Abstract
Recent genetic evidence has established a pathogenetic role for NF-κB signaling in cancer. NF-κB signaling is engaged transiently when normal B lymphocytes respond to antigens, but lymphomas derived from these cells accumulate genetic lesions that constitutively activate NF-κB signaling. Many genetic aberrations in lymphomas alter CARD11, MALT1, or BCL10, which constitute a signaling complex that is intermediate between the B-cell receptor and IκB kinase. The activated B-cell-like subtype of diffuse large B-cell lymphoma activates NF-κB by a variety of mechanisms including oncogenic mutations in CARD11 and a chronic active form of B-cell receptor signaling. Normal plasma cells activate NF-κB in response to ligands in the bone marrow microenvironment, but their malignant counterpart, multiple myeloma, sustains a variety of genetic hits that stabilize the kinase NIK, leading to constitutive activation of the classical and alternative NF-κB pathways. Various oncogenic abnormalities in epithelial cancers, including mutant K-ras, engage unconventional IκB kinases to activate NF-κB. Inhibition of constitutive NF-κB signaling in each of these cancer types induces apoptosis, providing a rationale for the development of NF-κB pathway inhibitors for the treatment of cancer.
Mutations in upstream signaling molecules such as CARD11 activate NF-κB transcription factors in many lymphomas. NF-κB inhibitors cause apoptosis and are therefore promising anticancer drugs.
Given the crucial role of NF-κB in signaling downstream of a multitude of surface receptors, cancer inevitably has found mechanisms to co-opt this pathway. NF-κB plays an important role in the initiation and promotion of cancer by fostering an inflammatory milieu in which various cytokines aid and abet malignant transformation (reviewed in Karin 2010; Karin et al. 2006). Some cancers are caused by viruses that encode activators of the NF-κB pathway, which block the cell death inherent in viral transformation (reviewed in Hiscott et al. 2006). In this article, I focus on mechanisms by which NF-κB is aberrantly and stably activated by genetic lesions in human cancer. The selective advantage imparted to a tumor cell on engagement of the NF-κB pathway derives in large measure from the ability of this pathway to block apoptosis. In a variety of lymphoid cancers, NF-κB is constitutively active owing to diverse somatic mutations, genomic amplifications and deletions, and chromosomal translocations. These abnormalities subvert the normal function of NF-κB in immune cell signaling. An oncogenic role for NF-κB has surfaced in epithelial cancers as well. This emerging genetic evidence shows that the NF-κB pathway is central to the pathogenesis of many cancer types, providing impetus for the development of therapeutics targeting this pathway.
NF-κB signaling can be dichotomized into a “classical” pathway in which IκB kinase β (IKKβ) phosphorylates IκBα and an “alternative” NF-κB pathway in which IKKα phosphorylates the p100 precursor of the NF-κB p52 subunit. The IKK complex in the classical pathway requires the regulatory IKKγ subunit, whereas the IKK complex in the alternative pathway does not. The result of these signaling events is the accumulation of the heterodimeric NF-κB transcription factors in the nucleus, with the classical pathway regulating mainly p50/p65 and p50/c-Rel dimers and the alternative pathway regulating p52/relB dimers. In addition, NF-κB can be activated by other kinases, including the unconventional IKK family members, IKKε and TBK1, although the exact mechanisms linking these kinases to NF-κB activation need clarification. Numerous signaling pathways converge on these NF-κB regulators, providing ample means by which cancers can aberrantly stimulate NF-κB.
NF-κB IN LYMPHOMA
As outlined in the following discussion, many subtypes of human lymphoma rely on constitutive activity of the NF-κB pathway for survival. This dependency likely has its roots in the pervasive role of the NF-κB pathway in normal B-cell maturation and activation. Genetic deletion of NF-κB subunits in B cells blocks B-cell differentiation at a variety of steps, depending on which subunit is ablated (reviewed in Vallabhapurapu et al. 2009). The alternative NF-κB pathway is activated in response to exposure of B cells to BAFF, a tumor necrosis factor (TNF) family member made by myeloid-derived cells in secondary lymphoid organs. Signals from BAFF are essential for development of mature follicular B cells from transitional B cells (Claudio et al. 2002). NF-κB is also required for the maintenance of all mature resting B cells because conditional deletion of the IKKβ or IKKγ subunits causes B cells to be lost from the follicular compartment (Pasparakis et al. 2002), and a small molecule inhibitor of IKKβ depletes the mature B-cell pool (Nagashima et al. 2006). During antigenic challenge, the classical NF-κB pathway is strongly activated by B-cell receptor signaling, via formation of the “CBM” signaling complex consisting of CARD11, MALT1, and BCL10 (Thome 2004). The CBM pathway is pathologically altered in several lymphoma subtypes.
Roughly 90% of human lymphomas arise from B lymphocytes at various stages of differentiation, with the remainder derived from T lymphocytes. The most prevalent type of non-Hodgkin’s lymphoma is diffuse large B-cell lymphoma, comprising ∼40% of cases. This diagnostic category harbors several molecularly and clinically distinct diseases, as originally defined by gene expression profiling (Alizadeh et al. 2000). The three well-delineated DLBCL subtypes are termed germinal center B-cell-like (GCB) DLBCL, activated B-cell-like (ABC) DLBCL, and primary mediastinal B-cell lymphoma (PMBL). These subtypes apparently arise from different stages of normal B-cell differentiation and use distinct oncogenic pathways (Staudt et al. 2005). The DLBCL subtypes respond differentially to standard chemotherapeutic regimens (Lenz et al. 2008) and to regimens incorporating newer, more targeted agents (Dunleavy et al. 2009).
ABC DLBCL
A hallmark of ABC DLBCL is constitutive activation of the NF-κB pathway (Davis et al. 2001). Gene expression profiling of tumor biopsy samples revealed preferential expression of NF-κB target genes in ABC DLBCL compared with GCB DLBCL, and this was also true for cell line models of ABC DLBCL. ABC DLBCLs engage the classical NF-κB pathways because they have rapid phosphorylation and turnover of IκBα and prominent nuclear accumulation of p50/p65 heterodimers with lesser accumulation of p50/c-rel heterodimers (Davis et al. 2001). Interruption of NF-κB signaling with an IκB super repressor or with a small molecule inhibitor of IKKβ induces apoptosis in ABC DLBCL but not GCB DLBCL cell lines (Davis et al. 2001; Lam et al. 2005).
A biological consequence of NF-κB signaling in ABC DLBCL is to propel the malignant cell forward toward the plasma cell stage of differentiation. An NF-κB target in ABC DLBCL is IRF4 (Lam et al. 2005), a key transcription factor that drives plasmacytic differentiation (Klein et al. 2006; Sciammas et al. 2006; Shaffer et al. 2008; Shaffer et al. 2009). In normal lymphocytes, IRF4 drives terminal differentiation by transactivating PRDM1, which encodes Blimp-1 (Sciammas et al. 2006; Shaffer et al. 2008), another master regulator of plasmacytic differentiation (Shaffer et al. 2002; Shapiro-Shelef et al. 2005; Shapiro-Shelef et al. 2003; Shapiro-Shelef et al. 2005; Turner et al. 1994). Together, IRF4 and Blimp-1 directly or indirectly promote expression of XBP-1, a transcription factor that specifies the secretory phenotype (Iwakoshi et al. 2003; Reimold et al. 2001; Shaffer et al. 2004). Interestingly, ABC DLBCLs express IRF4 and XBP-1, but frequently have genetic lesions that inactivate Blimp-1 (Iqbal et al. 2007; Pasqualucci et al. 2006; Shaffer et al. 2000; Tam et al. 2006). These observations suggest a model in which NF-κB activation in ABC DLBCL transactivates IRF4 and initiates plasmacytic differentiation, but full plasmacytic differentiation is blocked by lesions that inactivate Blimp-1. In this way, the tumor escapes the cell cycle arrest that typifies normal plasma cells, which is due in part to repression of MYC by Blimp-1 (Lin et al. 1997).
Another important influence of NF-κB signaling on ABC DLBCL biology is the production of the cytokines IL-6 and IL-10 (Ding et al. 2008; Lam et al. 2008). Both cytokines are secreted by ABC DLBCL cells and signal in an autocrine fashion, activating the transcription factor STAT3. A signature of STAT3 target genes typifies a subset of ABC DLBCL tumors, and these tumors have high STAT3 protein levels and have phosphorylated STAT3 in the nucleus (Lam et al. 2008). In contrast, GCB DLBCL biopsies lack both STAT3 target gene expression and phosphorylated STAT3. ABC DLBCLs with STAT3 activation also have higher expression of NF-κB target genes, in keeping with the fact that STAT3 physically interacts with NF-κB factors, thereby increasing their ability to transactivate their targets (Yang et al. 2007). As a consequence of this interaction, a JAK kinase inhibitor, which extinguished STAT3 phosphorylation, synergized with an IKKβ inhibitor in killing ABC DLBCL cells (Lam et al. 2008).
