estimates of linkage disequilibrium (LD) between SNPs were determined by calculating pairwise D′ and r2 statistics. As a first step in investigating the association between the SNPs in candidate genes and circulating FAs, we used a measured genotype analysis (19), as implemented in SOLAR. This approach extends the classic variance components-based biometrical model to account for both the random effects of kinship and the main effects of SNP genotypes. For each SNP, we compared this saturated model with a null model in which the main effect of the SNP is constrained to zero. The test statistic, twice the difference in loge(likelihood) between the saturated model and the SNP-specific null, is distributed as a chi-square with 1 df.
Measured genotype analysis was conducted on each SNP to calculate a marginal P value. To compute a gene-based association (20), a gene-centric P value correction was used that utilizes the effective number of SNPs per defined gene region, allows for nonindependence among family members, and is therefore better able to determine significance under multiple testing (21). We calculated the effective number of SNPs (ie, informative markers) by using the method of Moskvina and Schmidt (22). The marginal P value and the effective number of SNPs was used to determine an adjusted P value to be used in multiple testing (21).
Bayesian quantitative trait nucleotide analysis
The measured genotype analysis described above was used to assess association between the trait and each SNP, analyzing one SNP at a time. However, multiple functional variants will often exist within a chromosomal locus, and joint analysis of multiple variants may be more powerful to detect their effects and to establish which of the many polymorphisms within a region are the most likely to be functional. Blangero et al (18) developed a Bayesian quantitative trait nucleotide (BQTN) approach, which is essentially a Bayesian method based on an underlying measured genotype model that permits joint analysis of multiple variants. The BQTN analysis was implemented in SOLAR (15) to statistically identify the most likely functional SNPs associated with a phenotype. This method has been described in detail elsewhere (18). In short, Bayesian model selection identifies the set of variants that optimally predicts the phenotype. In a Bayesian framework, 2 competing hypotheses (models) are compared by evaluation of the Bayes factor, which is the ratio of the integrated likelihoods of the competing models (23). The Bayesian information criterion estimates whether the QTN model explains sufficient variation in the phenotype to justify the number of parameters included and is used in reference to a null model. A Bayesian information criterion difference >2 units provides support for the estimated model over the null model with a posterior probability >75%, and a Bayesian information criterion difference >6 units between the 2 models represent strong support for the estimated model with a posterior probability >99% (18).The BQTN approach also accounts for model uncertainty and thus provides the posterior probability that each variant is associated with the phenotype of interest. This variant-specific posterior probability is a measure of the evidence that a particular variant is likely to be functional or in high linkage disequilibrium with a functional variant.
RESULTS
The mean and SDs of age, BMI, waist circumference, percentage body fat, triglycerides, and plasma FA concentrations are shown in Table 1. Women had higher values for BMI and percentage body fat as compared with men. There were no significant differences between sexes in age, waist circumference, and serum triglycerides. With respect to FAs, women had significantly higher percentages of plasma SFAs, whereas men had higher percentages of PUFAs. However, monounsaturated FAs (MUFAs) were not significantly different between men and women (Table 1).
TABLE 1.
Sex-specific characteristics and plasma fatty acids (n = 761)1
| Phenotype | All | Women | Men | P value2 |
| Age (y) | 42.5 ± 16.0 | 42.9 ± 16.2 | 42.0 ± 15.7 | 0.33 |
| BMI (kg/m2) | 27.62 ± 5.7 | 28.51 ± 6.2 | 26.47 ± 4.9 | 0.0001 |
| Waist circumference (cm) | 87.91 ± 13.2 | 88.32 ± 14.0 | 87.40 ± 12.4 | 0.24 |
| Percentage body fat (%) | 38.11 ± 8.4 | 43.25 ± 5.7 | 31.51 ± 6.5 | 0.0001 |
| Triglycerides (mg/dL) | 124. 45 ± 67.2 | 125.09 ± 66.1 | 123.63 ± 68.7 | 0.72 |
| Total saturated fatty acids (%)3 | 37.52 ± 5.4 | 38.22 ± 5.5 | 36.66 ± 5.3 | 0.0001 |
| Total monounsaturated fatty acids (%)3 | 27.20 ± 5.2 | 27.22 ± 5.3 | 27.19 ± 8.4 | 0.93 |
| Total polyunsaturated fatty acids (%)3 | 33.83 ± 8.8 | 32.80 ± 9.5 | 35.15 ± 7.7 | 0.0001 |
All values are means ± SDs.
SOLAR version 4.0 (Southwest Foundation for Biomedical Research, San Antonio, TX) was used to identify the significant differences between men and women (15).
Percentages of total fatty acids.
Estimation of heritability
To assess the additive genetic component of the variation in plasma FAs, we conducted a variance-component based heritability analysis. We used age, sex, their higher order terms, and interactions as covariates, irrespective of their significance. In another model we used dietary FAs as an additional covariate along with age, sex, their higher order terms, and interactions. Heritabilities were significant for total FAs as well as PUFAs, MUFAs, and SFAs (Table 2) for both models. The heritabilities ranged from 0.35 to 0.55. We also carried out heritability analyses for all fractions of plasma FAs and found all of them to be significantly heritable (data not shown).
TABLE 2.
