Abstract
A plant's roots system determines both the capacity of a sessile organism to acquire nutrients and water, as well as providing a means to monitor the soil for a range of environmental conditions. Since auxins were first described, there has been a tight connection between this class of hormones and root development. Here we review some of the latest genetic, molecular, and cellular experiments that demonstrate the importance of generating and maintaining auxin gradients during root development. Refinements in the ability to monitor and measure auxin levels in root cells coupled with advances in our understanding of the sources of auxin that contribute to these pools represent important contributions to our understanding of how this class of hormones participates in the control of root development. In addition, we review the role of identified molecular components that convert auxin gradients into local differentiation events, which ultimately defines the root architecture.
Auxin generated in shoots and roots drives root development. Coordination of its synthesis, transport, and degradation establishes phytohormone gradients that determine local differentiation events.
The early characterization of auxins as “root forming hormones of plants” established a long-standing link between this class of small molecules and root development (Went 1929; Thimann and Went 1934). As with the aerial portion of the plant body, a series of iterative modules produces the overall root architecture; the root, which is established during embryogenesis, gives rise to new lateral roots in a continuous, indeterminate manner. Evidence from many studies highlights the central role of auxins in orchestrating the final root architecture. Defining the role of auxins as a component of endogenous developmental programs as well as in mediating environmental stimuli to shape the final root architecture remains at the heart of many active research programs. Here we review some recent discoveries that demonstrate the importance of auxin gradients and the conversion of this information into molecular responses that coordinate root development.
CELLULAR ORGANIZATION OF A ROOT
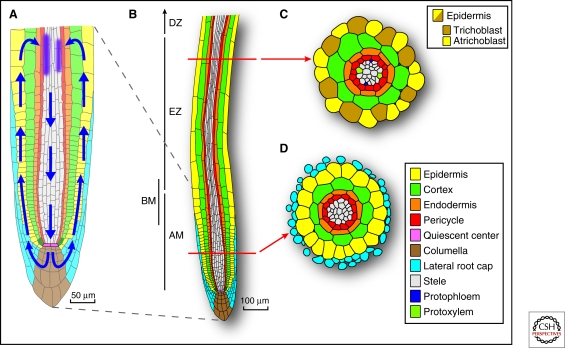
A single root contains a number of cell types, which can be discerned by visual and molecular markers (Dolan et al. 1993; Birnbaum et al. 2005; Laplaze et al. 2005; Brady et al. 2007). The development, organization, and patterning of these cell types is typically described using terminology that encompasses the circumferential, radial, and longitudinal structure of an individual root (Fig. 1). Along the proximal–distal axis a root is characterized by a series of developmental zones (Fig. 1) (Ishikawa and Evans 1995). The quiescent center (QC) promotes its neighboring cells to continuously produce initial cells that give rise to cell files. Through time, cells arising near the QC undergo additional rounds of division and become displaced from the root meristematic zone. The region of growing root where the rate of cell division slows and cell expansion begins is known as the basal meristem (Fig. 1) (Beemster et al. 2003; De Smet et al. 2007; Nieuwland et al. 2009). Subsequently, cells become part of the elongation zone and then the differentiation zone. As such, the longitudinal axis of a root represents a constantly renewing gradient of cell differentiation. Although many of the developmental events that regulate patterning and the capacity to form lateral roots are not observable, the epidermal root surface bears easy detectable markers of the transition between these distinct zones: An increased length of epidermal cells demarcates the transition between the meristematic and elongation zones and the appearance of root hairs marks the start of the differentiation zone.
Figure 1.
Cellular organization and the “inverted fountain” of auxin movement in the root tip of Arabidopsis. (A) Magnified view of the meristematic zone of the root tip that highlights the transport-mediated “reflux” of auxin from the lateral root cap through the epidermis to the basal meristem and then back towards the root tip. (B) The proximal–distal organization of the root arises from the iterative process of cell division and expansion of four types of stem cell initials, which are immediately adjacent to the mitotically inactive quiescent center cells: The epidermal/lateral root cap initials, which give rise to the epidermis and lateral root cap, the collumella initials, which contribute to the central portion of the root cap, the cortex/endodermal initials sustain the formation of the ground tissue, and the vascular initials give rise to the vascular tissues and the pericycle. Indicated on the left of the root are the root apical meristem (AM), elongation zone (EZ), differentiation zone (DZ), and the basal meristem (BM). (C) Cross section of root in the elongation zone highlighting the circumferential and radial organization of the root. The position of trichoblast and atrichoblast cell files are indicated. Also shown is the location of the protoxylem and protophloem that give rise to the diarch symmetry of the mature Arabidopsis root. (D) Cross section of an immature root showing the radial organization of cell files.
SHOOT-DERIVED AUXIN CONTRIBUTES TO ROOT ORGANIZATION
Auxins, a class of essential plant hormones, orchestrate almost every aspect of plant growth and development (Woodward and Bartel 2005; Vanneste and Friml 2009). In roots, the most well-characterized auxin-associated phenotypes are the dose-dependent increase in the length of epidermal-derived root hairs, the bimodal effect of auxin concentration on primary root length, the dose-dependent increase in number of lateral root primordia, and the response to gravity (Pitts et al. 1998; Rahman et al. 2002; Ishida et al. 2008; Peret et al. 2009). Proper output of auxin signaling depends on the interplay of the transport, biosynthesis, conjugation, and signaling of this class of small molecules. Because the role of auxin during embryogenesis is covered in another monograph of this series (Möller and Weijers 2009), we focus here on recent findings related to auxin and postembryonic root development.
Auxin can be synthesized in young leaves and cotyledons (Ljung et al. 2001). The same long-distance pathway that carries carbohydrates from “source” to “sink” also facilitates the bulk flow of auxin and other hormones such as ABA and cytokinins as well as mRNA and proteins (Robert and Friml 2009). Because bulk flow provides the driving force through membraneless phloem channels, the rate of auxin movement is rapid, reaching speeds of up to 7 cm/h in the roots of Populus tremula and Vicia faba (Eliasson 1972; Tsurumi and Wada 1980). The symplastic isolation of the sieve elements and companions cells along this transport route likely drives the movement of IAA towards the root tip, which represents the major sink tissue.
Although auxin is transported over long distances, our understanding of its role during development comes primarily from studies focused on sink tissues such as root tips, where cells are largely undifferentiated and where auxin movement through the apoplast would not be impeded by secondary cell walls. Similarly, the protonation of auxin in the acidic environment of the cell wall facilitates its movement by diffusion across cell membranes. It is not until the molecule becomes ionized in the less acidic cytoplasm that movement through sink tissue becomes limited and requires the involvement of integral membrane transport proteins to control directional movement. Commonly referred to as “polar auxin transport” (PAT) this mechanism of directing auxin distribution clearly differs from the long-distance transport described earlier. The rates of PAT in roots of Phaseolus vulgaris, Pisum sativum, and Arabidopsis thaliana are 10 mm/h, which is at least significantly slower than auxin movement through the phloem. PAT is the outcome of the coordinated activities of several membrane transport protein families that facilitate auxin influx (e.g., AUXIN RESISTANT1 [AUX1] and LIKE-AUX1 [LAX]) (Bennett et al. 1996; Swarup et al. 2001) and efflux (e.g., members of the P-GLYCOPROTEIN ABC transporter family [Geisler and Murphy 2006] and the PIN-FORMED [PIN] proteins [Petrasek et al. 2006]). The molecular, cellular, and biochemical aspects of these proteins have been the focus of a number of excellent reviews (Paponov et al. 2005; Petrasek and Friml, 2009; Vanneste and Friml, 2009).
AUXIN GRADIENTS ARE CONVERTED TO LOCAL DIFFERENTIATION EVENTS
The local control of auxin levels generates regional concentration gradients and local maxima that are crucial for establishing and maintaining a root primordium (reviewed by Benjamins and Scheres 2008). The cellular levels of auxin, in turn contribute to the regulation of gene expression that defines cell fate and pharmacological or genetic disruptions of auxin movement dramatically impacts root patterning (Aida et al. 2004; Galinha et al. 2007). A cell differentiation process in roots that strongly relies on cellular auxin gradients is the formation of root hairs. During this process the auxin-driven cell elongation of root hairs is supported by AUX1-directed auxin import into nonroot hair forming epidermal cells (Jones et al. 2009).