Engagement of the CARD11/BCL10/MALT1 Signaling Module in ABC DLBCL
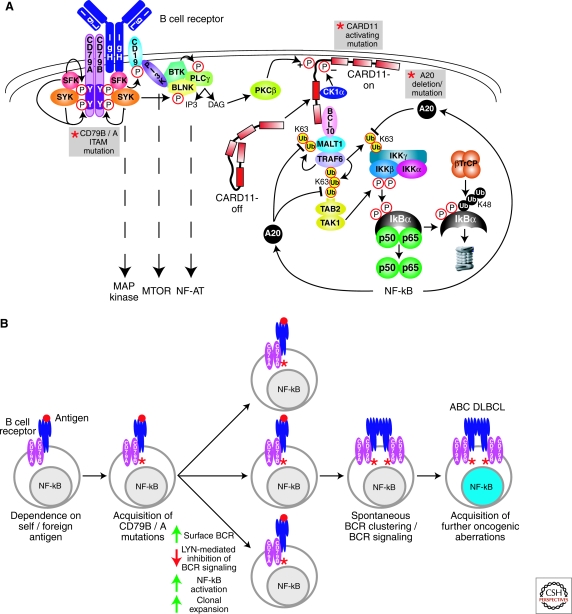
The mechanisms responsible for constitutive NF-κB signaling in ABC DLBCL have been elucidated by RNA interference genetic screens (Ngo et al. 2006). In these so-called “Achilles heel” screens, shRNAs are screened for those that block cancer cell proliferation and survival. An initial Achilles heel screen revealed toxicity of shRNAs targeting CARD11, MALT1, and BCL10 for ABC but not GCB DLBCL cell lines (Ngo et al. 2006). In normal lymphocytes, these three proteins form a “CBM” signaling complex that is required for NF-κB signaling downstream of the antigen receptors (reviewed in Blonska et al. 2009; Rawlings et al. 2006; Thome 2004) (Fig. 1A). CARD11 is a multidomain signaling scaffold protein, consisting of an amino-terminal CARD domain, a coiled-coil domain, and a carboxy-terminal MAGUK domain. In a resting lymphocyte, CARD11 resides in a latent form in the cytoplasm. On antigen receptor engagement, CARD11 is phosphorylated in a “linker” region residing between its coiled-coil and MAGUK domains (Matsumoto et al. 2005; Sommer et al. 2005). In B cells, PKCβ carries out this function, whereas PKCθ is responsible in T cells. Phosphorylated CARD11 relocalizes to the plasma membrane, where it recruits a multiprotein complex consisting of MALT1, BCL10, TRAF6, TAK1, caspase 8, and c-Flip (McCully et al. 2008). A series of ubiquitin-dependent interactions leads to IKK activation (reviewed in Chen 2005). On recruitment to the CBM complex, TRAF6 becomes active as a ubiquitin ligase, attaching K63-linked polyubiquitin chains to itself and MALT1 (Oeckinghaus et al. 2007; Sun et al. 2004). The IKK complex is subsequently recruited, most likely using its ubiquitin binding domain to attach to ubiquitinated TRAF6 and MALT1. TRAF6 ubiquitinates IKKγ, which is a required step in its activation as a kinase. Ubiquitinated TRAF6 also interacts with TAB2, leading to activation of the associated kinase TAK1. TAK1 carries out the second required step in IKK activation, phosphorylation of IKKβ in its activation loop.
Figure 1.
Role of BCR signaling to NF-κB in ABC DLBCL. (A) Schematic of BCR signaling to NF-κB. Recurrent genetic alterations in ABC DLBCL that result in constitutive NF-κB activation are indicated by the gray boxes. Proximal signaling by the BCR is initiated by SRC-family kinases (SFK; e.g., LYN, FYN, FGR, and BLK), which phosphorylate the ITAM motifs in the CD79A and CD79B components of the BCR receptor. SYK is recruited to the phosphorylated ITAMs and activated to phosphorylate many downstream proteins. The PI(3) kinase pathway is activated by SRC-family kinase phosphorylation of the BCR coreceptor CD19. The generation of PIP3 by PI(3) kinase recruits BTK and associated BLNK and phospholipase Cγ2 (PLCγ2) to the plasma membrane. PLCγ2 generates inositol triphosphate (IP3), which leads to opening of the capacitative calcium channel, thereby activating the NF-AT pathway. Diacylglycerol (DAG) is also generated, which activates protein kinase Cβ (PKCβ). PKCβ phosphorylates the latent form of CARD11 (CARD-off) in the cytoplasm, causing it to adopt an active conformation (CARD11-on), translocate to the plasma membrane, and recruit the signaling adapters BCL10 and MALT1. MALT1 binds TRAF6, causing TRAF6 to catalyze K63-linked polyubiquitination of the IKKγ subunit, MALT1 and itself. TAB2 recognizes the polyubiquitin chains on TRAF6, leading to phosphorylation of IKKβ in its activation loop by TAK1 kinase. Ubiquitination of IKKγ and phosphorylation of IKKβ activate IKKβ to phosphorylate IκBα. Phosphorylated IκBα is ubiquitinated by the ubiquitin ligase βTrCP, leading to its proteasomal degradation. Nuclear NF-κB heterodimers activate multiple target genes, including A20. A20 terminates NF-κB signaling by removing K63-linked ubiquitin chains from IKKγ, TRAF6, and MALT1, and attaching K48-linked ubiquitin chains. (B) The role of chronic active BCR signaling in the pathogenesis of ABC DLBCL. Genetic mutations in CD79B and CD79A ITAM regions in ABC DLBCLs suggest that BCR signaling is key to the pathogenesis of ABC DLBCL. CD79B and CD79A ITAM mutations in the mouse cause hyperactive BCR signaling, suggesting that CD79 ITAM mutations in ABC DLBCL may amplify antigen-stimulated BCR signaling. CD79 ITAM mutations increase surface BCR expression and decrease activation of LYN kinase, a negative regulator of BCR signaling, potentially resulting in increased signaling to NF-κB and greater clonal expansion. A separate step leading to chronic active BCR signaling in ABC DLBCL is the acquisition of spontaneous BCR clustering, which is not caused by the ITAM mutations. The BCR clustering phenotype could theoretically be acquired either before or after the CD79 mutations. Finally, ABC DLBCLs must acquire additional oncogenic hits to become fully malignant.
RNA interference screening in DLBCL also revealed casein kinase 1α (CK1α) as a new component of the CBM pathway (Bidere et al. 2009). shRNAs targeting CK1α were toxic for ABC but not GCB DLBCL cell lines and blocked the formation of the CBM complex in activated lymphocytes. CK1α performs two opposing functions in this pathway. First, CK1α interacts with CARD11 and promotes the formation of the CBM complex; this function does not require its kinase activity. Second, CK1α phosphorylates serine 608 in the CARD11 linker region, decreasing NF-κB signaling, perhaps by enhancing turnover of CARD11. Given these opposing activities, the only effective way in which to target CK1α therapeutically would be to inhibit its interaction with CARD11.
Oncogenic CARD11 Mutations
Whereas the function of the CBM complex in normal antigen receptor signaling is transient, being limited by several negative feedback loops, the CBM complex is continuously required for the survival of ABC DLBCL cells. An explanation of this paradox was revealed by the discovery of somatic CARD11 mutations in DLBCL (Lenz et al. 2008). CARD11 mutations are present in ∼10% of ABC DLBCLs, and all mutations are confined to the coiled-coil domain. At a lower frequency, CARD11 mutations are present in GCB DLBCLs, and these rare cases have high NF-κB target gene expression, unlike most GCB DLBCLs. The CARD11 mutants can constitutively activate NF-κB when introduced into heterologous cells. Moreover, these mutants potentiated the activation of NF-κB following antigen receptor stimulation.
The oncogenic activation of CARD11 by mutations DLBCL is likely explained by their effect on CARD11 subcellular localization (Lenz et al. 2008). Whereas wild-type CARD11 localizes diffusely in the cytoplasm, the CARD11 mutations form prominent cytoplasmic aggregates. These aggregates colocalize with other components of the CBM signalosome, including MALT1 and phosphorylated IKK, suggesting that they are sites of active signaling. The quantitative degree of aggregation by the various CARD11 mutations correlates directly with the degree of IKK kinase activity, again suggesting the aggregates are functional.
The coiled-coil domain mediates several essential functions in CARD11 (Tanner et al. 2007). Point mutations in the coiled-coil can destroy the function of the protein in lymphocyte activation (Jun et al. 2003). Certain mutations in the coiled-coil domain impair its self association, whereas others block membrane localization of CARD11 (Tanner et al. 2007). The coiled-coil domain appears to be under the negative influence of the linker domain. Deletion of the linker regions results in a constitutively active form of CARD11 (McCully et al. 2008; Sommer et al. 2005), suggesting that the linker region may fold back on the coiled-coil domain, inhibiting its function. Indeed, an isolated linker peptide can associate with a peptide consisting of the amino-terminal CARD and coiled-coil domains (McCully et al. 2008). Certain phosphorylations of the linker domain activate CARD11 (Matsumoto et al. 2005; Sommer et al. 2005), and these presumably interfere with the association of the linker regions with the coiled-coil domain, although this needs to be directly tested experimentally. Once relieved of the negative influence of the linker region, the coiled-coil domain contributes to binding of BCL10, Caspase-8, TRAF6, and IKKγ (McCully et al. 2008).
These results suggest a simple model in which the CARD11 mutations in DLBCL prevent association of the coiled-coil and linker domains, creating an isoform that binds several NF-κB pathway components constitutively. Less clear is why the CARD11 mutations cause the protein to form such prominent cytosoloic aggregates. Presumably, this represents uncontrolled lattice formation involving both the coiled-coil domain and the carboxy-terminal MAGUK domain. MAGUK proteins form large protein interaction lattices that contribute to the postsynaptic density in neurons and tight junctions (Funke et al. 2005). By analogy, the CARD11 aggregates may be signaling lattices that massively nucleate components of the NF-κB signaling cascade. Therapeutic targeting of CARD11 coiled-coil domain interactions should be considered because overexpression of an isolated coiled-coil domain abrogates NF-κB signaling and kills ABC DLBCL cell lines with either wild-type or mutant CARD11 (Lenz et al. 2008). A structural analysis of the CARD11 coiled-coil domain will be required to determine how a small molecule might interfere with the function of the mutant coiled-coil domains.
Chronic Active B-cell Receptor Signaling
In the majority of ABC DLBCLs, CARD11 is not mutated, yet the tumors have high expression of NF-κB target genes. Further, ABC DLBCL cell lines with wild-type CARD11 are killed by CARD11 knockdown. These observations led to the hypothesis that an upstream signaling pathway activates CARD11 and NF-κB in these cases (Davis et al. 2010). An RNA interference screen revealed that the kinase BTK is required for NF-κB signaling in ABC DLBCLs with wild-type CARD11. Mutations in BTK cause a failure of B-cell production in Bruton’s X-linked agammaglobulinemia (Satterthwaite et al. 2000). BTK is also a key kinase required to connect B-cell receptor (BCR) signaling to NF-κB (Bajpai et al. 2000; Petro et al. 2000). Following BCR signaling, BTK forms a complex with the adapter BLNK and phospholipase Cγ2 (PLCγ2). PLCγ2 then produces the second messenger diacyl glycerol, which activates PKCβ, leading to CARD11 phosphorylation and NF-κB signaling. In keeping with this model, mouse B-cells deficient in BLNK, PLCγ2, and PKCβ are all defective in NF-κB activation (Leitges et al. 1996; Petro et al. 2001; Saijo et al. 2002; Tan et al. 2001).
ABC DLBCLs with wild-type CARD11 activate BTK as a consequence of signals emanating from the BCR itself. In these lymphoma cells, knockdown of the immunoglobulin heavy or light chains is lethal, as is knockdown of the essential signaling subunits of the BCR, CD79A (Ig-α) and CD79B (Ig-β) (Davis et al. 2010). BCR disruption blocks several downstream signaling pathways in these cells, including NF-κB, AKT/mTOR, ERK MAP kinase, and NF-AT. Given that BCR signaling engages these potent cell growth and survival pathways, it is perhaps not surprising that lymphomas have found ways to use BCR signaling pathologically.