Heritabilities and genome-wide linkage scan results for plasma fatty acid distribution in Alaska Natives (n = 761)1
| Fatty acids | h2 ± SE | P value | LOD score | Chromosome | Location (cM) | Nearest marker |
| SFAs2 | 0.38 ± 0.11 | 2.8 × 10minus5 | 1.68 | 12 | 146 | D12S79 |
| SFAs3 | 0.35 ± 0.11 | 2.0 × 10minus4 | 1.44 | 2 | 95 | D2S2368 |
| MUFAs2 | 0.48 ± 0.12 | 1.5 × 10minus6 | 3.82 | 8 | 50 | Between D8S1771 and D8S505 |
| MUFAs3 | 0.46 ± 0.12 | 9.0 × 10minus6 | 3.57 | 8 | 59 | Between D8S1771 and D8S505 |
| PUFAs2 | 0.42 ± 0.12 | 5.2 × 10minus5 | 2.93 | 6 | 170 | Between D6S308 and D6S441 |
| PUFAs3 | 0.40 ± 0.12 | 2.2 × 10minus4 | 2.88 | 6 | 170 | Between D6S308 and D6S441 |
| TFAs2 | 0.55 ± 0.12 | 1.4 × 10minus6 | 1.94 | 10 | 97 | D10S537 |
| TFAs3 | 0.53 ± 0.12 | 4.8 × 10minus6 | 2.51 | 10 | 97 | D10S537 |
SOLAR version 4.0 (Southwest Foundation for Biomedical Research, San Antonio, TX) was used to conduct the linkage analysis (15). SFAs, saturated fatty acids; h2, heritability; LOD, logarithm of odds; MUFAs, monounsaturated fatty acids; PUFAs, polyunsaturated fatty acids; TFAs, total fatty acids.
Adjusted for age, sex, and their higher order and interaction terms.
Adjusted for age, sex, and their interaction terms and dietary fatty acids.
Localization of quantitative trait loci through genome-wide scan
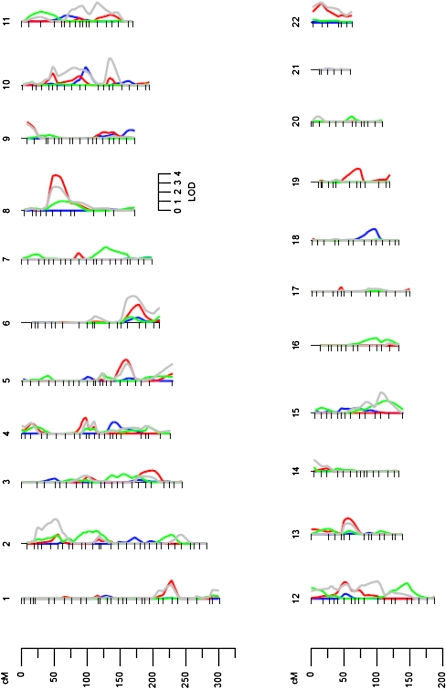
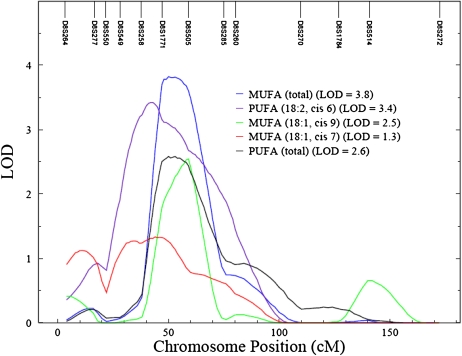
A total of 867 relative pairs are represented in this analysis. Of these, most were siblings (n = 194) followed by half-siblings (n = 192), parent-offsprings (n = 156), and half-first cousins (n = 125). Genome-wide scans for the 4 FA types are shown in Figure 1. The genome-wide scan for FAs showed several interesting chromosomal regions harboring quantitative trait loci (QTLs), with the evidence of suggesting significant linkage (Table 2). The most significant linkage was found for total MUFAs on chromosome 8p12–p21 between markers D8S1771 and D8S505 [logarithm of odds (LOD) score = 3.8]. The one-LOD support interval for this QTL spanned ≈20 cM (44–64 cM). This region of chromosome 8 showed evidence of suggestive linkage not only for the sum of all PUFAs and MUFAs but also for fractions of PUFAs and MUFAs such as PUFAs (18:2n−6) (LOD = 3.4) and MUFAs (18:1n−9) (LOD = 2.5) (Figure 2). Evidence of suggestive linkage (LOD > 2) was found on chromosome 6q for total PUFAs. For the sum of all measured FAs, suggestive linkage was observed on chromosome 10q (Table 2). Circulating FAs are partly reflective of dietary FA intake (8). To adjust for that, we conducted genetic analyses using dietary FAs as an additional covariate. Linkage signals did not differ significantly, except for SFAs. This suggests that these signals are specific for plasma FAs and are not affected by dietary intakes.
FIGURE 1.
Genome-wide results by each chromosome. Chromosomal location (cM) is represented on the x axis, and the logarithm of odds (LOD) score is shown on the y axis. Blue represents total fatty acids, green represents saturated fatty acids, gray represents polyunsaturated fatty acids, and red represents monounsaturated fatty acids. SOLAR version 4.0 (Southwest Foundation for Biomedical Research, San Antonio, TX) was used to conduct the linkage analysis (15).
FIGURE 2.
Evidence of significant linkage for plasma unsaturated fatty acids on chromosome 8. Chromosomal location (cM) is represented on the x axis, and logarithm of odds (LOD) score is shown on the y axis. SOLAR version 4.0 (Southwest Foundation for Biomedical Research, San Antonio, TX) was used to conduct the linkage analysis (15). MUFA, monounsaturated fatty acid; PUFA, polyunsaturated fatty acid.
Bivariate genetic analysis
Bivariate genetic analysis showed significant genetic correlations between plasma FAs and adiposity-related traits (Table 3). The SFA myristic acid (C-14) was positively correlated with BMI, percentage fat, waist circumference, 2-h insulin, and glucose; triglycerides and palmitic acid (C-16) were correlated with 2-h glucose and triglycerides, and the long-chain SFA stearic acid was associated negatively with the same parameters. MUFAs, in general, showed a negative correlation with fasting glucose, triglycerides, and glycated hemoglobin. Total and LDL cholesterol were positively correlated with total FAs or the sum of FAs (genetic and phenotypic).
TABLE 3.