Modeling of auxin distribution based solely on shoot-derived auxin moving by diffusion and PIN-directed efflux through structured sink tissue predicts the formation of a strong auxin maximum in the distal meristem (Grieneisen et al. 2007). In computer-based models, the source of IAA, whether simulated as being applied or produced endogenously, does not affect the maintenance of the auxin maximum (Laskowski et al. 2008). Experimentally, the analysis of cell-sorted root cells provides the most direct evidence of auxin gradients in the root, including the predicted maximum in the QC (Petersson et al. 2009). Importantly, genetic or pharmacological manipulation of PAT that disturbs the maintenance of the auxin maximum dramatically alters the patterning of the root tip (Reed et al. 1998; Sabatini et al. 1999; Benjamins et al. 2001; Friml et al. 2002; Lewis et al. 2007; Benjamins and Scheres 2008).
The PAT-installed auxin maximum in the QC creates an organizing center that organizes the root tip through contact with neighboring stem cell initials (van den Berg et al. 1997; Sabatini et al. 1999). Stem cells divide asymmetrically to produce two daughter cells: one that retains stem cell identity and the other that undergoes additional divisions and moves toward differentiation. The maintenance of stems cells ensures the continued contribution of new cells to the apical meristem and underlies the indeterminate growth of the root (Sena et al. 2009). The availability of a stem cell pool has also been associated with the high regeneration capacity of roots (Xu et al. 2006). The presence of a stem cell population, however, is not a prerequisite for regenerating a root from differentiating cells because Arabidopsis lines that do not properly maintain a stem cell niche are still capable of regenerative reprogramming of the root apex after removal of the tip including the QC (Sena et al. 2009). Furthermore, the early events towards reestablishing an organized root meristem in this system likely relies on reestablishing auxin gradients through PAT as many of the genes whose RNAs are up-regulated in the first 24 h are auxin responsive while blocking auxin transport inhibits root regeneration (Sena et al. 2009).
Although it is clear that the proper movement of auxin in the root tip is required for root patterning, our understanding of how cells interpret this information to create a molecular response that leads to developmental changes remains in its infancy. The PLETHORA (PLT1-4) genes represent a subclade of AP2 transcription factors that are required to define the root-stem cell niche (Aida et al. 2004). The promoter activities of these genes are strongly correlated with auxin levels, suggesting that PLT protein levels directly reflect the underlying auxin gradients. Furthermore, although these genes are differentially expressed in a graded fashion in the root tip, their overlapping expression patterns leads to maximal PLT protein levels in the stem cell zone (Galinha et al. 2007). This expression pattern also coincides with that of the radial patterning genes SCR and SHR, which provide parallel inputs in specifying root stem cells (Aida et al. 2004). Based on several lines of evidence, including the ability of ectopically expressed PLT2 to direct the formation of root meristems in shoots and the inability of plt1/plt2 plants to specify root stem cells, even in the presence of high exogenous auxin, it appears that high PLT levels promote stem cell identity and maintenance, that intermediate PLT levels promote mitotic activity of stem cell daughters, and that cells with the lowest PLT levels are capable of differentiation (Aida et al. 2004; Galinha et al. 2007). Both experimental data (Blilou et al. 2005) and computer modeling (Grieneisen et al. 2007; Laskowski et al. 2008) highlight the importance of auxin distribution and the accompanying gradients in PLT gene expression. Since the PLT genes are also required for PIN gene transcription, this feed-forward mechanism would help stabilize the auxin maximum in the QC (Blilou et al. 2005).
Root-generated Auxin Contributes to Root Development
Despite being the first plant hormone identified, experimental evidence detailing multiple auxin biosynthetic pathways has only recently been reported (Zhao et al. 2001; Zhao et al. 2002; Woodward and Bartel 2005; Cheng et al. 2006; Seidel et al. 2006; Normanly 2009). In addition to being made in the shoots, auxin is also made in the roots (Muller et al. 1998; Ljung et al. 2001; Ljung et al. 2005; Stepanova et al. 2005; Stepanova et al. 2008; Ikeda et al. 2009; Yamada et al. 2009). Because many of the proteins involved belong to multigene families and several chemical intermediates have yet to be identified, the contribution of each pathway to developmental processes remains to be determined (reviewed by Woodward and Bartel 2005; Normanly 2009). What has become clear, however, is that many of the genes involved in the synthesis of auxin are expressed in roots and that root-generated auxin contributes to the maintenance of the gradients and maxima required for normal root development. (Ljung et al. 2005; Ikeda et al. 2009; Petersson et al. 2009). Many auxin biosynthesis genes are expressed in the root tip and high-resolution auxin-measurements in specific cell types after fluorescent activated cell sorting revealed the sites of auxin biosynthesis in the root apex (Petersson et al. 2009). These sites of synthesis contribute to the pools of auxin available for PAT and are consistent with previously described auxin gradients and regional maxima.
Several distinct lines of experimental evidence supports the idea that root-derived auxin contributes to the maintenance of a proper hormone gradient in the root. First, evidence for the role of auxin biosynthesis routes in roots came from the characterization of the WEAK ETHYLENE INSENSITIVE (wei2 and wei7) and TRANSPORT INHIBITOR RESPONSE 7 (tir7-1) mutants (Ljung et al. 2005; Stepanova et al. 2005). These mutants suppress the high auxin phenotypes of the auxin-overproducing SUPERROOT (sur1 or sur2) and disrupt one of the two subunits of anthranilate synthase, an enzyme that catalyzes the rate limiting formation of anthranilate from chorismate during tryptophan synthesis. The failure of specific cells to produce tryptophan lowers their capacity to produce indole-3-acetic acid, which impairs root growth (Ljung et al. 2005; Stepanova et al. 2005).
Second, the pdx1 mutants are unable to produce pyridoxal phosphate, vitamin B6 (Chen and Xiong 2005), which in turn impairs the capacity of these plants to convert tryptophan to IAA. These plants have a short primary root with a reduced root meristem size. Reciprocal grafting experiments established that shoot-derived auxin could not rescue these phenotypes and that root-derived auxin is required for proper postembryonic root growth (Chen and Xiong 2009). Third, a mutation in the protein kinase domain of the Raf-like kinase, CONSTITUTIVE TRIPLE RESPONSE 1 (CTR1) leads to increased IAA synthesis in roots, indicating that it is likely a negative regulator of auxin formation. The higher auxin levels in the ctr1 plants disrupt the auxin gradient in the root tip and alter the proximal–distal placement of root hairs, which is a hallmark of root epidermal cell polarity (Ikeda et al. 2009). Based on these observations, it appears that root-generated auxin plays a critical role in maintaining the planar polarity of root epidermal cells.
Finally, three groups working on independent mutants, wei8 (Stepanova et al. 2008), sav3 (Tao et al. 2008), and tir2 (Yamada et al. 2009), with auxin-related phenotypes showed that their mutants fall in an aminotransferase (TRYPTOPHAN AMINOTRANSFERASE of Arabidopsis, TAA). Biochemical studies established that the encoded enzyme catalyzes the transamination of tryptophan to form indole pyruvic acid, which in turn can be converted to IAA. In taa mutants, the defects in gravitropism and vascular tissue differentiation correlate with the reduced IAA levels found in these plants. Furthermore, the enhanced expression of this gene on the lower side of a bending root might contribute to a positive regulatory loop that is active during root gravitropism (Yamada et al. 2009).
These examples clearly establish the importance of root-derived auxin pools and future studies will have to deal with this extra level of complexity to obtain a more realistic view on root development.
AUXIN HOMEOSTASIS IN ROOTS
The majority of IAA in Arabidopsis is found conjugated through ester or amide linkages to sugars, amino acids, or peptides (Ljung et al. 2005). In addition to de novo biosynthesis, the formation or hydrolysis of these conjugates provides an additional mechanism to finely control the cellular level of free auxin (Woodward and Bartel, 2005). Disturbance of the auxin homeostasis can have dramatic effects on root growth and development as is illustrated by the IAA-ALANINE RESISTANT4 (iar4) mutants that have a twofold higher level of IAA-Glu conjugates and have strong reduction in the number of lateral roots combined with other low-auxin phenotypes such as diminished root hair initiation and elongation (Quint et al. 2009). The authors provide evidence that the loss of ability to release free IAA from conjugates, especially in root tissue where the IAR4 is most strongly expressed, leads to altered root responses, which can be rescued by auxin application. The IAR4 gene encodes a putative mitochondrial pyruvate dehydrogenase E1α-subunit and the biochemical role of this protein in IAA deconjugation and root morphogenesis awaits further work.
Although conjugation of IAA is thought to reduce the pool of active IAA, a recent study provided evidence that the Trp conjugate of IAA (and also of jasmonic acid) can act as independent signaling compounds. This antagonism of the Trp-IAA conjugate relies on TIR1 functioning, but the mode of action is not clear, as the conjugates did not interfere with IAA in the pull-down assays (Staswick 2009).