Total internal reflection microscopy revealed that the BCRs in these lymphoma cells form prominent clusters in the plasma membrane that have low diffusion (Davis et al. 2010). These are likely to be the sites of active BCR signaling because the clusters colocalize with phosphotyrosine. In contrast, the BCRs in cell lines representing GCB DLBCL, Burkitt’s lymphoma, and mantle cell lymphoma are distributed diffusely in the plasma membrane. In normal B cells, interaction with a membrane-bound antigen causes the BCRs to form immobile clusters within seconds of contact (Tolar et al. 2009). Hence, the clustered BCRs in ABC DLBCLs could represent the influence of an antigen. Alternatively, these lymphoma cells may have defects in the regulation of BCR assembly in the plasma membrane.
The constitutive BCR activity in these ABC DLBCLs has been termed “chronic active BCR signaling” to deliberately distinguish it from “tonic” BCR signaling. Tonic BCR signaling was initially defined based on experiments in which the BCR was conditionally ablated in mouse B cells (Kraus et al. 2004; Lam et al. 1997). In these mice, all mature B cells disappeared over the course of 1–2 weeks, demonstrating an ongoing requirement for BCR signaling in the survival of mature B cells. Tonic BCR signaling most likely does not require antigen engagement by the BCR because artificial BCR mimics that lack immunoglobulin components can rescue the development and survival of B cells that lack immunoglobulin (Monroe 2006). Recent experiments implicate PI(3) kinase signaling as an important survival signal that is delivered by tonic BCR signaling (Srinivasan et al. 2009). An activated allele of the p110 subunit of PI(3) kinase could rescue B cells following BCR ablation but an activated IKKβ allele could not. Nonetheless, NF-κB may still contribute to tonic signaling because conditional ablation of IKKβ or IKKγ in mouse B-cells impairs their survival (Pasparakis et al. 2002).
Chronic active BCR signaling can be distinguished from tonic BCR signaling by two main criteria. First, tonic BCR signaling does not appear to require the CBM complex because mice deficient in CBM components have relatively normal numbers of follicular B cells (Thome 2004), unlike mice in which tonic signaling has been blocked. Because the BCR signaling in ABC DLBCL acts through the CBM complex, it cannot be characterized as “tonic.” Second, tonic BCR signaling in the mouse is required to maintain resting mature B cells, which have unclustered BCRs. In contrast, the BCRs in ABC DLBCLs are clustered, more akin to antigen-stimulated B cells than to resting B cells. Although it remains possible that tonic BCR signaling is required for the survival of some lymphoma subtypes, it does not appear to play a role in the pathogenesis of ABC DLBCL.
Cancer gene resequencing has revealed recurrent somatic mutations in BCR components in ABC DLBCL, providing a genetic “smoking gun” implicating BCR signaling in the pathogenesis of this lymphoma subtype (Davis et al. 2010). In over one fifth of ABC DLBCL specimens, somatic mutations are present in CD79B and less frequently in CD79A, but such mutations are rare or absent in other lymphoma subtypes. All mutations affect the critical “ITAM” signaling motifs of CD79A and CD79B. The ITAM motif is an evolutionarily conserved signaling module in several immune receptors that includes two invariant tyrosine residues (Reth 1989). SRC-family tyrosine kinases phosphorylate the ITAM tyrosines and then become further active as kinases when they bind the phosphorylated ITAMs through their SH2 domains. SYK is also recruited via its tandem SH2 domains to phosphorylated ITAMs, becoming active as a kinase in the process. One sixth of ABC DLBCLs have mutations that change the first ITAM tyrosine of CD79B to a variety of other amino acids (Davis et al. 2010). One ABC DLBCL tumor was identified that has a surgical three-base-pair deletion that removed this tyrosine, further accentuating its importance. At a lower frequency, ABC DLBCL tumors have point mutations affecting other conserved CD79B ITAM residues or deletions that remove all or part of the ITAM regions of CD79B or CD79A.
The CD79 mutations in ABC DLBCL are not loss-of-function because they sustain survival of ABC DLBCL cells and promote signaling to NF-κB (Davis et al. 2010). Unlike CARD11 coiled-coil mutants, however, the CD79 mutants do not activate NF-κB when introduced into a heterologous cell type. Clues to the function of the CD79 mutations come from studies of knockin mice with loss or mutation of CD79A or CD79B ITAMs (Gazumyan et al. 2006; Kraus et al. 1999; Torres et al. 1999). Unexpectedly, mature B cells develop in these animals despite disruption of one or the other CD79 ITAM motif. In fact, B cells from these mice are hyperresponsive to certain antigenic stimuli. For example, mice with both CD79B ITAM residues changed to alanine have higher serum IgM levels and respond with 10-fold higher antibody titers to immunization with a T cell-independent antigen (Gazumyan et al. 2006). Further, B cells from these mice have higher expression of the BCR on the cell surface because of decreased receptor internalization. This latter phenotype is also a feature of the CD79 mutants from ABC DLBCL tumors: Reconstitution of the BCR with a CD79B mutant in the first ITAM tyrosine yields higher BCR expression than reconstitution with wild-type CD79B (Davis et al. 2010). In contrast, a CD79B mutant affecting the second ITAM tyrosine, which has not been observed in ABC DLBCL tumors, does not raise surface BCR expression, highlighting the functional specificity of the human CD79B mutants. Chronic active BCR signaling lowers surface expression of the BCRs containing wild-type CD79B but not those with mutant CD79B, suggesting that CD79B mutations may have been selected in part to prevent signaling-induced receptor internalization (Davis et al. 2010).
A second functional attribute of the CD79 mutations in ABC DLBCL is to block negative autoregulation by the kinase LYN. LYN performs a dual function in B cells (Gauld et al. 2004; Xu et al. 2005). Like other SRC-family kinases, LYN participates in the initial activation of BCR signaling by phosphorylating CD79 ITAMs. However, LYN is unique among SRC-family kinases in initiating a feedback loop that attenuates BCR signaling. LYN can phosphorylate the “ITIM” modules in CD22 and the Fc γ-receptor, allowing recruitment of the phosphatase SHP1 to the BCR, leading to ITAM tyrosine dephosphorylation. LYN can also phosphorylate SYK at a negative regulatory site, decreasing SYK kinase activity. Because of these negative regulatory mechanisms, mice deficient in LYN have hyperactive BCR signaling that leads to a severe and sometimes fatal autoimmune disease (Chan et al. 1997). ABC DLBCL cells reconstituted with mutant CD79B have consistently less LYN kinase activity than those reconstituted with wild-type CD79B (Davis et al. 2010). Because LYN is an important regulator of BCR internalization (Ma et al. 2001; Niiro et al. 2004), the inhibition of LYN kinase activity might explain the higher surface BCR levels in ABC DLBCL cells with mutant CD79B.
These observations suggest a model in which the CD79 mutations are selected in ABC DLBCLs to avoid negative influences on BCR signaling (Fig. 1B). This could occur at an early stage of lymphomagenesis in which a self or foreign antigen-dependent B cell might acquire a CD79 ITAM mutation, allowing it to undergo greater clonal expansion, enhance activation of NF-κB, and avoid cell death. An independent step in the formation of ABC DLBCL appears to be the acquisition of BCR clustering, which is independent of the CD79 mutations (Davis et al. 2010), but is likely responsible for the strong constitutive BCR signaling that typifies these lymphomas. Finally, ABC DLBCLs accumulate a number of other oncogenic hits that are required to create a fully malignant clone (Lenz et al. 2008).
Other Genetic Alterations Deregulating NF-κB in ABC DLBCL
Taken together, the CARD11 and CD79 mutations could potentially account for the NF-κB activation in roughly one third of ABC DLBCLs, implying that many more genetic alterations will be discovered that affect NF-κB in this lymphoma subtype. One common target of genetic inactivation in ABC DLBCL is A20, a negative regulator of NF-κB signaling. ABC DLBCLs have a variety of nonsense mutations and genomic deletions that inactivate A20 in roughly one-quarter of cases, but these events have been observed in only 2% of GCB DLBCLs (Compagno et al. 2009). Reintroduction of wild-type A20 into an ABC DLBCL cell line with an A20 mutation extinguished NF-κB signaling and caused apoptosis. A20 serves as a negative regulator of NF-κB signaling by deubiquitinating TRAF2, TRAF6, MALT1, and the IKKγ subunit (Boone et al. 2004; Lin et al. 2008; Mauro et al. 2006; Wertz et al. 2004). An important point to emphasize is that A20 serves as a brake on NF-κB signaling, but its loss most likely cannot by itself cause NF-κB signaling. This concept is illustrated by A20-deficient mice, which die from a lethal inflammatory disease (Boone et al. 2004). The inflammatory disease in these mice depends on homeostatic MyD88 signaling in response to commensal gut flora (Turer et al. 2008). In ABC DLBCLs, it is likely that A20 mutations augment but do not cause constitutive NF-κB signaling. A20 mutations may cooperate with events that activate NF-κB signaling, such as CARD11 mutations or chronic active BCR signaling. For example, one cell line with an A20 mutation, RC-K8 (Compagno et al. 2009), also harbors inactivating mutations in IκBα (Kalaitzidis et al. 2002).
One TRAF2 mutant was isolated that can activate NF-κB when overexpressed (Compagno et al. 2009). However, the mutant protein appeared to be significantly less stable than wild-type TRAF2 protein, making the interpretation of this mutant complicated. Other NF-κB modulators reported to be mutated in DLBCLs include RANK, TRAF5, and TAK1, but the effect of these mutations on NF-κB signaling was not studied (Compagno et al. 2009).
GCB DLBCL
GCB DLBCL likely derives from the rapidly proliferating centroblasts within the germinal center. Following activation by antigen, B-cells acutely activate NF-κB via the CBM complex. Thereafter, by mechanisms that are poorly understood, the B cell adopts a new gene expression program that is strikingly distinct from that of an acutely activated B cell. In particular, the NF-κB pathway is essentially silent in germinal center B cells, as measured by expression of NF-κB target genes and by nuclear accumulation of NF-κB heterodimers (Basso et al. 2004; Shaffer et al. 2001). However, it is likely that NF-κB signaling is periodically activated in germinal center B cells because CD40, a known activator of the classical and alternative NF-κB pathway, is continuously required to sustain the germinal center reaction (Han et al. 1995). A small fraction of germinal center B cells have nuclear NF-κB, indicating that they may be receiving stronger signals either from CD40 ligand on the surface of follicular helper T cells or from antigen (Basso et al. 2004). This stronger NF-κB signal can promote transcription of the NF-κB target gene IRF4, which encodes a key inducer of plasmacytic differentiation.