Significant genetic correlations between circulating fatty acids and anthropometric and biochemical markers (n = 761)1
| Trait 1 | Trait 2 | rhoG ± SE | rhoP ± SE |
| Myristic acid | BMI | 0.552 ± 0.172 | 0.257 ± 0.043 |
| Waist circumference | 0.433 ± 0.172 | 0.241 ± 0.043 | |
| Percentage fat | 0.601 ± 0.172 | 0.260 ± 0.043 | |
| Insulin, 2 h | 0.690 ± 0.263 | 0.244 ± 0.053 | |
| HDL cholesterol | −0.36 ± 0.153 | −0.350 ± 0.032 | |
| Triglycerides | 0.732 ± 0.132 | 0.563 ± 0.032 | |
| Palmitic acid | Glucose, 2 h | 0.565 ± 0.233 | 0.193 ± 0.04 |
| Triglycerides | 0.329 ± 0.133 | 0.362 ± 0.032 | |
| Stearic acid | Percentage fat | −0.357 ± 0.143 | −0.165 ± 0.04 |
| Fasting glucose | −0.39 ± 0.193 | −0.123 ± 0.04 | |
| HDL cholesterol | 0.41 ± 0.173 | 0.124 ± 0.04 | |
| Insulin, 2 h | −0.543 ± 0.253 | −0.156 ± 0.05 | |
| Triglycerides | −0.57 ± 0.122 | −0.316 ± 0.042 | |
| MUFAs, 18:1n−9 | Fasting glucose | −0.387 ± 0.183 | −0.057 ± 0.05 |
| Triglycerides | −0.399 ± 0.163 | −0.154 ± 0.05 | |
| PUFAs, 20:3n−6 | LDL cholesterol | −0.526 ± 0.252 | −0.259 ± 0.053 |
| Total fatty acids | LDL cholesterol | 0.51 ± 0.152 | 0.141 ± 0.04 |
| Total cholesterol | 0.418 ± 0.153 | 0.131 ± 0.04 | |
| Total MUFAs | Fasting glucose | −0.455 ± 0.183 | −0.025 ± 0.04 |
| Glycated hemoglobin | −0.311 ± 0.153 | −0.119 ± 0.04 | |
| Total PUFAs | Fasting glucose | 0.495 ± 0.203 | 0.029 ± 0.04 |
SOLAR version 4.0 (Southwest Foundation for Biomedical Research, San Antonio, TX) was used to conduct the bivariate analysis (15). MUFAs, monounsaturated fatty acids; PUFAs, polyunsaturated fatty acids; rhoG, genetic correlations; rhoP, phenotypic correlations.
P < 0.005.
P < 0.05.
Measured genotype analysis
For association analyses, 231 of 239 SNPs were used after accounting for departures from the Hardy-Weinberg equilibrium. All these SNPs were tested for an association with traits that attained their strongest evidence for linkage on chromosome 8 as well as with adiposity-related traits. Polymorphisms in APOJ showed an association with total FAs and MUFAs. Similarly, significant associations were also observed with polymorphisms in TNFRSF10B, LPL, and MSR1. However, no association was found between plasma FAs and ADRB3 variants (Table 4). Some polymorphisms in the candidate genes were also associated with BMI, percentage fat, waist circumference, and plasma triglycerides. All results are shown uncorrected for multiple testing (P < 0.05).
TABLE 4.
P values for the association of polymorphisms in candidate genes on chromosome 8 and plasma fatty acid distribution and adiposity-related phenotypes (n = 761)1
| Gene | SNP | TFAs | MUFAs | PUFAs | SFAs | 18:1n–7 | 18:1n−9 | 18:2n−6 | BMI | % Fat | Waist | Triglycerides |
| ADRB3 | rs35361594 | 0.226 | 0.267 | 0.371 | 0.767 | 0.304 | 0.413 | 0.206 | 0.025 | 0.009 | 0.043 | 0.545 |
| APOJ | rs10503814 | 0.173 | 0.028 | 0.017 | 0.619 | 0.727 | 0.381 | 0.037 | 0.414 | 0.165 | 0.799 | 0.742 |
| rs11136000 | 0.604 | 0.364 | 0.802 | 0.382 | 0.509 | 0.611 | 0.368 | 0.002 | 0.005 | 0.003 | 0.04 | |
| rs1982229 | 0.992 | 0.409 | 0.742 | 0.916 | 0.097 | 0.041 | 0.388 | 0.252 | 0.555 | 0.126 | 0.135 | |
| rs538181 | 0.033 | 0.076 | 0.274 | 0.584 | 0.197 | 0.841 | 0.279 | 0.161 | 0.065 | 0.600 | 0.784 | |
| rs569205 | 0.032 | 0.110 | 0.263 | 0.505 | 0.512 | 0.910 | 0.385 | 0.095 | 0.022 | 0.390 | 0.859 | |
| rs7812347 | 0.874 | 0.016 | 0.722 | 0.657 | 0.080 | 0.024 | 0.883 | 0.128 | 0.032 | 0.057 | 0.502 | |
| rs9331891 | 0.008 | 0.188 | 0.057 | 0.331 | 0.295 | 0.218 | 0.230 | 0.780 | 0.688 | 0.986 | 0.073 | |
| LPL | rs1059611 | 0.172 | 0.086 | 0.828 | 0.975 | 0.376 | 0.994 | 0.041 | 0.718 | 0.517 | 0.253 | 6.4 × 10minus6 |
| rs1121923 | 0.380 | 0.199 | 0.467 | 0.565 | 0.863 | 0.615 | 0.177 | 0.024 | 0.011 | 0.034 | 0.636 | |
| rs13702 | 0.745 | 0.023 | 0.558 | 0.565 | 0.166 | 0.501 | 0.032 | 0.861 | 0.575 | 0.394 | 6.9 × 10minus5 | |
| rs1534649 | 0.398 | 0.144 | 0.546 | 0.331 | 0.807 | 0.674 | 0.017 | 0.605 | 0.593 | 0.693 | 3.3 × 10minus4 | |