Auxin can also be found as a side chain–lengthened indole-3-butyric acid (IBA). IBA, initially described as a synthetic auxin that induces rooting in woody plant cuttings (Cooper 1935), is an endogenous plant compound (Epstein et al. 1989; Ludwig-Müller 2000). Genetic studies led to the identification of peroxisomal enzymes that are most likely involved in the conversion of IBA to free IAA (Zolman et al. 2000; Zolman et al. 2007; Zolman et al. 2008). The 50% reduction in lateral root formation seen when the IBA-RESISTANT genes contain mutations demonstrates that the IBA-derived IAA pool contributes to the regulation of root architecture. Moreover, interference with IBA efflux from roots by mutation of the PLEIOTROPIC DRUG RESISTANCE8/ABCG36 ATP Binding Cassette Transporter (PDR8) gene leads to higher IBA content in the root tip that, in turn, increases lateral root production, suggesting that PDR8 normally inhibits lateral root formation by transporting away part of the IBA pool in root tips (Strader and Bartel 2009). These new insights concerning IBA-transport and conversion show the importance of this compound for root architecture. Many questions, however, remain unanswered, including how, when, and where IBA is synthesized, whether IBA can serve as a signaling molecule on its own, what pathway(s) are involved in its conversion to IAA, and what components regulate IBA distribution in roots.
LATERAL ROOT DEVELOPMENT RELIES ON BOTH ROOT- AND SHOOT-DERIVED AUXIN POOLS
The roles of multiple plant hormones and the effects of environmental signals demonstrate that auxin is the dominant regulator of lateral root development (reviewed by Benkova and Hejatko 2009; Fukaki and Tasaka 2009). In many dicot plants including Arabidopsis, lateral roots are initiated from root pericycle cells adjacent to the protoxylem poles of the parent root (Beeckman et al. 2001). Although there remains a paucity of data that defines the molecular machinery responsible for generating a lateral root, recent observations indicate that the process progresses through at least four recognizable phases: priming, initiation, patterning, and emergence (Malamy 2009; Peret et al. 2009). Each of these phases is controlled or at least influenced by auxin.
The first visible events of lateral root formation are the divisions of a few pericycle cells situated adjacent to a protoxylem pole. These cell division events are commonly designated as “lateral root initiation.” Because the entire protoxylem pole pericycle shows a strong cell proliferation competence (Beeckman et al. 2001) and every pericyle cell adjacent to a xylem pole shows the capacity to divide in response to elevated auxin levels (Himanen et al. 2002; Himanen et al. 2004; Dubrovsky et al. 2008), it is believed that important control mechanisms exist to restrict lateral root initiation to certain sites and time-points during root growth. Before pericycle cell division many cell cycle-related genes such as cyclins and cyclin-dependent kinases (CDKs) are activated in auxin-treated Arabidopsis roots, suggesting that auxin promotes lateral root initiation by activating cell cycle-related genes. Inhibition of the G1-to-S transition by overexpression of KIP-RELATED PROTEIN2 (KRP2), a CDK inhibitor, decreased the number of lateral roots (Himanen et al. 2002). In addition, disruption of the D-type cyclin CYCD4;1 results in lower lateral root density, which can be rescued by low concentrations of exogenous auxin (Nieuwland et al. 2009). These data argue for the existence of a local, finely tuned, and cell-specific regulation of the cell cycle before lateral root initiation.
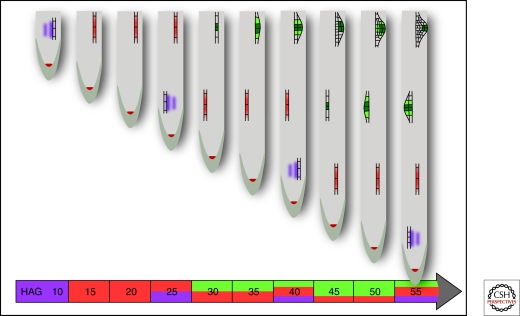
The regular spacing and positioning of lateral root primordia correlates with a priming event that targets only a few pericycle cells as they exit the basal meristem (Fig. 1) (De Smet et al. 2007). These founder cells become primed due to an auxin response maximum that arises in the neighboring protoxylem cells. The auxin response maximum in the basal meristem and the concomitant priming is not continuous but oscillates with a period of 15 h, which is in turn reflected in the regular spacing of lateral root along the root axis (Fig. 2).
Figure 2.
Spatial and temporal factors govern the formation of lateral root primordia. The regular spacing of lateral root primordia arises from pulses of auxin signaling in the basal meristem. The basal meristem encompasses the set of cells that are transitioning from the meristematic region into the elongation zone and thus includes cells that are undergoing division as well as elongation. In seedlings that are grown in constant light, pulses of auxin signaling, highlighted in purple, occur with a periodicity of 15 h. The response to auxin signaling in the xylem cells primes the adjacent pericycle cells (shown in red) so that they are competent to become lateral root founder cells (shown in green) upon a second, auxin-dependent signal in the differentiation zone (De Smet et al. 2007). As such, the pulses of auxin in the basal meristem coupled with the continuous growth of the root leads to the observed regular spacing of lateral root primordia. HAG, hours after germination.
After priming, initiation of new lateral roots occurs through the characteristic asymmetric, anticlinal cell divisions of two adjacent protoxylem pericycle cells of the same cell file. This results in two pairs of shorter and longer daughter cells (Beeckman et al. 2001; De Smet et al. 2007). After the daughter cells replicate, periclinal cell divisions establish a lateral root primordium with two cell layers. Subsequently, additional cell divisions and cell differentiation lead to the patterning of a mature lateral root with the radial and distal–basal axis and its root meristem (Malamy and Benfey, 1997).
The formation of lateral roots involves both shoot- and root-derived auxin pools. Phloem-based transport of IAA from the leaf to the root at the seedling stage is essential for the emergence, but not the initiation, of lateral roots in Arabidopsis and genetic or pharmacological manipulation of this auxin flow disrupts lateral root formation (Reed et al. 1998; Bhalerao et al. 2002; Wu et al. 2007). Furthermore, experiments with the IAA transport inhibitor NPA show that IAA movement through the root tip is essential for lateral root initiation, whereas shoot-derived transport is required for lateral root emergence (Casimiro et al. 2001). In contrast, stimulation of the shoot-to-root transport by growing rice plants in saturated humidity increased the number of emerged lateral roots (Chhun et al. 2007). Unfortunately, in the latter study, the authors did not make a distinction between emerged and nonemerged lateral roots thus leaving open the question of whether in rice the shoot-derived auxin was stimulatory specifically for emergence or rather served to promote the entire lateral root formation process, including initiation.
Because the root is continuously elongating, lateral root priming, initiation and emergence are not only separated in time, but also in space (Fig. 2). These temporal and spatial differences likely impact the combinations of auxin signaling components that are available to mediate the subsequent developmental steps. Recently, additional auxin-mediated control of lateral root emergence has been identified through the characterization of the lax3 mutant (Swarup et al. 2008). The LAX3 (LIKE AUX1 3) gene encodes an auxin influx transporter and its expression is up-regulated in the cells of the cortex and epidermis that overly a developing primordium. Induction of LAX3-expression occurs along with the formation of an auxin maximum within the growing primordia and triggers a positive feedback loop in the adjacent cells by stimulating LAX3-dependent auxin uptake. The increased auxin levels in turn stimulate the expression of cell wall remodeling genes that are crucial for regulated local cell separation and passage of the primordia through the outer cell layers (Swarup et al. 2008).
In general, mutations or transgenes affecting auxin biosynthesis, auxin metabolism, auxin transport, and auxin signaling influence the ability to form lateral roots. The existence of several types of mutations specifically inhibiting lateral root initiation, lateral root morphogenesis, or lateral root emergence, indicate that auxin is necessary not only for lateral root initiation but also for lateral root primordium development and emergence, respectively (Fukaki and Tasaka 2009). Mutations in multiple PIN family members disrupt auxin-induced lateral root primordium development, which results in the accumulation of layers of divided pericycle cells (Benkova et al. 2003). In addition, functional analysis of AUX1/LAX family members revealed that AUX1 is required for lateral root initiation because of its role in the basipetal auxin transport from the root tip toward the basal region through outer cell layers (lateral root cap and epidermis) (Swarup et al. 2001; De Smet et al. 2007), whereas LAX3 promotes lateral root emergence by affecting auxin influx of the outer endodermis and cortex cells (Swarup et al. 2008). These studies show that the regulation of distinct auxin transport systems are active at different stages of lateral root formation.