Because of their germinal center derivation, GCB DLBCLs have generally lower expression of NF-κB target genes than other DLBCL subtypes. A confounding issue in these analyses is that NF-κB target genes can be expressed by nonmalignant immune cells (macrophages and T cells) that infiltrate the lymphoma. However, GCB DLBCL cell lines have much lower expression of NF-κB target genes than ABC DLBCL cell lines (Davis et al. 2001). Moreover, in situ analysis of nuclear NF-κB abundance confirms that the majority of GCB DLBCLs have little NF-κB activation (Compagno et al. 2009), and thus inherit from their nonmalignant counterparts a low NF-κB set point. Accordingly, treatment of GCB DLBCL cell lines with inhibitors of IκB kinase has no effect (Lam et al. 2005).
A minority of GCB DLBCLs may activate NF-κB, as evidenced by the presence of oncogenic CARD11 mutations in 3.7% of cases (Lenz et al. 2008). These rare GCB DLBCLs retain expression of germinal center B cell signature genes, on which is layered the expression of NF-κB target genes (Lenz et al. 2008). An occasional GCB DLBCL may also use the chronic active BCR signaling mechanism because 3% of these cases had genetic alterations of the CD79B ITAM (Davis et al. 2010).
Curiously, a recurrent genetic abnormality in GCB DLBCL is amplification of the locus on chromosome 2 encoding c-rel (Lenz et al. 2008). This amplicon occurs in 27% of GCB DLBCLs but in only 5% of ABC DLBCLs. These GCB DLBCLs are not associated with nuclear c-Rel (Feuerhake et al. 2005), nor with higher expression of the NF-κB target gene signature (Davis et al. 2001). Therefore, the selective pressure for GCB DLBCLs to amplify this locus may relate to another gene in this genomic interval. Alternatively, the amplification of the c-rel locus could have been selected early in the evolution of the tumor to enhance responsiveness to extracellular stimuli that engage the NF-κB pathway.
PRIMARY MEDIASTINAL B-CELL LYMPHOMA
Primary mediastinal B-cell lymphoma (PMBL) is a subtype of DLBCL that is clinically distinct, occurring most commonly in young women under 40. As the name implies, this lymphoma subtype typically arises in the mediastinum and most likely originates from a rare thymic B-cell subpopulation (Copie-Bergman et al. 2002). Gene expression profiling revealed that PMBL has a distinctive gene expression signature that can be used to differentiate this entity from other DLBCL subtypes (Rosenwald et al. 2003; Savage et al. 2003).
Unexpectedly, PMBL shares an extensive gene expression signature with the malignant cells of Hodgkin lymphoma, known as Hodgkin Reed-Sternberg (HRS) cells (Rosenwald et al. 2003; Savage et al. 2003). Included in the shared gene expression signature are NF-κB target genes (Rosenwald et al. 2003; Savage et al. 2003). Inhibition of NF-κB signaling in a PMBL cell line, using either an IKKβ inhibitor or an IκBα super-repressor, extinguished NF-κB target gene expression and induced cell death (Feuerhake et al. 2005; Lam et al. 2005).
Clues regarding the molecular nature of constitutive NF-κB signaling in PMBL are beginning to emerge. Inactivating mutations and deletions of A20 are recurrent in PMBL, occurring in roughly one third of cases (Schmitz et al. 2009). As in ABC DLBCL, the removal of the negative influence of A20 may potentiate other mechanisms that activate NF-κB in these cells. Roughly one fifth of PMBL cases have amplification of the c-rel locus (Lenz et al. 2008), and this is associated with nuclear c-rel (Feuerhake et al. 2005). Overexpression of c-rel likely augments the constitutive NF-κB activation in these cases. Despite these clues, the root cause of constitutive IKKβ activation in PMBL remains elusive.
HODGKIN LYMPHOMA
Hodgkin lymphoma is characterized by tumors in which the malignant HRS cells are imbedded in a sea of inflammatory cells from a variety of hematopoietic lineages. In roughly one half of Hodgkin lymphomas, the HRS cell harbors the Epstein-Barr virus (EBV), which encodes LMP1, a potent activator of NF-κB. Although LMP1 may account for NF-κB signaling in EBV-positive cases, analysis of primary HRS cells and cell lines from EBV-negative cases also revealed constitutive nuclear p50/p65 heterodimers, consistent with classical NF-κB signaling (Bargou et al. 1996). Inhibition of NF-κB using an IκBα super-repressor was toxic to Hodgkin cell lines (Bargou et al. 1997).
In ∼20% of Hodgkin lymphoma cases, the HRS cells have inactivating mutations or deletions of NFKBIA, encoding IκBα (Cabannes et al. 1999; Emmerich et al. 1999; Jungnickel et al. 2000; Lake et al. 2009). Less commonly, genetic events inactivate NFKBIE, encoding IκBε (Emmerich et al. 2003). Notably, genetic inactivation of IκBα occurs preferentially in EBV-negative Hodgkin lymphomas, presumably because the LMP1-mediated NF-κB eliminates the selective pressure for IκBα abnormalities (Lake et al. 2009).
In some EBV-negative Hodgkin lymphoma cell lines, IKK constitutively phosphorylates and degrades wild-type IκBα (Krappmann et al. 1999). Although the molecular underpinnings of this constitutive IKK activity are not fully understood, HRS cells from 44% of Hodgkin lymphomas sustain genomic mutations and deletions that inactivate A20, the majority of which are in EBV-negative cases (Schmitz et al. 2009). Reconstitution of A20-deficient Hodgkin cell lines with A20 causes cell death by shutting down NF-κB signaling. However, as in ABC DLBCL, loss of the negative regulator A20 may be insufficient to activate NF-κB, and additional mechanisms may contribute in positively activating IKKβ in Hodgkin lymphoma in a cell autonomous fashion.
Within the inflammatory milieu of the Hodgkin lymphoma tumor, various TNF receptor signaling pathways could conceivably contribute to NF-κB activation of the HRS cell. For example, activated T cells in the microenvironment of the HRS cells often express CD40 ligand (Carbone et al. 1995; Gruss et al. 1994), which may active the classical and alternative pathways in HRS cells. Additionally, the TNF family receptor CD30 is expressed highly in Hodgkin lymphoma (Schwab et al. 1982) and its ligand is expressed on mast cells and eosinophils, which are present in Hodgkin lymphoma tumors (Molin et al. 2002; Pinto et al. 1996).
A less common form of Hodgkin lymphoma, termed lymphocyte-predominant Hodgkin lymphoma, has a cellular composition resembling a normal germinal center reaction, including the presence of follicular dendritic cells and T-follicular helper cells (Schmitz et al. 2009). This Hodgkin lymphoma subtype also has constitutive NF-κB activation as evidenced by NF-κB target gene expression and nuclear p65 (Brune et al. 2008). The mechanisms responsible are unknown but do not include A20 inactivation.
MALT LYMPHOMA
Mucosa-associated lymphoid tissue (MALT) lymphoma, a form of marginal zone lymphoma, is an often indolent disease that occurs most commonly in the stomach but can also arise at a variety of anatomical sites near mucosal surfaces (Isaacson et al. 2004). In most cases, gastric MALT lymphoma is associated with persistent infection with Helicobacter pylori. Other marginal zone lymphomas have been associated with Chlamydia psittaci, Campylobacter jejuni, Borellia burgdorferi, and hepatitis C virus infections (Ferreri et al. 2007). Ex vivo culture experiments suggest that H. pylori antigens trigger T-cell proliferation but do not affect the malignant MALT lymphoma clone (Hussell et al. 1996). Instead, the BCRs of the malignant B-cell clone often possess rheumatoid factor activity (Bende et al. 2005). This suggests a scenario in which nonmalignant B cells in the inflammatory microenvironment produce polyclonal IgG antibodies specific for H. pylori that generate immune complexes, which then bind to the BCR of the malignant B-cell clone owing to its rheumatoid factor activity. These gastric MALT lymphomas presumably activate NF-κB via BCR signaling although this has not been directly tested. Remarkably, eradication of H. pylori by antibiotic treatment causes sustained complete remissions in many of these patients with gastric MALT lymphoma (Wotherspoon et al. 1993).
However, antibiotic therapy is ineffective in a substantial proportion of gastric MALT cases, suggesting a different pathogenesis. Approximately 25% of gastric MALT lymphoma harbor a t(11:18) translocation that creates a fusion protein between c-IAP2 and MALT1 (Akagi et al. 1999; Dierlamm et al. 1999; Morgan et al. 1999). These cases do not appear to have BCRs with rheumatoid factor activity (Bende et al. 2005) and are not responsive to antibiotic treatment (Liu et al. 2002). Less commonly, MALT1 can be overexpressed in MALT lymphoma as a consequence of a t(14;18) translocation linking it to the immunoglobulin heavy chain (IgH) locus or by chromosomal amplification of the MALT1 locus (Sanchez-Izquierdo et al. 2003). In rare cases, MALT lymphomas are associated with a t(1;14) translocation that juxtaposes BCL10 translocation with the IgH locus (Willis et al. 1999; Zhang et al. 1999).
Transgenic mice expressing either the c-IAP2-MALT1 fusion protein or BCL10 under control of immunoglobulin heavy chain enhancer elements develop splenic marginal zone hyperplasia (Baens et al. 2006; Li et al. 2009). These findings underscore the relationship between CARD11/BCL10/MALT1 signaling and marginal zone B-cell differentiation, which is also evident from the defective marginal zone development in mice deficient in MALT1 or BCL10 (Thome 2004). The low frequency and delayed onset of lymphoma in these mice suggests that uncontrolled NF-κB activation is insufficient for lymphomagenesis and that additional cooperating oncogenic events are necessary.