| rs17091738 | 0.041 | 0.519 | 0.164 | 0.493 | 0.674 | 0.355 | 0.154 | 0.624 | 0.637 | 0.607 | 0.741 | |
| rs1801177 | 0.041 | 0.521 | 0.165 | 0.494 | 0.676 | 0.357 | 0.271 | 0.627 | 0.639 | 0.609 | 0.745 | |
| rs248 | 0.921 | 0.022 | 0.033 | 0.417 | 0.154 | 0.145 | 0.041 | 0.492 | 0.962 | 0.430 | 0.347 | |
| rs249 | 0.095 | 0.130 | 0.482 | 0.235 | 0.822 | 0.651 | 0.054 | 0.652 | 0.620 | 0.885 | 2.8 × 10minus4 | |
| rs255 | 0.366 | 0.293 | 0.558 | 0.293 | 0.904 | 0.668 | 0.047 | 0.998 | 0.918 | 0.492 | 0.005 | |
| rs256 | 0.331 | 0.272 | 0.621 | 0.319 | 0.932 | 0.735 | 0.039 | 0.967 | 0.877 | 0.460 | 0.003 | |
| rs263 | 0.522 | 0.305 | 0.458 | 0.157 | 0.867 | 0.583 | 0.055 | 0.827 | 0.842 | 0.645 | 0.003 | |
| rs285 | 0.091 | 0.017 | 0.395 | 0.705 | 0.053 | 0.374 | 0.006 | 0.722 | 0.664 | 0.796 | 0.006 | |
| rs328 | 0.140 | 0.076 | 0.756 | 0.934 | 0.357 | 0.979 | 0.031 | 0.749 | 0.524 | 0.263 | 4.5 × 10minus6 | |
| rs3735964 | 0.141 | 0.080 | 0.744 | 0.912 | 0.347 | 0.991 | 0.026 | 0.796 | 0.592 | 0.315 | 3.6 × 10minus6 | |
| rs3916027 | 0.654 | 0.017 | 0.394 | 0.842 | 0.139 | 0.476 | 0.010 | 0.832 | 0.496 | 0.319 | 9.8 × 10minus5 | |
| rs8176337 | 0.309 | 0.087 | 0.824 | 0.293 | 0.984 | 0.674 | 0.013 | 0.740 | 0.682 | 0.989 | 3.4 × 10minus4 | |
| MSR1 | rs10503573 | 0.609 | 0.546 | 0.509 | 0.033 | 0.308 | 0.780 | 0.302 | 0.243 | 0.080 | 0.045 | 0.236 |
| rs10503574 | 0.872 | 0.079 | 0.049 | 0.465 | 0.695 | 0.028 | 0.546 | 0.159 | 0.141 | 0.058 | 0.721 | |
| rs10503575 | 0.366 | 0.155 | 0.311 | 0.965 | 0.843 | 0.304 | 0.401 | 0.163 | 0.038 | 0.033 | 0.737 | |
| rs12114599 | 0.091 | 0.006 | 0.011 | 0.191 | 0.736 | 0.021 | 0.119 | 0.001 | 0.004 | 0.001 | 0.558 | |
| rs12216764 | 0.340 | 0.106 | 0.922 | 0.056 | 0.010 | 0.702 | 0.132 | 0.146 | 0.089 | 0.047 | 0.209 | |
| rs12675698 | 0.228 | 0.062 | 0.044 | 0.380 | 0.457 | 0.232 | 0.119 | 0.042 | 0.026 | 0.018 | 0.758 | |
| rs12678503 | 0.083 | 0.004 | 0.010 | 0.217 | 0.561 | 0.014 | 0.088 | 0.003 | 0.009 | 0.001 | 0.844 | |
| rs12680619 | 0.138 | 0.003 | 0.007 | 0.212 | 0.381 | 0.009 | 0.107 | 0.022 | 0.038 | 0.013 | 0.594 | |
| rs17620929 | 0.518 | 0.262 | 0.754 | 0.418 | 0.042 | 0.471 | 0.165 | 0.326 | 0.320 | 0.216 | 0.267 | |
| rs17677191 | 0.331 | 0.026 | 0.017 | 0.234 | 0.713 | 0.122 | 0.048 | 0.196 | 0.240 | 0.177 | 0.504 | |
| rs351554 | 0.310 | 0.122 | 0.954 | 0.067 | 0.014 | 0.772 | 0.127 | 0.114 | 0.062 | 0.041 | 0.225 | |
| rs918 | 0.158 | 0.081 | 0.056 | 0.387 | 0.610 | 0.276 | 0.103 | 0.031 | 0.035 | 0.019 | 0.802 | |
| TNFRSF10B | rs1001792 | 0.676 | 0.037 | 0.146 | 0.676 | 0.219 | 0.905 | 0.190 | 0.355 | 0.232 | 0.287 | 0.135 |
| rs10090860 | 0.845 | 0.160 | 0.750 | 0.467 | 0.917 | 0.035 | 0.443 | 0.334 | 0.173 | 0.193 | 0.234 | |
| rs10095594 | 0.815 | 0.953 | 0.616 | 0.818 | 0.592 | 0.200 | 0.498 | 0.127 | 0.025 | 0.130 | 0.577 | |
| rs1047266 | 0.727 | 0.761 | 0.371 | 0.298 | 0.744 | 0.151 | 0.031 | 0.618 | 0.984 | 0.593 | 0.083 | |
| rs10866818 | 0.012 | 0.026 | 0.750 | 0.233 | 0.101 | 0.013 | 0.989 | 0.665 | 0.757 | 0.646 | 0.730 | |
| rs1105944 | 0.006 | 0.041 | 0.992 | 0.164 | 0.180 | 0.024 | 0.701 | 0.672 | 0.688 | 0.602 | 0.601 | |
| rs12541497 | 0.026 | 0.159 | 0.286 | 0.155 | 0.632 | 0.751 | 0.214 | 0.875 | 0.897 | 0.280 | 0.717 | |
| rs12541697 | 0.003 | 0.036 | 0.801 | 0.086 | 0.115 | 0.013 | 0.371 | 0.932 | 0.778 | 0.849 | 0.671 | |
| rs35974498 | 0.989 | 0.901 | 0.439 | 0.411 | 0.002 | 0.704 | 0.755 | 0.064 | 0.125 | 0.557 | 0.145 | |
| rs4872046 | 0.155 | 0.277 | 0.443 | 0.824 | 0.035 | 0.459 | 0.804 | 0.488 | 0.565 | 0.649 | 0.485 | |
| rs6557612 | 0.071 | 0.130 | 0.271 | 0.456 | 0.191 | 0.040 | 0.717 | 0.624 | 9,464 | 0.774 | 0.630 | |
| rs7836280 | 0.606 | 0.038 | 0.156 | 0.693 | 0.181 | 0.915 | 0.194 | 0.373 | 0.250 | 0.324 | 0.155 |
SOLAR version 4.0 (Southwest Foundation for Biomedical Research, San Antonio, TX) was used to conduct the association analysis (15). MUFAs, monounsaturated fatty acids; PUFAs, polyunsaturated fatty acids; SNP, single nucleotide polymorphism; TFAs, total fatty acids; SFAs, saturated fatty acids. P values <0.05 indicate significance.