AUXIN SIGNALING IN ROOT CELLS
The combination of auxin transport, production, degradation, and conjugation in the cells of a plant ultimately define, over time, the “auxin status” of a given cell. Superimposed on the auxin status is the capacity of a given cell to generate a biochemical, physiological, or molecular response to the given level of auxin. These responses could be continuous or defined by a threshold level of auxin depending on the level and characteristics of the signaling components present in a given set of cells. In this way, the same auxin concentration can promote cell expansion in hypocotyls while inhibiting cell elongation in favor of cell division in the root meristem.
A number of the molecular components involved in auxin signaling have been identified. Auxin signaling occurs through its ability to stabilize the interaction of the TRANSPORT INHIBITOR RESPONSE 1 (TIR1) protein, or the closely related AUXIN F-BOX PROTEINs (AFBs), with members of the Aux/IAA protein family, which is encoded by 29 genes. The TIR1 and AFBs provide substrate specificity for the E3 ubquitin-ligase activity of Skp1-Cul1-F-box (SCF) complexes (Chapman and Estelle 2009). The IAA-stabilized interaction of SCFTIR or AFB leads to Aux/IAA ubiquitination and the removal of these proteins by the 26S proteasome (Maraschin et al. 2009).
The majority of Aux/IAA proteins contain four conserved domains (I, II, III, and IV) and based on the analysis of representative members of this family, these domains contribute discrete surfaces that enable distinct protein–protein interactions. Domain I facilitates the interaction with TOPLESS1, a corepressor of transcription (Szemenyei et al. 2008), domain II creates a cooperative auxin-binding site with TIR1 (Tan et al. 2007), and domains III and IV mediate interactions among members of the Aux/IAA protein family as well as DNA-binding proteins that belong to the AUXIN RESPONSE FACTOR (ARF) transcription factors (Kim et al. 1997; Ulmasov et al. 1997), which are encoded by a 23-member gene family. As such, although direct evidence for capacity of each domain to mediate identical interactions is lacking, the scaffolding function of the Aux/IAA proteins and the regulation of specificity and/or affinity for distinct interacting partners provides a molecular basis for the fine-tuned, cell-specific responses to endogenous and applied auxin (Lokerse and Weijers 2009). The high degree of redundancy provided by multiple genes that encode components of the auxin signaling pathway continues to provide challenges to a refined understanding of the unique and overlapping function of individual, or subsets of, these genes during plant growth and development (Okushima et al. 2005; Overvoorde et al. 2005).
Aux/IAA Proteins Contribute to Root Development
The control of postembryonic root growth and lateral root formation is tightly regulated by auxin and represents a robust developmental system for studying auxin signaling. Gain-of-function mutations in IAA1/AXR5, IAA3/SHY2, IAA12/BDL, IAA14/SLR, IAA18/CRANE, IAA19/MSG2, and IAA28 genes lead to plants with altered capacity to form lateral roots (Tian and Reed, 1999; Rogg et al. 2001; Fukaki et al. 2002; Tatematsu et al. 2004; Yang et al. 2004; Uehara et al. 2008; Ploense et al. 2009). Although a detailed, direct comparison of the root-related phenotypes of these mutants has not been performed, the slr-1/iaa14 and iaa28 gain-of-function plants show the strongest reduction in lateral root formation and likely play a prominent role in lateral root formation (Rogg et al. 2001; Fukaki et al. 2002). Particularly, the slr-1/iaa14 mutant has no lateral roots because of the inhibition of anticlinal cell divisions for lateral root initiation at the protoxylem pericycle (Fukaki et al. 2002; Vanneste et al. 2005). These observations indicate that the Aux/IAA proteins function as negative regulators during the lateral root process by inactivating ARF-mediated transcription, which is required for lateral root formation.
The phenotypic differences among gain-of-function mutants in Aux/IAA proteins are partly caused by the differential expression patterns of Aux/IAA members. When the Aux/IAA is expressed in tissues where lateral roots are initiated, the gain-of-function mutation results in a defect in lateral root formation (e.g., slr-1/iaa14, shy2/iaa3, msg2/iaa19, axr5/iaa1, crane/iaa18, and iaa28). In contrast, stabilizing mutations in Aux/IAA genes that are expressed in the root epidermis alter gravitropic responses and reduce the number of root hairs that form (e.g., axr2/iaa7, axr3/iaa17, slr-1/iaa14). Further support for these generalizations come from the observation that ectopic expression of the slr-1/iaa14 protein from the stele-specific SHR promoter results in the strong reduction in lateral root formation but does not affect root gravitropism and root hair formation (Fukaki et al. 2005). In contrast, the SLR/IAA14 promoter drives expression in both the root epidermis and the stele and the slr-1/iaa14 mutant, shows defects in lateral root formation, root gravitropism, and root hair formation (Fukaki et al. 2005; Vanneste et al. 2005). Although the tissue-specific expression profiles of all Aux/IAAs have not been examined, Aux/IAA members have distinct and overlapping functions in the regulation of auxin-mediated root growth and development (Overvoorde et al. 2005).
In addition to differences in expression profiles, several reports exploit the distinct root phenotypes—root hair formation, gravitropism, lateral root formation—conferred by shy2/iaa3, axr3/iaa17, and slr-1/iaa14 gain-of-function mutations to assess the specificity of the encoded proteins for controlling root growth and patterning (Knox et al. 2003; Weijers et al. 2005; Muto et al. 2007). The results of these promoter-swap experiments provide additional evidence that distinct patterns of expression impart one level of regulating the contribution of an Aux/IAA protein to a developmental response. More intriguing, however, are the observations that when different Aux/IAA proteins are expressed in the same developmental context, they generate nonoverlapping responses, which indicates that unique protein–protein interactions likely determine the role of specific polypeptides encoded by this large gene family (Knox et al. 2003; Weijers et al. 2005; Muto et al. 2007). The final cellular response appears to be determined by a combination of gene expression pattern as well as biochemical properties of the encoded proteins.
The stabilized forms of the Aux/IAA repressors likely exert their effect through domain III- and IV-mediated interactions with the ARF proteins. Intragenic suppressors of axr3/iaa17 (Rouse et al. 1998) or axr2/iaa7 (Nagpal et al. 2000) phenotypes are caused by point mutations that alter domain III of these stabilized proteins and impairs their capacity to homo- and hetero-dimerize (Ouellet et al. 2001). Recently, the observation that ARF1 and ARF7/NPH4 interact in vivo and in vitro with MYB77, one of 125-proteins belonging to the MYB R2R3-subfamily of transcription factors, and that IAA19/MSG2 interacts with MYB77 in coimmunoprecipitation experiments indicates that a larger number of protein–protein interactions may need to be considered (Shin et al. 2007).
ARF Proteins Contribute to Root Development
The combination of forward and reverse genetic approaches also highlight the importance of ARF proteins during lateral root formation (Tatematsu et al. 2004; Okushima et al. 2005; Wilmoth et al. 2005). Loss-of-function mutations in either ARF7/NPH4 or ARF19 lead to the mild reductions in lateral roots formed, where as the double arf7/arf19 mutant shows a dramatic delay in lateral root formation, indicating these genes are capable of performing similar functions in the root (Okushima et al. 2005; Wilmoth et al. 2005). In yeast two-hybrid assays, the ARF7 and ARF19 proteins interact with a number of Aux/IAA proteins, including IAA3/SHY2, IAA12/BDL, IAA14/SLR, IAA17/AXR3, IAA18/CRANE, and IAA19/MSG2 (Ouellet et al. 2001; Tatematsu et al. 2004; Fukaki et al. 2005; Uehara et al. 2008). The extent to which these heterotypic interactions occur in the root remains to be determined. It is likely that additional factors participate in the lateral root formation process as lateral roots do form after 14 d in an arf7/arf19 background (Fukaki et al. unpublished results). In addition, exogenous auxin can restore the production of low numbers of lateral roots in these double mutants (Wilmoth et al. 2005), but not in the iaa14/arf7/arf19 triple mutant (Fukaki et al. unpublished results), suggesting that additional ARFs play a role during lateral root formation.
Recently, ARF5/MP and its interacting partner IAA12/BDL, which play a well-characterized role during embryogenesis (Hamann et al. 2002; Weijers et al. 2006), were also shown to be involved in postembryonic lateral root formation (De Smet et al. 2010). Both gain-of-function bdl and loss-of-function mp mutants displayed lateral root defects such as severe reduction in emerged lateral roots and clustering of primordia. As lateral root initiation is still possible in these mutants and because ectopic expression of ARF5/MP resulted in the induction of lateral roots in a iaa14/slr mutant background, the ARF5/MP- IAA12/BDL pair defines an additional auxin response module that is active after, but not downstream of, the ARF7/ARF19- IAA14/SLR dependent control of lateral root initiation.