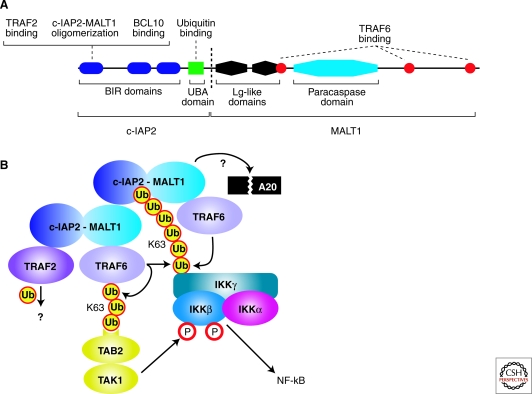
Although the chromosomal breakpoints in t(11;18) translocations are somewhat variable (Isaacson et al. 2004), in most cases the c-IAP2-MALT1 fusion protein includes 3 BIR domains of c-IAP2 and the paracaspase domain of MALT1 (Fig. 2A). Missing is the c-IAP2 RING finger domain, presumably because it mediates c-IAP2 autoubiquitination and destabilization. Also missing is the death domain of MALT1, which is required for BCL10 interaction. During normal antigen receptor signaling, BCL10 is required to oligimerize MALT1 to activate NF-κB (Lucas et al. 2001). The role of BCL10 in c-IAP2-MALT1 signaling to NF-κB is controversial: Two studies found that BCL10 is not required for NF-κB activation by c-IAP2-MALT1 (Noels et al. 2007; Ruland et al. 2003), whereas another report found that BCL10 knockdown abrogated NF-κB activation by the fusion protein in HeLa cells (Hu et al. 2006). This latter study reported that c-IAP2 binds BCL10 through its BIR domains (Hu et al. 2006), but this was not observed in another study (Noels et al. 2007). BCL10 protein is more abundant in MALT lymphomas with the t(11;18) translocation than in cases lacking this translocation (Hu et al. 2006), perhaps because of the ability of c-IAP2-MALT1 to dominantly interfere with the ability of c-IAP2 to ubiquitinate and degrade BCL10 (Hu et al. 2006). Some of the discrepancies in the aforementioned studies may be traced to different model systems and conditions, most of which use overexpression of proteins in heterologous cell types. Clearly lacking in these studies is an analysis of these biochemical interactions in MALT lymphoma cells.
Figure 2.
Pathogenesis of MALT lymphoma. (A) Schematic of the c-IAP2-MALT1 fusion oncoprotein produced by the t (11;18) translocation in MALT lymphomas. Shown is one common fusion breakpoint (dashed line), but other breakpoints occur less frequently (for review, see Isaacson et al. 2004). (B) Molecular mechanisms of NF-κB activation by the c-IAP2-MALT1 fusion protein. c-IAP2-MALT1 forms multimers by heterotypic interactions between its BIR and MALT1 regions. c-IAP2-MALT1 binds TRAF6, activating the K63-linked ubiquitin ligase activity of TRAF6 for IKKγ and itself. Polyubiquitated TRAF6 binds TAB2, thereby activating the associated TAK1 kinase to phosphorylate IKKβ. The ubiquitin-binding domain of c-IAP2-MALT1 stabilizes its interaction with ubiquitinated IKK. The proteolytic cleavage of a substrate protein, potentially A20, by the paracaspase domain of MALT1 is required for the function of c-IAP2-MALT1. TRAF2 is also required for c-IAP2-MALT1 activity, but its substrate is unknown.
The c-IAP2-MALT1 fusion protein activates NF-κB when overexpressed (Lucas et al. 2001; Ruland et al. 2003; Uren et al. 2000). The carboxy-terminal BIR1 domain appears to mediate two functions required for NF-κB activation. First, this domain binds TRAF2, which contributes to NF-κB activation by the c-IAP2-MALT1 fusion protein by an unknown mechanism (Garrison et al. 2009; Lucas et al. 2007; Samuel et al. 2006). Second, the BIR1 domain mediates oligomerization of the c-IAP2-MALT1 fusion protein by interacting heterotypically with the carboxy-terminal region from MALT1 (Lucas et al. 2007; Zhou et al. 2005). In the IAP family of proteins, the BIR domains interact with caspase domains, thereby inhibiting their activation and blocking apoptosis (Wright et al. 2005). Hence, it is conceivable that the BIR1 domain may bind the paracaspase domain in an analogous fashion, although this has not been tested experimentally. Because artificial dimerization of wild-type MALT1 activates NF-κB (Lucas et al. 2001; Sun et al. 2004; Zhou et al. 2004), BIR1-mediated oligomerization of c-IAP2-MALT1 may play an important role in the oncogenic activity of this fusion protein.
In normal lymphocytes, activation of IKK downstream of antigen receptor signaling requires lysine-63 polyubiquitination of the IKKγ subunit, which may be mediated by TRAF6 recruitment by the CBM complex (Sun et al. 2004; Zhou et al. 2004). c-IAP2-MALT1 activation of NF-κB also requires ubiquitination of IKKγ (Zhou et al. 2005). This activity may require TRAF6 (Noels et al. 2007) and/or the ability of the fusion protein to bind an E2 ubiquitin conjugating enzyme containing Ubc13 (Zhou et al. 2005). TRAF2 binding to the BIR domains could also contribute to IKKγ ubiquitination, but this has not been explicitly shown (Garrison et al. 2009; Lucas et al. 2007; Samuel et al. 2006).
c-IAP2 also contains a ubiquitin binding domain (UBD) that is retained in most of the c-IAP1-MALT1 fusion proteins from MALT lymphomas (Gyrd-Hansen et al. 2008) (Fig. 2A). Mutations that prevent ubiquitin binding by this domain attenuate NF-κB activation by the c-IAP1-MALT1 fusion protein. As mentioned earlier, lysine-63 ubiquitination of IKKγ is required for IKK activation by c-IAP2-MALT1. Notably, the c-IAP2 UBD interacts with ubiquitinated IKKγ (Gyrd-Hansen et al. 2008).
The paracaspase domain of MALT1 has recently been shown to have proteolytic activity directly toward arginine residues of its substrates A20 and BCL10 (Coornaert et al. 2008; Rebeaud et al. 2008). A c-IAP2-MALT1 fusion protein that is proteolytically inactive is less efficient in stimulating NF-κB than the corresponding wild-type fusion protein (Coornaert et al. 2008; Uren et al. 2000). Activation of caspases is mediated by induced proximity (Yang et al. 1998), raising the possibility that the MALT1 paracaspase proteolytic activity is induced by c-IAP2-MALT1 oligomerization. Conceivably, the c-IAP2-MALT1 might cleave and inactivate A20, thereby preserving IKKγ ubiquitination and potentiating NF-κB signaling. It is possible that an inhibitor of MALT paracaspase activity (see the following) might promote apoptosis in MALT lymphoma cells, but this has not yet been directly tested.
Figure 2B presents a plausible model for NF-κB activation by c-IAP2-MALT1. Oligomerization of the fusion protein may provide a multivalent platform for the recruitment of the ubiquitin ligases TRAF6 and TRAF2. Although it is unclear how IKK is recruited initially to this complex, once it is ubiquitinated, the ubiquitin binding domain of the fusion protein can help retain IKK in the complex, possibly facilitating its phosphorylation by TAK1. The c-IAP2-MALT1 oligomers may have active proteolytic activity for A20, thereby augmenting the NF-κB signaling output.
MULTIPLE MYELOMA
Multiple myeloma is a malignancy derived from plasmacytic cells dwelling in the bone marrow. Its progression is often slow, but all patients eventually need treatment. Multimodality therapy, including chemotherapy and more targeted agents such as bortezomib, can delay disease progression for years but is not usually curative. The search for mechanisms of therapy resistance has focused on cell survival signals contributed by stromal cells in the bone marrow microenvironment (Dalton 2003; Hideshima et al. 2002). Eventually, myeloma cells may sustain genetic hits that confer independence from the bone marrow microenvironment.
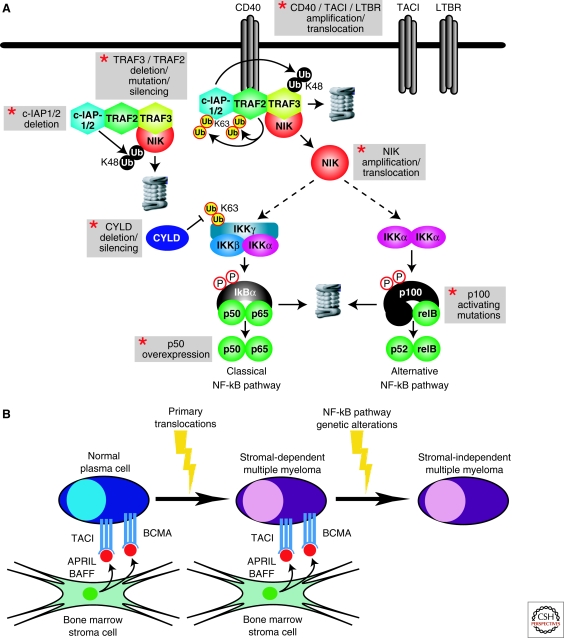
In this scenario, the NF-κB pathway appears to play a pivotal role. Two complementary studies uncovered recurrent genetic alterations that cause constitutive NF-κB signaling in multiple myeloma (Annunziata et al. 2007; Keats et al. 2007) (Fig. 3A). In one study, a substantial fraction of multiple myeloma cell lines were found to be sensitive to a highly specific IKKβ inhibitor, suggesting that the classical NF-κB pathway was active (Annunziata et al. 2007). These cell lines had phosphorylated unstable IκBα, nuclear NF-κB complexes containing p65, and expression of NF-κB target genes, whereas the IKKβ-resistant cell lines did not. The NF-κB target gene signature was evident in over 80% of primary samples of multiple myeloma, and these malignant cells had nuclear p65. Thus, a substantial fraction of multiple myeloma cases engage the classical NF-κB pathway. In addition, myeloma cells with NF-κB pathway activation have nuclear p52 and RelB, which has been taken as evidence for alternative NF-κB pathway activation, although the detailed mechanism responsible has not been clarified (see later discussion).
Figure 3.
Constitutive NF-κB pathway activation in multiple myeloma. (A) Genetic abnormalities that activate NF-κB in multiple myeloma. Gray boxes highlight recurrent genetic aberrations in multiple myeloma involving NF-κB pathway components. The kinase NIK is tethered to the ubiquitin ligases c-IAP1 and/c-IAP2 (c-IAP1/2) by TRAF2 and TRAF3, leading to rapid turnover of NIK protein because of c-IAP1/2-catalyzed K46-linked polyubiquitination. On recruitment to a subset of TNF receptor-family proteins—notably CD40, TACI, and lymphotoxin-β receptor (LTBR)—TRAF2 ubiquitin ligase activity is induced, leading to K63-linked polyubiquitination of c-IAP1/2. The K46-linked ubiquitin ligase activity of c-IAP1/2 is directed toward TRAF3, leading to its proteasomal degradation. NIK is liberated and stabilized in the process. NIK overexpression stimulates both the classical and alternative NF-κB pathways. The deubiquitinase CYLD negative regulates this signaling pathway by removing K63-linked polyubiquitin chains from IKKγ. (B) Model of NF-κB activation during the genesis of multiple myeloma. Normal plasma cells receive signals from BAFF and APRIL, two TNF family ligands in the bone marrow microenvironment. Their receptors, TACI and BCMA, are highly expressed in multiple myeloma and signal to NF-κB through both the classical and alternative NF-κB pathways. Initial transformation is often caused by oncogenic translocations, but the myeloma cell may still remain dependent on the bone marrow microenvironment to receive prosurvival NF-κB signals. Myeloma cells that acquire mutations that cause constitutive NF-κB pathway activation are selected because they allow the malignant cells to survive and proliferate without being limited by the bone marrow microenvironment.