BQTN analysis
Because BQTN (18) analysis identifies the SNPs most likely to have a functional effect on the trait and is robust to multiple testing, the SNPs were next tested by BQTN analysis. In the current analyses, 115 SNPs were used after removing redundant SNPs that were in LD (>90%) with other SNPs. The variants in 3 genes (APOJ, LPL, and TNFRSF10B) had posterior probabilities ≥75%, which indicated significant support for a functional effect on plasma FAs. The SNP rs285 in LPL showed a posterior probability of 1, which indicated that this SNP is most likely to have a functional effect on the PUFA 18:2n−6. All SNPs with posterior probability of a functional effect ≥0.5 are shown in Table 5. Of all SNPs, rs328 in LPL accounted for the maximum proportion of variance in circulating triglycerides, followed by rs1059611 in LPL. Both of these SNPs were associated with lower concentrations of triglycerides. Polymorphisms in rs285 were associated with higher concentrations of the PUFA 18:2 n−6, whereas the 2 other SNPs with strong posterior probabilities, rs9331891 and rs12541697, were associated with lower concentrations of total FAs.
TABLE 5.
Bayesian quantitative trait nucleotide (BQTN) analysis of plasma fatty acid distribution and polymorphisms in candidate genes on chromosome 81
| Trait | Gene | SNP | Minor allele/frequency | Mean effect size2 | Probability of a functional effect (BQTN)3 | Location of the SNP in the gene |
| % | ||||||
| Total fatty acids | APOJ | rs569205 | A/0.484 | 0.74 | 0.68 | Flanking 5′ UTR |
| rs9331891 | A/0.002 | 1.11 | 0.79 | Intron | ||
| TNFRSF10B | rs12541697 | A/0.109 | 1.38 | 0.75 | Intron | |
| PUFAs, 18:2n−6 | LPL | rs285 | A/0.448 | 1.43 | 1.00 | Intron |
| MUFAs, 18:1n−7 | TNFRSF10B | rs10090860 | A/0.210 | 0.005 | 0.62 | Intron |
| MUFAs, 18:1n−9 | APOJ | rs9314349 | G/0.210 | 0.82 | 0.54 | Flanking 5′ UTR |
| Percentage body fat | APOJ | rs11136000 | A/0.463 | 0.87 | 0.62 | 3′ UTR |
| MSR1 | rs12114599 | G/0.033 | 0.86 | 0.67 | Intron | |
| Waist circumference | MSR1 | rs12114599 | G/0.033 | 1.20 | 0.57 | Intron |
| Triglycerides | LPL | rs1059611 | G/0.226 | 2.50 | 0.50 | Intron |
| rs328 | G/0.227 | 2.61 | 0.50 | Coding (nonsynonymous) S474 |
SOLAR version 4.0 (Southwest Foundation for Biomedical Research, San Antonio, TX) was used to conduct the BQTN analysis (15). MUFAs, monounsaturated fatty acids; PUFAs, polyunsaturated fatty acids; UTR, untranslated region; SNP, single nucleotide polymorphism.
Mean effect size: proportion of total variance explained by the SNP.
Probability of functional effect ≥0.50 is depicted in the table.
DISCUSSION
In this study, we showed that there is a strong genetic component to the variation in plasma FA distribution. In addition, we identified a QTL on chromosome 8 (8p12–p21) that appears to significantly modulate the variation in the proportion of unsaturated FAs. We also found evidence of a significant association between SNPs in candidate genes in this region and plasma FAs and identified an SNP that may have a functional effect on PUFA concentrations. To the best of our knowledge, this is the only study thus far to report a strong genetic influence on all circulating FA types and a significant association between plasma FAs and SNPs in positional candidate genes, with evidence of a functional effect of these SNPs on plasma FAs.
Plasma FA distribution showed higher plasma SFAs than MUFAs or PUFAs in the study participants; women had higher percentages of plasma SFAs and lower percentages of PUFAs than did men. Historically, Alaskan Eskimos have consumed more n−3 FAs because of their traditional diets. It was hypothesized that this explained the low CVD mortality that has been observed among this population (8). However, recent trends have shown an increase in CVD mortality (9). A shift toward nontraditional foods rich in saturated fats and lower rates of physical activity may have been major factors influencing this change in CVD profile (8).
Bivariate analysis was conducted to investigate whether there is a genetic influence on the relation between plasma FAs and adiposity traits. Significant genetic correlations between FAs and adiposity traits indicate that a common set of genes might be influencing FAs as well as the adiposity traits. Although several studies have found positive correlations between FAs and adiposity traits, this is the first study to identify genetic correlations between them. In addition, the phenotypic correlations of plasma FAs with adiposity traits observed in the current study are consistent with previous studies (5, 24).