ARF7 and ARF19 positively regulate the expression of many auxin-responsive genes (Okushima et al. 2005). Among the direct targets of ARF7 and ARF19 are LATERAL ORGAN BOUNDARIES-DOMAIN (LBD)/ASYMMETRIC LEAVES2-like (ASL) genes LBD16/ASL18 and LBD29/ASL16, which encode plant specific nuclear proteins that are important for lateral root initiation (Okushima et al. 2007). Overexpression of LBD16 and LBD29 partially rescued the arf7arf19 lateral root phenotype whereas overexpression of LBD16-fused to the transcriptional-repressor domain (SRDX) inhibited lateral root formation, indicating that LBD16 and other auxin-inducible LBD proteins regulate lateral root initiation downstream of ARF7/19 (Okushima et al. 2007). Although the molecular function of these LBD proteins remains unknown, these studies reinforce the importance of auxin-mediated gene expression cascades during the process of lateral root formation.
Auxin also plays a role during lateral root primordium establishment, likely through the activity of undefined ARF protein(s). The PUCHI gene, encodes a putative transcription factor that contains an AP2 domain, which is necessary for both correct lateral root primordium establishment and floral primordium development (Hirota et al. 2007; Karim et al. 2009). Application of auxin induces PUCHI expression in roots and this expression is dependent on the auxin-responsive elements in the PUCHI promoter, which suggests that undefined ARF proteins regulate PUCHI expression during lateral root primordium development (Hirota et al. 2007).
In addition to activating transcription, transient expression assays suggest that ARF proteins can also function as transcriptional repressors (Ulmasov et al. 1999; Tiwari et al. 2003). The root phenotype of the arf10/arf16 double mutants provides genetic evidence that some ARFs function as negative regulators. The arf10/arf16 knockout line produces more lateral roots, whereas the arf10 or arf16 single mutants show no root phenotypes (Wang et al. 2005). Furthermore, the overexpression of microRNA160, which targets ARF10, ARF16, and ARF17 leads to an increased number of lateral roots and a reduction in primary root growth (Mallory et al. 2005; Wang et al. 2005). Interestingly, the expression of a microRNA resistant form of ARF17 leads to a dose-dependent reduction in lateral root numbers (Mallory et al. 2005).
Multiple ARFs are also subject to posttranscriptional regulation via transacting small-interfering RNA (Allen et al. 2005; Williams et al. 2005). The microRNA390 participates the generation of mature TAS3 tasiRNAs, which target ARF3 and ARF4 transcripts (Montgomery et al. 2008). The expression of one of the two MIR390 genes, MIR390b is specifically up-regulated by auxin and not affected by other plant hormones. Given the recent discovery that tasiRNAs targeting ARFs act in a noncell autonomous manner (Chitwood et al. 2009), defining the regulation and role of these molecules during lateral root formation will add to our understanding of this dynamic process. Taken together these results highlight the importance of posttranscriptional regulation of ARFs to fine-tune the complex web of protein–protein interactions that govern root development.
MULTIPLE PATHWAYS OF AUXIN PERCEPTION FUNCTION IN ROOTS
The many examples of lateral root defects due to impaired auxin response suggest that multiple signaling pathways function to regulate this developmental process. For instance, the observations that loss of TIR1 leads to a ∼50% reduction in lateral roots density, implies that related AFB's or undefined pathways also regulate this process (Gray et al. 2001). In addition to our understanding of the TIR1/AFB-Aux/IAA-ARF mediated auxin signaling, recent reports also indicate roles for the AUXIN BINDING PROTEIN 1 (ABP1) (Tromas et al. 2009) and the Arabidopsis heterotrimeric G-proteins in mediating auxin responses (Mudgil et al. 2009).
Despite the many lines of evidence that point towards auxin signaling as the dominant regulator of lateral root formation, there are reports that suggest auxin-independent mechanisms might be sufficient to induce lateral root initiation. For instance an examination of the effect of mechanically stimulating a root demonstrated a closely linked influx of Ca2+ into pericycle cells, which was capable of initiating lateral root primordia even in auxin transporter and receptor/response mutants including tir1 (Richter et al. 2009).
In addition, signaling pathways that involve other plant hormones also influences the role of auxin in roots. For instance, there is accumulating evidence for a rate-limiting role of the brassinosteroid pathway in the auxin response (Nemhauser et al. 2004; Hardtke et al. 2007; Kuppusamy et al. 2008; Vert et al. 2008). Global gene expression analyses have indicated that auxin-responsive gene expression is impaired in brevis radix (brx) mutants (Mouchel et al. 2006). brx mutants display impaired root growth because of decreased cell proliferation in the root meristem and vasculature and generally reduced cell elongation (Mouchel et al. 2004). Interestingly, the brx stunted root phenotype and impaired auxin-responsive gene expression can be partially restored by application brassinosteroids to the medium (Mouchel et al. 2006). Furthermore, expression of BRX is itself strongly induced by auxin (Mouchel et al. 2006). Consistently, BRX expression is no longer auxin-responsive in brx mutants, suggesting that auto-regulatory feedback exists. BRX is localized to the plasma membrane and raising the auxin content in cells and/or auxin fluxes through cells results in a translocation of the protein to the nucleus (Scacchi et al. 2009). As such, BRX could represent a cellular component that helps translate auxin levels or fluxes recorded at the plasma membranes into transcriptional responses in the nucleus. Although outside the scope of this review, readers interested in additional examples of plant hormone cross-talk in root patterning, growth, and development are referred to a number of more comprehensive reviews (Benkova and Hejatko 2009; Fukaki and Tasaka 2009; Perilli et al. 2009).
ACKNOWLEDGMENTS
Given the breadth of this topic, we apologize to those whose exciting work has been excluded. We thank Gert Van Isterdael for assistance with the artwork. The authors are supported by the University of Ghent Bijzonder Onderzoeksfonds and the U.S. National Science Foundation award OISE 0838812 (P.J.O.), and the Interuniversity Attraction Poles Programme (P6/33 and P5/13), initiated by the Belgian State, Science Policy Office (BELSPO) (T.B.).