In part, NF-κB activation in multiple myeloma may be caused by signals from the bone marrow microenvironment (Fig. 3B). In a survey of normal B-cell subpopulations, bone marrow-derived plasma cells had the highest expression of NF-κB target genes (Annunziata et al. 2007). Quite conceivably, NF-κB could be induced in these cells by two TNF family ligands in the bone marrow microenvironment, BAFF and APRIL (Hideshima et al. 2002; Marsters et al. 2000; Moreaux et al. 2005; O’Connor et al. 2004). Indeed, interference with BAFF signaling leads to the disappearance of plasma cells from the bone marrow (O’Connor et al. 2004). Plasma cells express two receptors for BAFF and APRIL highly—BCMA and TACI—and multiple myeloma cells variably retain these receptors (Moreaux et al. 2005). Multiple myeloma cases with high TACI expression have a gene expression signature of mature plasma cells and a more favorable prognosis, whereas those with low TACI expression have a more plasmablastic phenotype and an inferior prognosis (Moreaux et al. 2005). Potentially, the TACI-high myeloma subset may retain their dependence on BAFF and APRIL the bone marrow microenvironment whereas the TACI-low subset may be less dependent on the microenvironment for NF-κB survival signals.
In many myeloma cases, however, a diverse array of genetic aberrations target regulators of NF-κB (Annunziata et al. 2007; Keats et al. 2007) (Fig. 3A). At various frequencies, multiple myeloma cells harbor amplifications and/or translocations of NIK, CD40, lymphotoxin-β receptor, and TACI, as well as deletions, inactivating mutations, or transcriptional silencing of TRAF3, TRAF2, and CYLD. Occasionally, myeloma acquire abnormalities that alter NF-κB transcription factors directly, including massive overexpression of NF-κB p50 and mutations of NFKB2 that produce truncated NF-κB p100 isoforms that do not need protolytic processing to enter the nucleus (Annunziata et al. 2007; Keats et al. 2007; Migliazza et al. 1994). Generally, these abnormalities are nonoverlapping, however much additional work needs to be performed to determine if additional genetic abnormalities affect NF-κB in this disease.
Many of the aberrations focus attention on the kinase NIK, a known regulator of both classical and alternative NF-κB signaling (Claudio et al. 2002; Coope et al. 2002; Ramakrishnan et al. 2004; Yin et al. 2001). In some cases, myelomas have high-level amplifications of the NIK genomic locus, resulting in overexpression. In others, NIK is deregulated by translocations, some of which lead to overexpression of NIK because of juxtaposition with strong immunoglobulin enhancer elements. One interesting translocation fuses the EFTUD and NIK genes, creating a fusion protein that lacks the amino-terminal 86 amino acids of NIK (Annunziata et al. 2007), which are required for TRAF3-mediated NIK degradation (Liao et al. 2004). The resultant fusion protein is highly stable, an observation that raised the possibility that TRAF3 might mediate NIK turnover in myeloma cells.
Indeed, roughly one eighth of myeloma cases sustain inactivating mutations or homozygous deletions of the TRAF3 gene (Annunziata et al. 2007; Keats et al. 2007). These genetic aberrations are scattered through the length of the protein, but include some carboxy-terminal events that affect the MATH domain, which is important for interaction of TRAF3 with NIK and TNF family receptors (Li et al. 2003; Liao et al. 2004; Ni et al. 2004; Ni et al. 2000). In cell lines that have inactivated TRAF3, NIK protein is stabilized. Less commonly, some myelomas have biallelic deletion of a genomic region housing BIRC2 and BIRC3, encoding c-IAP1 and c-IAP2, respectively. Myeloma cell lines with this deletion also have high protein expression of NIK and TRAF3, which provided the first clue that c-IAP1 and c-IAP2 are linked in a biochemical pathway with NIK and TRAF3.
Myelomas also recurrently inactivate CYLD, a negative regulator of NF-κB signaling, either by deletion, mutation, or transcriptional silencing (Annunziata et al. 2007, Keats et al. 2007). CYLD is capable of deubiquitinating several NF-κB regulators, including the IKKγ subunit, TRAF2, TRAF6, and BCL3 (Brummelkamp et al. 2003; Kovalenko et al. 2003; Massoumi et al. 2006; Regamey et al. 2003; Trompouki et al. 2003), but the functionally relevant target of CYLD in multiple myeloma is not known.
By a number of criteria, these diverse genetic abnormalities activate both the classical and alternative NF-κB pathways in multiple myeloma. Both NIK overexpression and TRAF3 deletion induce IκBα kinase activity in myeloma cells and the nuclear accumulation of p50/p65 (Annunziata et al. 2007). This finding is in keeping with the sensitivity of these cells to an IKKβ inhibitor and shows classical NF-κB activation. However, p52/RelB heterodimers also accumulate in the nucleus as a result of these genetic aberrations, consistent with alternative NF-κB activation (Annunziata et al. 2007; Keats et al. 2007). In normal cells, the alternative NF-κB pathway relies on activated IKKα to phosphorylate p100, leading to its processing by the proteasome into p52 (Senftleben et al. 2001). However, a strong knockdown of IKKα by RNA interference was not toxic to myeloma cells with NIK overexpression, yet knockdown of NIK was toxic. Hence, p100 processing in these cells may depend on other IKKα-independent mechanisms in these myeloma cells.
Recent elegant biochemical studies have integrated these genetic clues from multiple myeloma into a signaling pathway regulating NIK stability (Matsuzawa et al. 2008; Vallabhapurapu et al. 2008; Zarnegar et al. 2008). In most cells, NIK protein is highly unstable, because of proteasomal degradation. This proteasomal targeting requires c-IAP-1 or c-IAP-2 (c-IAP1/2) to add K48-liked polyubiquitin chains to NIK. c-IAP1/2 are brought into complex with NIK via a molecular bridge in which TRAF3 binds to the NIK N-terminus, recruiting TRAF2 and c-IAP1/2. On ligand engagement of certain TNF family receptors, notably CD40, the TRAF3/TRAF2/c-IAP1/2 complex is recruited to the receptor, which stimulates K48-linked ubiquitination of TRAF3 by c-IAP1/2 and rapid TRAF3 degradation. Subsequently, NIK is stabilized, presumably because TRAF3 depletion dissociates c-IAP1/2 from NIK. It is not completely clear why c-IAP1/2 ubiquitination of TRAF3 is stimulated by receptor engagement, but TRAF2-mediated K63-linked ubiquitination of c-IAP1/2 may stimulate its activity as an E3 ligase for TRAF3 (Vallabhapurapu et al. 2008).
The fact that the genetic aberrations in myeloma target a pathway regulating NIK stability has led to a suggestion that these aberrations are selected to activate the alternative NF-κB pathway in myeloma cells (Keats et al. 2007). This is a natural conclusion given that in normal cells, NIK stabilization primarily stimulates IKKα-mediated NFKB2 processing into p52, leading to the nuclear accumulation of p52/RelB heterodimers. However, TRAF3 knockout fibroblasts also have elevated classical NF-κB signaling, as indicated by higher IκBα kinase activity and nuclear accumulation of the p65 subunit, possibly as a result of autocrine stimulation of NF-κB by cytokines (Vallabhapurapu et al. 2008; Zarnegar et al. 2008). Moreover, in cells stimulated with lymphotoxin-β, classical NF-κB activation depends on both NIK and IKKα (Zarnegar et al. 2008).
These considerations lead to the view that pathological overexpression of NIK, as occurs in multiple myeloma, may directly activate IKKβ and the classical NF-κB pathway (Annunziata et al. 2007). Indeed, NIK can activate classical NF-κB signaling when overexpressed (Malinin et al. 1997; O’Mahony et al. 2000; Woronicz et al. 1997) and certain TNF family members (CD40 ligand and BAFF) can activate the classical NF-κB pathway through NIK (Ramakrishnan et al. 2004). Another possibility is that alternative NF-κB signaling in myeloma cells causes the synthesis of cytokines that stimulate the classical NF-κB pathway in an autocrine fashion. As mentioned earlier, the role of IKKα in this process in myeloma cells is not supported by knockdown studies (Annunziata et al. 2007), but cannot be ruled out given the role of IKKα in stimulating the classical NF-κB pathway in cells treated with LTβ (Zarnegar et al. 2008). Irrespective of the mechanism, it is important to appreciate that myeloma cells activate the classical and alternative NF-κB pathways in concert and that therapeutic targeting of the classical pathway with IKKβ inhibitors could be developed as a strategy to treat this disease (Annunziata et al. 2007; Hideshima et al. 2002; Hideshima et al. 2006).
NF-κB IN EPITHELIAL CANCERS
A rich literature shows the central role of NF-κB signaling in the inflammatory milieu that fosters many epithelial cancers (Karin 2010; Karin et al. 2006). In colon cancer, NF-κB expression in the premalignant epithelium prevents apoptosis and promotes transformation (Greten et al. 2004). However, it has been less clear that fully formed epithelial cancers depend on NF-κB. Unlike lymphomas, solid tumors often genetically alter receptor tyrosine kinases, which provide prosurvival signals through the PI(3) kinase pathway. Perhaps as a result, genetic lesions in the NF-κB pathway have not been uncovered in most instances.
CYLD as a Tumor Suppressor
Familial cylindromatosis is a malignancy of the hair follicle stem cell in which CYLD functions as a classic tumor suppressor (reviewed in Sun 2009). Individuals with this syndrome inherit one loss-of-function CYLD allele and the other CYLD allele is somatically inactivated in the cylindromas. A related disorder, known as familial skin appendage tumors, also involves CYLD inactivation. The CYLD gene was positionally cloned and had no known function until three groups showed that it negatively regulated the NF-κB pathway (Brummelkamp et al. 2003; Kovalenko et al. 2003; Trompouki et al. 2003). CYLD can deubiquitinate a wide range of cellular proteins, including the NF-κB regulators TRAF2, TRAF6, TRAF7, BCL3, TAK1, and the IKKγ subunit (Sun 2009). CYLD may function as a tumor suppressor by negatively regulating classical NF-κB pathway because ubiquitination of the IKKγ is required for IKK activation. The importance of NF-κB signaling in familial cylindromatosis is highlighted by a recent clinical trial in which salicylic acid, an NF-κB inhibitor, was used topically to treat the cylindromas (Kovalenko et al. 2003). Although the study was small and not blinded, several cylindromas in one patient regressed completely with treatment.