Heritabilities for all plasma FAs ranged from 0.35 to 0.55, which indicates a significant additive genetic contribution. The heritability estimates for PUFAs in the study by Tanaka et al (6) ranged between 0.28 and 0.38, which is similar to our findings. Given the importance of plasma FAs in the pathogenesis of CVD and T2DM, the search for a QTL influencing the variation in plasma FA is a critical first step toward identifying candidate genes. The strongest evidence for a QTL was obtained for total MUFAs on chromosome 8p, with a one-LOD support interval of ≈20 cM. The QTL coincides with a QTL for total PUFAs, which makes this region unique for unsaturated FA concentrations. This region contains several strong positional candidate genes of potential interest. Most promising are β3-adrenergic receptor (ADRB3), lipoprotein lipase (LPL), apolipoprotein J or clusterin (APOJ or CLU), macrophage scavenger receptor 1(MSR1), and tumor necrosis factor superfamily member 10B (TNFRSF10B). ADRB3 encodes a G-protein coupled cell surface receptor (ADRB3) that plays an important role in lipolysis and energy expenditure (25). Associations between human obesity and the Trp26Arg polymorphism in ADRB3 have been consistently reported by several studies across populations (26, 27). Although SNPs in ADRB3 were not associated with circulating FAs in our current study, SNP rs35361594 showed an association with BMI, percentage body fat, and waist circumference. This SNP, located in the flanking 5′ untranslated region of the ADRB3, has not been previously associated with obesity.
LPL variation has been associated with hypertriglyceridemia, BMI, and body fat (28). LPL encodes an enzyme that hydrolyzes chylomicron triglycerides to free FAs (29). LPL variant Asn291Ser was found to increase the risk of dyslipidemia with advancing age and weight gain (30). Furthermore, in the Mediterranean population, Corella et al (27) also found a gene-gene interaction between ADRB3 and LPL SNPs influencing BMI in women. In our study, SNP rs285 was found to have strong statistical evidence for a functional effect and was associated with higher concentrations of the PUFA 18:2n−6. This polymorphism is in the sixth intron and generates a PvuII restriction enzyme site (31). Although this SNP (located in an intron) may not have a direct effect on the protein structure, it potentially could mediate an effect on gene transcription or it could be in strong LD with a functional variant. Another LPL SNP, rs328, results in a nonsense substitution at Ser447. In our study, this SNP was strongly associated with plasma triglycerides, with its minor allele (G) being associated with lower concentrations of triglycerides. Not only was this variant previously associated with a lower risk of coronary heart disease, but it was also associated with lower concentrations of triglycerides and higher HDL concentrations (36). Moreover, the same polymorphism was associated with lower blood pressure in adults (32). APOJ, another gene in our one-LOD support interval, codes for a glycoprotein that exists in plasma with HDL subfractions and has antiinflammatory properties. Polymorphisms of APOJ have been associated with tumorigenesis and Alzheimer disease (33–35). In addition, it plays a role in lipid transport, and its polymorphisms have been associated with high HDL concentrations (36, 37). In our study, the SNP rs9331891, which had a strong probability of functional effect on total FAs, was associated with higher concentrations of total FAs. Given its antiinflammatory properties and its role in lipid transport, APOJ appears to play an important part in atherosclerosis. In a study conducted in angiography patients, individuals with coronary stenosis had higher APOJ concentrations than those without stenosis (36).
Also within this one-LOD support interval are TNFRSF10B and MSR1. Also known as death receptor 5 (DR5)/tumor necrosis factor (TNF)–related apoptosis-inducing ligand-receptor 2 (TRAIL-R2), it encodes for TNFSRF10B, which mediates apoptosis through an intracellular adaptor molecule FADD (38). Free FAs are known to induce steatosis in liver and subsequently the apoptosis of hepatocytes. It has been shown that free FAs induced the apoptosis of hepatocytes, and adipocytes occur primarily through a TNFRSF10B-mediated mechanism (39, 40). Thus, TNFRSF10B appears to play a key role in FA metabolism. The TRAIL, a ligand for TNFRSF10B, has been associated with vascular injury, atherosclerosis, and diabetes (both type 1 and 2) (41). In a study conducted in healthy adults, serum TRAIL had significant positive correlations with total body fat in men and total and LDL cholesterol in men. Also, there was a negative correlation between lean body mass and TRAIL in men, which suggests an association between TRAIL and adiposity (42). TNFRSF10B variants have been previously shown to be associated with various cancers (43), but not with plasma FAs or adiposity traits, as shown in our study. Another interesting candidate gene in this region is macrophage scavenger receptor 1 (MSR1). MSR1 encodes a surface receptor that mediates the uptake of modified LDLs and results in the accumulation of lipid droplets and foam cell formation (44). MSR1 has been associated with the risk of atherosclerosis and prostate cancer. The association of MSR1 SNPs with prostate cancer has been reported extensively (45); however, to date, none of the MSR1 variants have been associated with plasma FAs or adiposity traits.
GWAS, conducted with blood lipids, have localized several associated genes, such as LPL, elongase of very-long fatty acids 2 (ELVOL2), fatty acid desaturases (FADS1, FADS2), apolipoprotein E (APOE), etc. Most consistent among them was LPL; SNP rs7007797, which is in complete LD with rs328, was associated with lower concentrations of triglycerides (46). Likewise, other LPL SNPs too have been associated with triglycerides (47, 48) and have been replicated in SNP-association studies (49). The only GWAS that used plasma FAs as phenotypes was focused on PUFAs and not all FAs (6). Most of the GWAS and other association studies with lipids have been conducted in whites, South Asians, Chinese, Japanese, and Hispanics. However, to date, no such studies have been conducted in Alaskan Eskimos or American Indians. With the increase in disorders such as T2DM and CVD in these populations, these results assume significance.