Footnotes
Editors: Mark Estelle, Dolf Weijers, Ottoline Leyser, and Karin Ljung
Additional Perspectives on Auxin Signaling available at www.cshperspectives.org
REFERENCES
- Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, Nussaume L, Noh YS, Amasino R, Scheres B 2004. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 119:109–120 [DOI] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Carrington JC 2005. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121:207–221 [DOI] [PubMed] [Google Scholar]
- Beeckman T, Burssens S, Inze D 2001. The peri-cell-cycle in Arabidopsis. J Exp Bot 52:403–411 [DOI] [PubMed] [Google Scholar]
- Beemster GT, Fiorani F, Inze D 2003. Cell cycle: the key to plant growth control? Trends in Plant Science 8:154–158 [DOI] [PubMed] [Google Scholar]
- Benjamins R, Quint A, Weijers D, Hooykaas P, Offringa R 2001. The PINOID protein kinase regulates organ development in Arabidopsis by enhancing polar auxin transport. Development 128:4057–4067 [DOI] [PubMed] [Google Scholar]
- Benjamins R, Scheres B 2008. Auxin: the looping star in plant development. Annu Rev Plant Biol 59:443–465 [DOI] [PubMed] [Google Scholar]
- Benkova E, Hejatko J 2009. Hormone interactions at the root apical meristem. Plant Mol Biol 69:383–396 [DOI] [PubMed] [Google Scholar]
- Benkova E, Michniewicz M, Sauer M, Teichmann T, Seifertova D, Jurgens G, Friml J 2003. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell 115:591–602 [DOI] [PubMed] [Google Scholar]
- Bennett MJ, Marchant A, Green HG, May ST, Ward SP, Millner PA, Walker AR, Schulz B, Feldmann KA 1996. Arabidopsis AUX1 gene: a permease-like regulator of root gravitropism. Science 273:948–950 [DOI] [PubMed] [Google Scholar]
- Bhalerao RP, Eklof J, Ljung K, Marchant A, Bennett M, Sandberg G 2002. Shoot-derived auxin is essential for early lateral root emergence in Arabidopsis seedlings. Plant J 29:325–332 [DOI] [PubMed] [Google Scholar]
- Birnbaum K, Jung JW, Wang JY, Lambert GM, Hirst JA, Galbraith DW, Benfey PN 2005. Cell type-specific expression profiling in plants via cell sorting of protoplasts from fluorescent reporter lines. Nat Methods 2:615–619 [DOI] [PubMed] [Google Scholar]
- Blilou I, Xu J, Wildwater M, Willemsen V, Paponov I, Friml J, Heidstra R, Aida M, Palme K, Scheres B 2005. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433:39–44 [DOI] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN 2007. A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318:801–806 [DOI] [PubMed] [Google Scholar]
- Casimiro I, Marchant A, Bhalerao RP, Beeckman T, Dhooge S, Swarup R, Graham N, Inze D, Sandberg G, Casero PJ, et al. 2001. Auxin transport promotes Arabidopsis lateral root initiation. Plant Cell 13:843–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman EJ, Estelle M 2009. Mechanism of auxin-regulated gene expression in plants. Ann Rev Gen 43:265–285 [DOI] [PubMed] [Google Scholar]
- Chen H, Xiong L 2005. Pyridoxine is required for post-embryonic root development and tolerance to osmotic and oxidative stresses. Plant J 44:396–408 [DOI] [PubMed] [Google Scholar]
- Chen H, Xiong L 2009. The short-rooted vitamin B6-deficient mutant pdx1 has impaired local auxin biosynthesis. Planta 229:1303–1310 [DOI] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y 2006. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes Dev 20:1790–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhun T, Uno Y, Taketa S, Azuma T, Ichii M, Okamoto T, Tsurumi S 2007. Saturated humidity accelerates lateral root development in rice (Oryza sativa L.) seedlings by increasing phloem-based auxin transport. J Exp Bot 58:1695–1704 [DOI] [PubMed] [Google Scholar]
- Chitwood DH, Nogueira FT, Howell MD, Montgomery TA, Carrington JC, Timmermans MC 2009. Pattern formation via small RNA mobility. Genes Dev 23:549–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper WC 1935. Hormones in relation to root formation on stem cuttings. Plant Physiol 10:789–794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I, Lau S, Voß U, Vanneste S, Benjamins R, Rademacher EH, Schlereth A, De Rybel B, Vassileva V, Grunewald W, et al. 2010. Bimodular auxin response controls organogenesis in Arabidopsis. Proc Natl Acad Sci 107:2705–2710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I, Tetsumura T, De Rybel B, Frey NF, Laplaze L, Casimiro I, Swarup R, Naudts M, Vanneste S, Audenaert D, et al. 2007. Auxin-dependent regulation of lateral root positioning in the basal meristem of Arabidopsis. Development 134:681–690 [DOI] [PubMed] [Google Scholar]
- Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, Scheres B 1993. Cellular-organization of the Arabidopsis thaliana root. Development 119:71–84 [DOI] [PubMed] [Google Scholar]
- Dubrovsky JG, Sauer M, Napsucialy-Mendivil S, Ivanchenko MG, Friml J, Shishkova S, Celenza J, Benkova E 2008. Auxin acts as a local morphogenetic trigger to specify lateral root founder cells. Proc Natl Acad Sci 105:8790–8794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliasson L 1972. Translocation of shoot-applied indolylacetic acid into the roots of Populus tremula. Physiologia Plantarum 27:412–416 [Google Scholar]
- Epstein E, Chen K-H, Cohen JD 1989. Identification of indole-3-butyric acid as an endogenous constituent of maize kernels and leaves. Plant Growth Regulation 8:215–223 [Google Scholar]
- Friml J, Benkova E, Blilou I, Wisniewska J, Hamann T, Ljung K, Woody S, Sandberg G, Scheres B, Jurgens G, et al. 2002. AtPIN4 mediates sink-driven auxin gradients and root patterning in Arabidopsis. Cell 108:661–673 [DOI] [PubMed] [Google Scholar]
- Fukaki H, Nakao Y, Okushima Y, Theologis A, Tasaka M 2005. Tissue-specific expression of stabilized SOLITARY-ROOT/IAA14 alters lateral root development in Arabidopsis. Plant J 44:382–395 [DOI] [PubMed] [Google Scholar]
- Fukaki H, Tameda S, Masuda H, Tasaka M 2002. Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. Plant J 29:153–168 [DOI] [PubMed] [Google Scholar]
- Fukaki H, Tasaka M 2009. Hormone interactions during lateral root formation. Plant Mol Biol 69:437–449 [DOI] [PubMed] [Google Scholar]
- Galinha C, Hofhuis H, Luijten M, Willemsen V, Blilou I, Heidstra R, Scheres B 2007. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 449:1053–1057 [DOI] [PubMed] [Google Scholar]
- Geisler M, Murphy AS 2006. The ABC of auxin transport: the role of p-glycoproteins in plant development. FEBS Lett 580:1094–1102 [DOI] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M 2001. Auxin regulates SCFTIR1-dependent degradation of AUX/IAA proteins. Nature 414:271–276 [DOI] [PubMed] [Google Scholar]
- Grieneisen VA, Xu J, Maree AF, Hogeweg P, Scheres B 2007. Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 449:1008–1013 [DOI] [PubMed] [Google Scholar]
- Hamann T, Benkova E, Baurle I, Kientz M, Jurgens G 2002. The Arabidopsis BODENLOS gene encodes an auxin response protein inhibiting MONOPTEROS-mediated embryo patterning. Genes Dev 16:1610–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke CS, Dorcey E, Osmont KS, Sibout R 2007. Phytohormone collaboration: zooming in on auxin-brassinosteroid interactions. Trends Cell Biol 17:485–492 [DOI] [PubMed] [Google Scholar]
- Himanen K, Boucheron E, Vanneste S, De Almeida Engler J, Inze D, Beeckman T 2002. Auxin-mediated cell cycle activation during early lateral root initiation. Plant Cell 14:2339–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himanen K, Vuylsteke M, Vanneste S, Vercruysse S, Boucheron E, Alard P, Chriqui D, Van Montagu M, Inze D, Beeckman T 2004. Transcript profiling of early lateral root initiation. Proc Natl Acad Sci 101:5146–5151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirota A, Kato T, Fukaki H, Aida M, Tasaka M 2007. The auxin-regulated AP2/EREBP gene PUCHI is required for morphogenesis in the early lateral root primordium of Arabidopsis. Plant Cell 19:2156–2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda Y, Men S, Fischer U, Stepanova AN, Alonso JM, Ljung K, Grebe M 2009. Local auxin biosynthesis modulates gradient-directed planar polarity in Arabidopsis. Nat Cell Biol 11:731–738 [DOI] [PubMed] [Google Scholar]
- Ishida T, Kurata T, Okada K, Wada T 2008. A genetic regulatory network in the development of trichomes and root hairs. Annu Rev Plant Biol 59:365–386 [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Evans ML 1995. Specialized zones of development in roots. Plant Physiol 109:725–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AR, Kramer EM, Knox K, Swarup R, Bennett MJ, Lazarus CM, Leyser HM, Grierson CS 2009. Auxin transport through non-hair cells sustains root-hair development. Nat Cell Biol 11:78–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karim MR, Hirota A, Kwiatkowska D, Tasaka M, Aida M 2009. A role for Arabidopsis PUCHI in floral meristem identity and bract suppression. Plant Cell 21:1360–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Harter K, Theologis A 1997. Protein-protein interactions among the Aux/IAA proteins. Proc Natl Acad Sci 94:11786–11791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox K, Grierson CS, Leyser O 2003. AXR3 and SHY2 interact to regulate root hair development. Development 130:5769–5777 [DOI] [PubMed] [Google Scholar]