Unconventional IKKs and Epithelial Cancers
Recent experiments suggest that two unconventional members of the IKK family, IKKε and TBK1, activate NF-κB in epithelial cancers. In the innate immune response to viruses, Toll receptors and cytosolic receptors for nucleic acids activate these unconventional IKKs, thereby engaging NF-κB and type-I interferon pathways (Clement et al. 2008; Hacker et al. 2006; Pichlmair et al. 2007). The mechanism by which these kinases regulate the interferon pathway is by phosphorylating IRF3 and IRF7, causing their nuclear translocation, but their mode of action in the NF-κB pathway is less well understood. One mechanism may be induction of TNFα by IRF3, thereby activating the classical NF-κB pathway in a feed-forward loop (Covert et al. 2005; Werner et al. 2005). It is also possible that the unconventional IKKs may play a more direct role in IκBα turnover. IKKε is part of a multiprotein complex that can target IκBα for degradation (Peters et al. 2000; Shimada et al. 1999). Although IKKε and TBK1 can promote IκBα degradation, they can only phosphorylate serine 36 of IκBα (Peters et al. 2000; Pomerantz et al. 1999; Tojima et al. 2000). Hence, another kinase must cooperate with these unconventional IKKs to phosphorylate serine 32 of IκBα, allowing it to be targeted for degradation (Brown et al. 1995). The unconventional IKKs have several other substrates, including the NF-κB subunits p65 and c-rel, which might contribute to their ability to activate NF-κB target genes (reviewed in Clement et al. 2008).
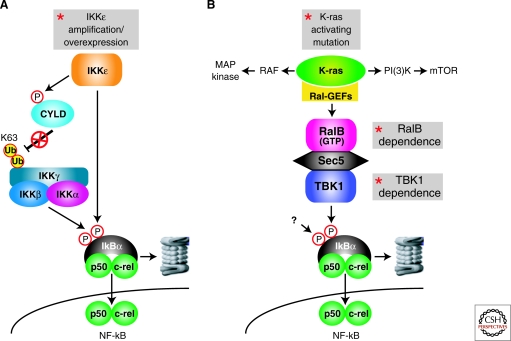
Functional genomics experiments nominated IKKε as an oncogene in breast cancer (Boehm et al. 2007) (Fig. 4A). In a screen of activated kinase alleles, activated IKKε took the place of activated AKT in a transformation assay using human embryonic kidney cells. In a complementary approach, roughly one sixth of breast cancer cases were found to harbor a gain or high-level amplification of a region on chromosome 1 encoding IKKε. Generally, the region of copy number gain spans many megabases, suggesting that several genes in this interval may play a role in breast cancer pathogenesis. However, IKKε appears to be one important actor because knockdown of IKKε was toxic to breast cancer cell lines harboring the IKKε amplicon. This finding is in keeping with the observation that a dominant negative IKKε isoform was toxic to breast cancer cell lines (Eddy et al. 2005). In breast cancer biopsies, IKKε expression was correlated with an increase in nuclear c-rel, suggesting activation of the NF-κB pathway may participate in the pathogenesis of these malignancies (Boehm et al. 2007). This finding is in keeping with previous work showing overexpression of IKKε in both human and mouse breast cancer in association with NF-κB activation (Eddy et al. 2005).
Figure 4.
Solid tumors activate the NF-κB pathway to maintain cell survival. (A) IKKε is frequently overexpressed in breast cancer in association with a gain or amplification of its genomic locus. IKKε phosphorylates a serine residue in CYLD, thereby lowering its deubiquitinase activity toward the IKKγ subunit, leading to a more active IKK enzyme. In addition, IKKε can directly phosphorylate serine-46 of IκBα. (B) Regulation of NF-κB by TBK1 downstream of mutant K-ras in lung cancer. Mutant K-ras signals to several downstream pathways, including the MAP kinase, PI(3) kinase, and RAL GTPase pathways. K-ras-associated RAL guanine nucleotide exchange factors (Ral-GEFs) promote the active, GTP-bound state of the RAL proteins. RalB interacts with Sec5, which in turn recruits TBK1, causing kinase activation. TBK1 triggers classical NF-κB pathway activation, as judged by the accumulation of nuclear p50/c-rel heterodimers. Because TBK1 can only phosphorylate one serine in IκBα, an as yet unknown kinase must cooperate with TBK1 to achieve dual IκBα phosphorylation.
In cell transformation experiments, the activated IKKε allele promoted IκBα degradation and the induction of NF-κB target genes (Boehm et al. 2007), but the exact mechanism by which IKKε achieves NF-κB activation has not been elucidated. A newly defined substrate of IKKε is the deubiquitinase CYLD (Peters et al. 2000; Shimada et al. 1999). Phosphorylation of serine 418 of CYLD diminishes its deubiquitinase action toward IKKγ and TRAF2. Thus, IKKε overexpression may relieve the negative influence of CYLD on the NF-κB pathway. Nonetheless, IKK requires phosphorylation of the IKKβ subunit in its activation loop in addition to IKKγ ubiquitination to become a functioning kinase, and it is currently unclear how IKKε overexpression promotes these posttranscriptional modifications.
Two complementary experimental approaches have revealed a connection between oncogenic signaling by mutant K-ras and NF-κB (Barbie et al. 2009; Meylan et al. 2009) ( . 4B). In a mouse model of lung cancer caused by mutant K-ras overexpression and p53 deletion, the classical NF-κB pathway is activated (Meylan et al. 2009). Lung cancer cell lines from these mice were killed when NF-κB was inhibited, and in vivo delivery of an IκBα super-repressor slowed growth of established lung tumors.
Mutant K-ras engages several downstream effector pathways by stimulating RAF and PI(3) kinases. Additional effectors of mutant K-ras are the RAL GTPases (RALA and RALB), which are converted into the active, GTP-bound state by K-ras-associated RAL guanine nucleotide exchange factors (RalGDS, Rgl, and Rlf). Recent experiments have implicated the unconventional IKK TBK1 in a signaling pathway that links mutant K-ras and NF-κB. TBK1 is activated by RALB in concert with SEC5, a component of the exocyst, and is required for K-ras-mediated transformation of mouse embryo fibroblasts (Chien et al. 2006). A synthetic lethal RNA interference genetic screen revealed that TBK1 shRNAs kill mutant K-ras-transformed cells but not cancer cells with wild-type K-ras (Barbie et al. 2009). TBK1 knockdown decreases viability of human lung cancer cell lines with mutant K-ras as well as lung epithelial cells acutely transformed with mutant K-ras. Knockdown of RALB, but not c-RAF, B-RAF, or AKT, is also synthetically lethal for mutant K-ras-transformed cells, suggesting a link between RALB and TBK1. Overexpression of mutant K-ras activates a set of K-ras signature genes, which includes a number of known targets of NF-κB signaling. Knockdown of TBK1 blocks expression of these NF-κB target genes in mutant K-ras-transformed cells and blocks nuclear accumulation of c-rel. Knockdown of c-rel or expression of an IκBα super-repressor is lethal to lung cancer cells with mutant K-ras, directly demonstrating the requirement for NF-κB signaling to block cell death. Notably, roughly three quarters of human lung adenocarcinomas with mutant K-ras coexpress the K-ras and NF-κB target gene signatures. Among lung adenocarcinomas with wild-type K-ras, one quarter of cases coexpress the K-ras and NF-κB signatures, suggesting that these cancers may have upstream signals that activate wild-type K-ras and engage TBK1 to activate NF-κB. Alternatively, these tumors may be an independent mechanism to activate NF-κB.
THERAPY OF NF-κB-DRIVEN CANCERS
Many prospects and hurdles can be envisaged when considering pharmacological interference with NF-κB signaling (reviewed in Baud et al. 2009; Karin et al. 2004). Inhibitors of IKKβ could have activity against ABC DLBCL, PMBL, MALT lymphoma, and multiple myeloma. However, long-term, full inhibition of IKKβ would be expected to impair the adaptive and innate immune system (Vallabhapurapu et al. 2009). Because IKKβ counteracts IL-1β processing and secretion (Greten et al. 2007), IKKβ inhibition might release IL-1β, causing febrile reactions and possibly inflammation. During development, IKKβ and p65 deficiency provokes massive apoptosis of hepatocytes, but the role of NF-κB in adult hepatocytes has not been fully resolved (reviewed in Schwabe et al. 2007). Conditional deficiency of IKKβ in adult hepatocytes causes little if any sensitivity to TNFα-dependent apoptosis (Luedde et al. 2005). However, deficiency in IKKγ or p65 makes adult hepatocytes markedly more sensitive to TNFα and other injuries to the liver (Beraza et al. 2007; Geisler et al. 2007). Moreover, conditional deficiency in IKKγ caused hepatic steatohepatitis and hepatocellular carcinoma (Luedde et al. 2007). Whether or not liver injury occurs may therefore depend on the extent and/or nature of NF-κB pathway inhibition as well as on the presence of concurrent infections that might elevate proapoptotic inflammatory stimuli such as TNFα.
Despite these cautions, IKKβ-directed therapy might find a role in cancer treatment if cancer cells are unusually addicted to NF-κB signaling and the right degree of NF-κB inhibition can be achieved. Not only might IKKβ inhibitors kill NF-κB-dependent cancer cells directly, but they could potentiate the apoptotic response to cytotoxic chemotherapy (Baldwin 2001; Wang et al. 1998). Many pharmaceutical companies have developed IKKβ inhibitors but none have entered into clinical trials in cancer, presumably because of the concerns expressed earlier. An indirect way to block IKK activity is by inhibiting HSP90. HSP90 is an intrinsic component of the IKK supramolecular complex, and its inhibition blocks the induction of IKK enzymatic activity by various stimuli as well as the biogenesis of IKKβ and IKKα (Broemer et al. 2004; Chen et al. 2002). HSP90 inhibitors are in clinical trials for a number of cancers (Workman et al. 2007) and might show particular activity for IKK-dependent tumors. In fact, the HSP90 inhibitor geldanimycin kills Hodgkin lymphoma lines with constitutive NF-κB activation (Broemer et al. 2004) as well as ABC DLBCL cells (unpubl. data).