In conclusion, our analysis provides strong evidence that a gene or genes on chromosome 8p influences susceptibility for plasma unsaturated FAs. This study not only reports an association of plasma FAs and adiposity traits with polymorphisms in genes that have not been previously associated with adiposity (APOJ, ADRB3, TNFRSF10B, and MSR1), but also confirms previously reported association of polymorphisms in LPL with adiposity traits.
Supplementary Material
Acknowledgments
We thank the participants of the GOCADAN study for their generous participation.
The authors’ responsibilities were as follows—VSV and AGC: performed or supervised all aspects of the statistical analyses; HHHG, SL, and SAC: helped with the performance or supervision of the statistical analysis; KH and SAC: responsible for the 10 cM short tandem repeat and SNP genotyping; JGU, SL, CRW, and MET: helped with the recruitment, data entry, and preparation of the manuscript; and SOEE, RBD, RRF, JWM, BVH, and AGC: responsible for the execution of the study and contributed to the preparation of the manuscript. None of the authors reported a conflict of interest.
REFERENCES
- 1.Saleh J, Sniderman AD, Cianflone K. Regulation of plasma fatty acid metabolism. Clin Chim Acta 1999;286:163–80 [DOI] [PubMed] [Google Scholar]
- 2.Paolisso G, Manzella D, Rizzo MR, et al. Elevated plasma fatty acid concentrations stimulate the cardiac autonomous nervous system in healthy subjects. Am J Clin Nutr 2000;72:723–30 [DOI] [PubMed] [Google Scholar]
- 3.Tiemeier H, van Tuijil HR, Hoffman A, Kiliaan AJ, Breteler MM. Plasma fatty acid composition and depression are associated in the elderly: the Rotterdam Study. Am J Clin Nutr 2003;78:40–6 [DOI] [PubMed] [Google Scholar]
- 4.Simon JA, Hodgkins ML, Browner WS. Serum fatty acids and the risk of coronary heart disease. Am J Epidemiol 1995;142:469–76 [DOI] [PubMed] [Google Scholar]
- 5.Wang L, Folsom AR, Zheng ZJ, Pankow JS, Eckfeldt JH. Plasma fatty acid composition and incidence of diabetes in middle-aged adults: the Atherosclerosis Risk in Communities (ARIC) study. Am J Clin Nutr 2003;78:91–8 [DOI] [PubMed] [Google Scholar]
- 6.Tanaka T, Shen J, Abecasis GR, et al. Genome-wide association study of plasma polyunsaturated fatty acids in the InCHIANTI study. PLoS Genet 2009;5:e1000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Middaugh JP. Cardiovascular deaths among Alaskan Natives, 1980-86. Am J Public Health 1990;80:282–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ebbesson SOE, Kennish J, Ebbesson L, Go O, Yeh J. Diabetes is related to fatty acid imbalance in Eskimos. Int J Circumpolar Health 1999;58:108–19 [PubMed] [Google Scholar]
- 9.Schumacher C, Davidson M, Ehrsam G. Cardiovascular disease among Alaska Natives: a review of the literature. Int J Circumpolar Health 2003;62:343–62 [DOI] [PubMed] [Google Scholar]
- 10.Howard BV, Devereux RB, Cole SA, et al. A genetic and epidemiological study of cardiovascular disease in Alaska Natives (GOCADAN): design and methods. Int J Circumpolar Health 2005;64:206–21 [DOI] [PubMed] [Google Scholar]
- 11.Ebbesson SOE, Laston S, Wenger CR, et al. Recruitment and community interactions in the GOCADAN study. Int J Circumpolar Health 2006;65:55–64 [DOI] [PubMed] [Google Scholar]
- 12.Sobel E, Lange K. Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker sharing statistics. Am J Hum Genet 1996;58:1323–37 [PMC free article] [PubMed] [Google Scholar]
- 13.Heath SC. Markov chain Monte Carlo segregation and linkage analysis for oligogenic models. Am J Hum Genet 1997;61:748–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kong A, Gudbjartsson DF, Sainz J, et al. A high-resolution recombination map of the human genome. Nat Genet 2002;31:241–7 [DOI] [PubMed] [Google Scholar]
- 15.Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet 1998;62:1198–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hopper JL, Mathews JD. Extensions to multivariate normal models for pedigree analysis. Ann Hum Genet 1982;46:373–83 [DOI] [PubMed] [Google Scholar]
- 17.Self S, Liang K. Asymptotic properties of maximum likelihood-ratio tests under nonstandard conditions. J Am Stat Assoc 1987;82:605–10 [Google Scholar]
- 18.Blangero J, Goring HHH, Kent JW, Jr, et al. Quantitative trait nucleotide analysis using bayesian model selection. Hum Biol 2005;77:541–59 [DOI] [PubMed] [Google Scholar]
- 19.Boerwinkle E, Chakraborty R, Sing CF. The use of measured genotype information in the analysis of quantitative phenotypes in man. I. Models and analytical methods. Ann Hum Genet 1986;50:181–94 [DOI] [PubMed] [Google Scholar]
- 20.Neale BM, Sham PC. The future of association studies: gene-based analysis and replication. Am J Hum Genet 2004;75:353–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Charlesworth JC, Peralta JM, Drigalenko E, et al. Toward the identification of causal genes in complex diseases: a gene-centric joint test of significance combining genomic and transcriptomic data. BMC Proc 2009;3:S92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moskvina V, Schmidt KM. Individual SNP allele reconstruction from informative markers selected by a non-linear Gauss-type algorithm. Hum Hered 2006;62:97–106 [DOI] [PubMed] [Google Scholar]
- 23.Kass RE, Raftery AE. Bayes factors. J Am Stat Assoc 1995;90:773–95 [Google Scholar]