- Kuppusamy KT, Walcher CL, Nemhauser JL 2008. Cross-regulatory mechanisms in hormone signaling. Plant Mol Biol 9:375–381 [DOI] [PubMed] [Google Scholar]
- Laplaze L, Parizot B, Baker A, Ricaud L, Martiniere A, Auguy F, Franche C, Nussaume L, Bogusz D, Haseloff J 2005. GAL4-GFP enhancer trap lines for genetic manipulation of lateral root development in Arabidopsis thaliana. J Exp Bot 56:2433–2442 [DOI] [PubMed] [Google Scholar]
- Laskowski M, Grieneisen VA, Hofhuis H, Hove CA, Hogeweg P, Maree AF, Scheres B 2008. Root system architecture from coupling cell shape to auxin transport. PLoS Biol 6:e307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis DR, Miller ND, Splitt BL, Wu G, Spalding EP 2007. Separating the roles of acropetal and basipetal auxin transport on gravitropism with mutations in two Arabidopsis multidrug resistance-like ABC transporter genes. Plant Cell 19:1838–1850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ljung K, Bhalerao RP, Sandberg G 2001. Sites and homeostatic control of auxin biosynthesis in Arabidopsis during vegetative growth. Plant J 28:465–474 [DOI] [PubMed] [Google Scholar]
- Ljung K, Hull AK, Celenza J, Yamada M, Estelle M, Normanly J, Sandberg G 2005. Sites and regulation of auxin biosynthesis in Arabidopsis roots. Plant Cell 17:1090–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokerse AS, Weijers D 2009. Auxin enters the matrix-assembly of response machineries for specific outputs. Curr Opin Plant Biol 12:520–526 [DOI] [PubMed] [Google Scholar]
- Ludwig-Müller J 2000. Indole-3-butyric acid in plant growth and development. Plant Growth Regulation 32:219–230 [Google Scholar]
- Malamy JE 2009. Lateral root formation. In Beeckman T., ed., Root development Annual Plant Reviews, Blackwell Publishing, November 2009, 352 pages, volume 37:83–126 [Google Scholar]
- Malamy JE, Benfey PN 1997. Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124:33–44 [DOI] [PubMed] [Google Scholar]
- Mallory AC, Bartel DP, Bartel B 2005. MicroRNA-directed regulation of Arabidopsis AUXIN RESPONSE FACTOR17 is essential for proper development and modulates expression of early auxin response genes. Plant Cell 17:1360–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraschin Fdos S, Memelink J, Offringa R 2009. Auxin-induced, SCFTIR1-mediated poly-ubiquitination marks AUX/IAA proteins for degradation. Plant J 59:100–109 [DOI] [PubMed] [Google Scholar]
- Möller B, Weijers D 2009. Auxin control of embryo patterning. Cold Spring Harbor Perspectives in Biology 1:a001545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery TA, Howell MD, Cuperus JT, Li D, Hansen JE, Alexander AL, Chapman EJ, Fahlgren N, Allen E, Carrington JC 2008. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell 133:128–141 [DOI] [PubMed] [Google Scholar]
- Mouchel CF, Briggs GC, Hardtke CS 2004. Natural genetic variation in Arabidopsis identifies BREVIS RADIX, a novel regulator of cell proliferation and elongation in the root. Genes Dev 18:700–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouchel CF, Osmont KS, Hardtke CS 2006. BRX mediates feedback between brassinosteroid levels and auxin signalling in root growth. Nature 443:458–461 [DOI] [PubMed] [Google Scholar]
- Mudgil Y, Uhrig JF, Zhou J, Temple B, Jiang K, Jones AM 2009. Arabidopsis N-MYC DOWNREGULATED-LIKE1, a positive regulator of auxin transport in a G protein-mediated pathway. Plant Cell 21:3591–3609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller A, Hillebrand H, Weiler EW 1998. Indole-3-acetic acid is synthesized from L-tryptophan in roots of Arabidopsis thaliana. Planta 206:362–369 [DOI] [PubMed] [Google Scholar]
- Muto H, Watahiki MK, Nakamoto D, Kinjo M, Yamamoto KT 2007. Specificity and similarity of functions of the Aux/IAA genes in auxin signaling of Arabidopsis revealed by promoter-exchange experiments among MSG2/IAA19, AXR2/IAA7, and SLR/IAA14. Plant Physiol 144:187–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal P, Walker LM, Young JC, Sonawala A, Timpte C, Estelle M, Reed JW 2000. AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol 123:563–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemhauser JL, Mockler TC, Chory J 2004. Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol 2:E258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwland J, Maughan S, Dewitte W, Scofield S, Sanz L, Murray JA 2009. The D-type cyclin CYCD4;1 modulates lateral root density in Arabidopsis by affecting the basal meristem region. Proc Natl Acad Sci 106:22528–22533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanly J 2009. Approaching cellular and molecular resolution of auxin biosynthesis and metabolism. Cold Spring Harbor Perspectives in Biology 1:a001594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y, Fukaki H, Onoda M, Theologis A, Tasaka M 2007. ARF7 and ARF19 regulate lateral root formation via direct activation of LBD/ASL genes in Arabidopsis. Plant Cell 19:118–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okushima Y, Overvoorde PJ, Arima K, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Lui A, Nguyen D 2005. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell 17:444–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellet F, Overvoorde PJ, Theologis A 2001. IAA17/AXR3: biochemical insight into an auxin mutant phenotype. Plant Cell 13:829–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overvoorde PJ, Okushima Y, Alonso JM, Chan A, Chang C, Ecker JR, Hughes B, Liu A, Onodera C, Quach H, et al. 2005. Functional genomic analysis of the AUXIN/INDOLE-3-ACETIC ACID gene family members in Arabidopsis thaliana. Plant Cell 17:3282–3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paponov IA, Teale WD, Trebar M, Blilou I, Palme K 2005. The PIN auxin efflux facilitators: evolutionary and functional perspectives. Trends in Plant Science 10:170–177 [DOI] [PubMed] [Google Scholar]
- Peret B, De Rybel B, Casimiro I, Benkova E, Swarup R, Laplaze L, Beeckman T, Bennett MJ 2009. Arabidopsis lateral root development: an emerging story. Trends in Plant Science 14:399–408 [DOI] [PubMed] [Google Scholar]
- Perilli S, Moubayidin L, Sabatini S 2010. The molecular basis of cytokinin function. Curr Opin Plant Biol 13:21–26 [DOI] [PubMed] [Google Scholar]
- Petersson SV, Johansson AI, Kowalczyk M, Makoveychuk A, Wang JY, Moritz T, Grebe M, Benfey PN, Sandberg G, Ljung K 2009. An auxin gradient and maximum in the Arabidopsis root apex shown by high-resolution cell-specific analysis of IAA distribution and synthesis. Plant Cell 21:1659–1668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrasek J, Friml J 2009. Auxin transport routes in plant development. Development 136:2675–2688 [DOI] [PubMed] [Google Scholar]
- Petrasek J, Mravec J, Bouchard R, Blakeslee JJ, Abas M, Seifertova D, Wisniewska J, Tadele Z, Kubes M, Covanova M, et al. 2006. PIN proteins perform a rate-limiting function in cellular auxin efflux. Science 312:914–918 [DOI] [PubMed] [Google Scholar]
- Pitts RJ, Cernac A, Estelle M 1998. Auxin and ethylene promote root hair elongation in Arabidopsis. Plant J 16:553–560 [DOI] [PubMed] [Google Scholar]
- Ploense SE, Wu MF, Nagpal P, Reed JW 2009. A gain-of-function mutation in IAA18 alters Arabidopsis embryonic apical patterning. Development 136:1509–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quint M, Barkawi LS, Fan KT, Cohen JD, Gray WM 2009. Arabidopsis IAR4 modulates auxin response by regulating auxin homeostasis. Plant Physiol 150:748–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman A, Hosokawa S, Oono Y, Amakawa T, Goto N, Tsurumi S 2002. Auxin and ethylene response interactions during Arabidopsis root hair development dissected by auxin influx modulators. Plant Physiol 130:1908–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed RC, Brady SR, Muday GK 1998. Inhibition of auxin movement from the shoot into the root inhibits lateral root development in Arabidopsis. Plant Physiol 118:1369–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter GL, Monshausen GB, Krol A, Gilroy S 2009. Mechanical stimuli modulate lateral root organogenesis. Plant Physiol 151:1855–1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robert HS, Friml J 2009. Auxin and other signals on the move in plants. Nat Chem Biol 5:325–332 [DOI] [PubMed] [Google Scholar]
- Rogg LE, Lasswell J, Bartel B 2001. A gain-of-function mutation in IAA28 suppresses lateral root development. Plant Cell 13:465–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse D, Mackay P, Stirnberg P, Estelle M, Leyser O 1998. Changes in auxin response from mutations in an AUX/IAA gene. Science 279:1371–1373 [DOI] [PubMed] [Google Scholar]
- Sabatini S, Beis D, Wolkenfelt H, Murfett J, Guilfoyle T, Malamy J, Benfey P, Leyser O, Bechtold N, Weisbeek P, Scheres B 1999. An auxin-dependent distal organizer of pattern and polarity in the Arabidopsis root. Cell 99:463–472 [DOI] [PubMed] [Google Scholar]
- Scacchi E, Osmont KS, Beuchat J, Salinas P, Navarrete-Gomez M, Trigueros M, Ferrandiz C, Hardtke CS 2009. Dynamic, auxin-responsive plasma membrane-to-nucleus movement of Arabidopsis BRX. Development 136:2059–2067 [DOI] [PubMed] [Google Scholar]
- Seidel C, Walz A, Park S, Cohen JD, Ludwig-Muller J 2006. Indole-3-acetic acid protein conjugates: novel players in auxin homeostasis. Plant Biol (Stuttg) 8:340–345 [DOI] [PubMed] [Google Scholar]
- Sena G, Wang X, Liu HY, Hofhuis H, Birnbaum KD 2009. Organ regeneration does not require a functional stem cell niche in plants. Nature 457:1150–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin R, Burch AY, Huppert KA, Tiwari SB, Murphy AS, Guilfoyle TJ, Schachtman DP 2007. The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell 19:2440–2453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE 2009. The tryptophan conjugates of jasmonic and indole-3-acetic acids are endogenous auxin inhibitors. Plant Physiol 150:1310–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Hoyt JM, Hamilton AA, Alonso JM 2005. A Link between ethylene and auxin uncovered by the characterization of two root-specific ethylene-insensitive mutants in Arabidopsis. Plant Cell 17:2230–2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Robertson-Hoyt J, Yun J, Benavente LM, Xie DY, Dolezal K, Schlereth A, Jurgens G, Alonso JM 2008. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133:177–191 [DOI] [PubMed] [Google Scholar]
- Strader LC, Bartel B 2009. The Arabidopsis PLEIOTROPIC DRUG RESISTANCE8/ABCG36 ATP binding cassette transporter modulates sensitivity to the auxin precursor indole-3-butyric acid. Plant Cell 21:1992–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarup K, Benkova E, Swarup R, Casimiro I, Peret B, Yang Y, Parry G, Nielsen E, De Smet I, Vanneste S, et al. 2008. The auxin influx carrier LAX3 promotes lateral root emergence. Nat Cell Biol 10:946–954 [DOI] [PubMed] [Google Scholar]
- Swarup R, Friml J, Marchant A, Ljung K, Sandberg G, Palme K, Bennett M 2001. Localization of the auxin permease AUX1 suggests two functionally distinct hormone transport pathways operate in the Arabidopsis root apex. Genes Dev 15:2648–2653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szemenyei H, Hannon M, Long JA 2008. TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319:1384–1386 [DOI] [PubMed] [Google Scholar]
- Tan X, Calderon-Villalobos LI, Sharon M, Zheng C, Robinson CV, Estelle M, Zheng N 2007. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 446:640–645 [DOI] [PubMed] [Google Scholar]
- Tao Y, Ferrer JL, Ljung K, Pojer F, Hong F, Long JA, Li L, Moreno JE, Bowman ME, Ivans LJ, et al. 2008. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133:164–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatematsu K, Kumagai S, Muto H, Sato A, Watahiki MK, Harper RM, Liscum E, Yamamoto KT 2004. MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana. Plant Cell 16:379–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimann KV, Went FW 1934. On the chemical nature of the root-forming hormone of plants. Proc Kon Akad Wetensch, Amsterdam37:456–458 [Google Scholar]
- Tian Q, Reed JW 1999. Control of auxin-regulated root development by the Arabidopsis thaliana SHY2/IAA3 gene. Development 126:711–721 [DOI] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle T 2003. The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15:533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tromas A, Braun N, Muller P, Khodus T, Paponov IA, Palme K, Ljung K, Lee JY, Benfey P, Murray JA, et al. 2009. The AUXIN BINDING PROTEIN 1 is required for differential auxin responses mediating root growth. PLoS One 4:e6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurumi S, Wada S 1980. Transport of shoot- and cotyledon-applied indole-3-acetic acid to Vicia faba root. Plant Cell Physiol 21:803–816 [DOI] [PubMed] [Google Scholar]
- Uehara T, Okushima Y, Mimura T, Tasaka M, Fukaki H 2008. Domain II mutations in CRANE/IAA18 suppress lateral root formation and affect shoot development in Arabidopsis thaliana. Plant Cell Physiol 49:1025–1038 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ 1997. ARF1, a transcription factor that binds to auxin response elements. Science 276:1865–1868 [DOI] [PubMed] [Google Scholar]
- Ulmasov T, Hagen G, Guilfoyle TJ 1999. Activation and repression of transcription by auxin-response factors. Proc Natl Acad Sci 96:5844–5849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg C, Willemsen V, Hendriks G, Weisbeek P, Scheres B 1997. Short-range control of cell differentiation in the Arabidopsis root meristem. Nature 390:287–289 [DOI] [PubMed] [Google Scholar]
- Vanneste S, De Rybel B, Beemster GT, Ljung K, De Smet I, Van Isterdael G, Naudts M, Iida R, Gruissem W, Tasaka M, et al. 2005. Cell cycle progression in the pericycle is not sufficient for SOLITARY ROOT/IAA14-mediated lateral root initiation in Arabidopsis thaliana. Plant Cell 17:3035–3050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste S, Friml J 2009. Auxin: a trigger for change in plant development. Cell 136:1005–1016 [DOI] [PubMed] [Google Scholar]
- Vert G, Walcher CL, Chory J, Nemhauser JL 2008. Integration of auxin and brassinosteroid pathways by Auxin Response Factor 2. Proc Natl Acad Sci U S A 105:9829–9834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JW, Wang LJ, Mao YB, Cai WJ, Xue HW, Chen XY 2005. Control of root cap formation by MicroRNA-targeted auxin response factors in Arabidopsis. Plant Cell 17:2204–2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D, Benkova E, Jager KE, Schlereth A, Hamann T, Kientz M, Wilmoth JC, Reed JW, Jurgens G 2005. Developmental specificity of auxin response by pairs of ARF and Aux/IAA transcriptional regulators. EMBO J 24:1874–1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weijers D, Schlereth A, Ehrismann JS, Schwank G, Kientz M, Jurgens G 2006. Auxin triggers transient local signaling for cell specification in Arabidopsis embryogenesis. Dev Cell 10:265–270 [DOI] [PubMed] [Google Scholar]
- Went FW 1929. On a substance causing root formation. Proc Kon Akad Wetensch, Amsterdam32:35–39 [Google Scholar]
- Williams L, Carles CC, Osmont KS, Fletcher JC 2005. A database analysis method identifies an endogenous trans-acting short-interfering RNA that targets the Arabidopsis ARF2, ARF3, and ARF4 genes. Proc Natl Acad Sci 102:9703–9708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilmoth JC, Wang S, Tiwari SB, Joshi AD, Hagen G, Guilfoyle TJ, Alonso JM, Ecker JR, Reed JW 2005. NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. Plant J 43:118–130 [DOI] [PubMed] [Google Scholar]
- Woodward AW, Bartel B 2005. Auxin: regulation, action, and interaction. Ann Bot 95:707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Lewis DR, Spalding EP 2007. Mutations in Arabidopsis multidrug resistance-like ABC transporters separate the roles of acropetal and basipetal auxin transport in lateral root development. Plant Cell 19:1826–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Hofhuis H, Heidstra R, Sauer M, Friml J, Scheres B 2006. A molecular framework for plant regeneration. Science 311:385–388 [DOI] [PubMed] [Google Scholar]
- Yamada M, Greenham K, Prigge MJ, Jensen PJ, Estelle M 2009. The TRANSPORT INHIBITOR RESPONSE2 gene is required for auxin synthesis and diverse aspects of plant development. Plant Physiol 151:168–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Lee S, So JH, Dharmasiri S, Dharmasiri N, Ge L, Jensen C, Hangarter R, Hobbie L, Estelle M 2004. The IAA1 protein is encoded by AXR5 and is a substrate of SCFTIR1. Plant J 40:772–782 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Christensen SK, Fankhauser C, Cashman JR, Cohen JD, Weigel D, Chory J 2001. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science 291:306–309 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Hull AK, Gupta NR, Goss KA, Alonso J, Ecker JR, Normanly J, Chory J, Celenza JL 2002. Trp-dependent auxin biosynthesis in Arabidopsis: involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev 16:3100–3112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Martinez N, Millius A, Adham AR, Bartel B 2008. Identification and characterization of Arabidopsis indole-3-butyric acid response mutants defective in novel peroxisomal enzymes. Genetics 180:237–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Nyberg M, Bartel B 2007. IBR3, a novel peroxisomal acyl-CoA dehydrogenase-like protein required for indole-3-butyric acid response. Plant Mol Biol 64:59–72 [DOI] [PubMed] [Google Scholar]
- Zolman BK, Yoder A, Bartel B 2000. Genetic analysis of indole-3-butyric acid responses in Arabidopsis thaliana reveals four mutant classes. Genetics 156:1323–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]