- 24.Zhou YE, Egeland GM, Meltzer SJ, Kubow S. The association of desaturase 9 and plasma fatty acid composition with insulin resistance—associated factors in female adolescents. Metabolism 2009;58:158–66 [DOI] [PubMed] [Google Scholar]
- 25.Taylor MRG.Pharmacogenetics of the human beta-adrenergic receptors. Pharmacogenomics J 2007;7:29–37 [DOI] [PubMed] [Google Scholar]
- 26.Mitchell BD, Blangero J, Comuzzie AG, et al. A paired sibling analysis of the beta-3 adrenergic receptor and obesity in Mexican Americans. J Clin Invest 1998;101:584–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Corella D, Guillén M, Portolés O, et al. Gender specific associations of the Trp64Arg mutation in the beta3-adrenergic receptor gene with obesity-related phenotypes in a Mediterranean population: interaction with a common lipoprotein lipase gene variation. J Intern Med 2001;250:348–60 [DOI] [PubMed] [Google Scholar]
- 28.Jemaa R, Tuzet S, Portos C, Betoulle D, Apfelbaum M, Fumeron F. Lipoprotein lipase gene polymorphisms: associations with hypertriglyceridemia and body mass index in obese people. Int J Obes Relat Metab Disord 1995;19:270–4 [PubMed] [Google Scholar]
- 29.Mattei MG, Etienne J, Chuat JC, et al. Assignment of the human lipoprotein lipase (LPL) gene to chromosome band 8p22. Cytogenet Cell Genet 1993;63:45–6 [DOI] [PubMed] [Google Scholar]
- 30.Hu Y, Liu W, Huang R, Zhang X. A systematic review and meta-analysis of the relationship between lipoprotein lipase Asn291Ser variant and diseases. J Lipid Res 2006;47:1908–14 [DOI] [PubMed] [Google Scholar]
- 31.Sagoo GS, Tatt I, Salanti G, et al. Seven lipoprotein lipase gene polymorphisms, lipid fractions, and coronary disease: a HuGE association review and meta-analysis. Am J Epidemiol 2008;168:1233–46 [DOI] [PubMed] [Google Scholar]
- 32.Clee SM, Loubser O, Collins J, Kastelein JJ, Hayden MR. The LPL S447X cSNP is associated with decreased blood pressure and plasma triglycerides, and reduced risk of coronary artery disease. Clin Genet 2001;60:293–300 [DOI] [PubMed] [Google Scholar]
- 33.Rizzi F, Bettuzzi S. The clusterin paradigm in prostate and breast carciongenesis. Endocr Relat Cancer 2010;17:R1–17 [DOI] [PubMed] [Google Scholar]
- 34.Harold D, Abraham R, Hollingworth P, et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer's disease. Nat Genet 2009;41:1088–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lambert JC, Heath S, Even G, et al. Genome-wide association study identifies variants at CLU and CR1 associated with Alzheimer's disease. Nat Genet 2009;41:1094–9 [DOI] [PubMed] [Google Scholar]
- 36.Poulakou MV, Paraskevas KI, Wilson MR, et al. Apolipoprotein J and leptin levels in patients with coronary heart disease. In Vivo 2008;22:537–42 [PubMed] [Google Scholar]
- 37.Nestlerode CS, Bunker CH, Sanghera DK, Aston CE, Ukoli FA, Kamboh MI. Apolipoprotein J polymorphisms and serum HDL cholesterol levels in African blacks. Hum Biol 1999;71:197–218 [PubMed] [Google Scholar]
- 38.Walczak H, Degli-Eposti MA, Johnson RA, et al. TRAIL-R2: a novel apoptosis-mediating receptor for TRAIL. EMBO J 1997;16:5386–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Malhi H, Barreyro FJ, Isomoto H, Brank SF, Gores GJ. Free fatty acids sensitize hepatocytes to TRAIL mediated cytotoxicity. Gut 2007;56:1124–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fischer-Posovszky P, Tornqvist H, Klaus-Michael D, Wabitsch M. Inhibition of death-receptor mediated apoptosis in human adipocytes by the insulin-like growth factor-1 (IGF-1)/IGF-1 receptor autocrine circuit. Endocinology 2004;145:1849–59 [DOI] [PubMed] [Google Scholar]
- 41.Kavurma MM, Bennett MR. Expression, regulation and function of trail in atherosclerosis. Biochem Pharmacol 2008;75:1441–50 [DOI] [PubMed] [Google Scholar]
- 42.Choi JW, Song JS, Pai SH. Associations of serum TRAIL concentrations, anthropometric variables and serum lipid parameters in healthy adults. Ann Clin Lab Sci 2004;34:400–4 [PubMed] [Google Scholar]
- 43.Teng MS, Brandwein-Gensler MS, Teixeira MS, Martignettie JA, Duffey DC. A study of TRAIL receptors in squamous cell carcinoma of the head and neck. Arch Otolaryngol Head Neck Surg 2005;131:407–12 [DOI] [PubMed] [Google Scholar]
- 44.Emi M, Asaoka H, Matsumoto A, et al. Structure, organization, and chromosomal mapping of the human macrophage scavenger receptor gene. J Biol Chem 1993;268:2120–5 [PubMed] [Google Scholar]
- 45.Bobryshev YV. Monocyte recruitment and foam cell formation on atherosclerosis. Micron 2006;37:208–22 [DOI] [PubMed] [Google Scholar]
- 46.Kathiresan S, Manning AK, Demissie S, et al. A genome-wise association study for blood lipid phenotypes in the Framingham Heart study. BMC Med Genet 2007;8:S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Willer CJ, Sanna S, Jackson AU, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet 2008;40:161–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chasman DI, Pare G, Mora S, et al. Forty-three loci associated with plasma lipoprotein size, concentration and cholesterol content in genome-wide analysis. PLoS Genet 2009;5:e100730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lanktree MB, Anand SS, Yusuf S, Hegele RA. SHARE investigators. Replication of genetic associations with plasma lipoprotein traits in a multiethnic sample. J Lipid Res 2009;50:1487–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.