Inhibitors of IKKα might potentiate the effects of IKKβ inhibition because IKKα can serve as an IκBα kinase, particularly under conditions of IKKβ inhibition (Lam et al. 2008). In an RNA interference genetic screen in ABC DLBCL, shRNAs targeting IKKα synergized with an IKKβ inhibitor in killing the lymphoma cells. However, knockdown of IKKα without IKKβ inhibition had no effect. Similar “compensatory” IKKα activity was observed in cells in which NF-κB was induced by PMA/ionomycin treatment, CARD11 pathway activation, or TNFα treatment. Because IKKα is in a macromolecular complex with IKKβ, it is conceivable that IKKα may be activated by the same upstream kinases as IKKβ, such as TAK1. Recombinant IKKβ is more active than IKKα as an IκBα kinase (Huynh et al. 2000). Hence, in an IKK complex containing both subunits, IKKβ would mediate most of the IκBα phosphorylation. But when IKKβ is pharmacologically inhibited, IKKα may take over this function, presumably relying on the high local concentration of IκBα.
Given the role of the proteasome in degrading IκBα, the proteasome inhibitor bortezomib might be harnessed to inactivate NF-κB in cancer. All cells require proteasome function and full inhibition of the proteasome is lethal to many cell types. Nonetheless, bortezomib was successfully developed as a cancer agent using a novel approach in which the dose-escalation end-point was not systemic toxicity but rather the degree of proteasome inhibition in vivo (Adams 2004).
Based on the constitutive activation of the NF-κB pathway in ABC DLBCL (Davis et al. 2001), a clinical trial was initiated in relapsed and refractory DLBCL that combined bortezomib with EPOCH, a type of multiagent cytotoxic chemotherapy (Dunleavy et al. 2009). All patients had tumor biopsies to allow the molecular diagnosis of ABC versus GCB DLBCL to be made. Initially, bortezomib was given as a single agent, but no responses were seen. However, the combination of bortezomib with EPOCH chemotherapy yielded complete and partial responses, and the vast majority of these were in patients with ABC DLBCL. This response difference translated into a significantly superior overall survival rate for patients with ABC DLBCL. This was notable because in all previous trials with chemotherapy alone, patients with ABC DLBCL had a decidedly inferior cure rate (Alizadeh et al. 2000; Hummel et al. 2006; Lenz et al. 2008; Monti et al. 2005; Rosenwald et al. 2002).
Bortezomib has been approved for the treatment of multiple myeloma because it delays disease progression (Richardson et al. 2005), but it is unclear whether its efficacy in this disease relates to NF-κB inhibition. In a trial comparing bortezomib and dexamethasone, myeloma cases with low TRAF3 expression had an increased response to bortezomib than those with high TRAF3 (Keats et al. 2007). However, patients stratified according to high or low expression of NF-κB target genes did not differ in their response to bortezomib (Keats et al. 2007), suggesting that this issue needs to be readdressed in a prospective fashion.
An intriguing new way to attack the NF-κB pathway is by interfering with the βTrCP ubiquitin ligase that targets IκBα for destruction. This SCF-type E3 ubiquitin ligase consists of a cullin subunit that serves as a scaffold to assemble SKP1, the RING finger E3 ubiquitin ligase ROC1, and the Fbox protein βTrCP, which is the receptor that recognizes phosphorylated IκBα (Ben-Neriah 2002; Yaron et al. 1998). The cullin subunit requires modification by a NEDD8 moiety for activity of the complex as an E3 ubiquitin ligase (Chiba et al. 2004; Podust et al. 2000; Read et al. 2000). NEDD8 is added by the concerted action of a series of enzymes that functionally mirror the ubiquitin conjugation machinery. The E1 enzyme in this system, termed NEDD8 activating enzyme (NAE), can be inhibited by the small molecule MLN4924 (Soucy et al. 2009). MLN4924 potently inhibits the βTrCP ubiquitin ligase by interfering with its neddylation, thereby causing the protein levels of βTrCP substrates to rise. One substrate CDT1, causes DNA re-replication during S phase, which triggers the S-phase DNA replication checkpoint to initiate apoptosis in cancer cells. MLN4924 is also expected to inhibit NF-κB by preventing IκBα degradation, making it an attractive candidate for clinical trials in cancers characterized by constitutive NF-κB activation.
Given the inherent risks of inhibiting NF-κB in all cell types, strategies that target specific upstream pathways leading to NF-κB in cancer are attractive. The CBM complex is a compelling target in this regard because mice with deficiency in CARD11 or MALT1 only have defects in the adaptive immune system (Thome 2004). Recently, the proteolytic activity of the MALT1 paracaspase domain has been directly shown and two of its proteolytic targets have been identified, BCL10 and A20 (Coornaert et al. 2008; Rebeaud et al. 2008). Unlike conventional caspases that cleave after aspartic acid residues, the paracaspase domain of MALT1 cleaves after arginine. Overexpression of a proteolytically inactive form of MALT1 is less active than wild-type MALT1 in enhancing IL-2 secretion by activated T cells and in activating NF-κB (Lucas et al. 2001; Rebeaud et al. 2008). Likewise, inhibition of MALT1 proteolytic activity with a cell-permeable peptide mimetic reduces NF-κB activation and IL-2 secretion in T cells stimulated through the antigen receptor (Rebeaud et al. 2008). However, the effect of MALT1 proteolytic activity on NF-κB signaling is quantitative rather than complete, suggesting that the proteolytic activity of MALT1 modulates a regulatory circuit engaged during NF-κB activation. Indeed, MALT1 cleavage inactivates A20, a negative regulator of NF-κB signaling (Coornaert et al. 2008).
Encouragingly, two studies have shown that the peptide inhibitors of MALT1 block NF-κB activity and kills ABC DLBCL cell lines but are not toxic for the NF-κB-independent GCB DLBCL cell lines (Ferch et al. 2009; Hailfinger et al. 2009). ABC DLBCL cell lines with wild-type or mutant forms of CARD11 are equally sensitive to MALT1 protease inhibition, in keeping with the dependence of both types of ABC DLBCL on MALT1 (Ngo et al. 2006). These studies pave the way for the development of small molecule peptidomimetic inhibitors of MALT1 for the therapy of ABC DLBCL, and possibly also MALT lymphomas with the c-IAP2-MALT1 fusion oncoprotein.
Chronic active BCR signaling in ABC DLBCL offers a wealth of therapeutic targets upstream of IKK (Davis et al. 2010). BTK kinase activity couples chronic active BCR signaling to NF-κB via IKK activation. Accordingly, a small molecule inhibitor of BTK, PCI-32765 (Pan et al. 2007), kills ABC DBLCL cells with chronic active BCR signaling but not other lymphoma lines (Davis et al. 2010). Dasatinib is a multikinase inhibitor that has especially high activity against SRC-family tyrosine kinases and BTK (Hantschel et al. 2007). This drug extinguished signaling to the NF-κB, AKT and ERK MAP kinase pathways and killed ABC DLBCL cells with chronic active BCR signaling (Davis et al. 2010). Clinical trials based on this observation can begin immediately because dasatinib is already approved as a second-line treatment of chronic myelogenous leukemia that is refractory to imatinib. Other kinases in the BCR pathway leading to NF-κB might also serve as therapeutic targets, such as PKCβ, which phosphorylates and activates wild-type CARD11 in ABC DLBCL. The tyrosine kinase SYK, although essential for normal BCR signaling, is only required by some ABC DLBCLs with chronic active BCR signaling, suggesting that drugs targeting this kinase may have a more restricted efficacy (Davis et al. 2010). By targeting BCR signaling in ABC DLBCL, it should be possible to achieve full inhibition of NF-κB in malignant (and normal) B cells while sparing other normal cells, providing an acceptable therapeutic window.
In multiple myeloma, an intriguing therapeutic target is the kinase NIK, because many genetic alterations in this cancer stabilize NIK protein expression, causing a critical dependence on this kinase (Annunziata et al. 2007; Keats et al. 2007). Although NIK-deficient mice have defects in the organization of secondary lymphoid organs (Miyawaki et al. 1994; Shinkura et al. 1999), this defect might not occur with short-term inhibition of NIK pharmacologically. In response to viral challenge, NIK-deficient B cells are impaired in isotype-switching, but NIK-deficient T cells are relatively unaffected (Karrer et al. 2000), suggesting that pharmacological inhibition of this kinase might only modestly impair adaptive immune responses. Some multiple myeloma cases may activate NIK and NF-κB in a quasi-physiological mechanism through BAFF and/or APRIL signaling in the bone marrow microenvonment. Disruption of this signaling by a TACI-Fc fusion protein should be tested for activity in this setting (Moreaux et al. 2005; Yaccoby et al. 2008).
As should be clear from this review, the wise deployment of NF-κB-based therapies in cancer will require molecular profiling to determine the subtype of cancer, the presence of genetic lesions in NF-κB regulators, and the activity of the NF-κB pathway. In the era of targeted cancer therapy, it is important to evaluate whether a targeted agent hits its mark. In trials of NF-κB inhibitors, this could be easily achieved by measuring the expression of NF-κB target genes before and during therapy, potentially by real-time RT-PCR or other quantitative methods. By this cautious and reasoned approach, I hope that NF-κB pathway inhibitors will find their way into the anticancer armamentarium.
ACKNOWLEDGMENTS
Supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
Footnotes
Editors: Louis M. Staudt and Michael Karin
Additional Perspectives on NF-κB available at www.cshperspectives.org
REFERENCES
- Adams J 2004. The proteasome: a suitable antineoplastic target. Nat Rev Cancer 4:349–360 [DOI] [PubMed] [Google Scholar]
- Akagi T, Motegi M, Tamura A, Suzuki R, Hosokawa Y, Suzuki H, Ota H, Nakamura S, Morishima Y, Taniwaki M, et al. 1999. A novel gene, MALT1 at 18q21, is involved in t(11;18) (q21;q21) found in low-grade B-cell lymphoma of mucosa-associated lymphoid tissue. Oncogene 18:5785–5794 [DOI] [PubMed] [Google Scholar]
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, et al. 2000. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403:503–511 [DOI] [PubMed] [Google Scholar]
- Annunziata CM, Davis RE, Demchenko Y, Bellamy W, Gabrea A, Zhan F, Lenz G, Hanamura I, Wright G, Xiao W, et al. 2007. Frequent engagement of the classical and alternative NF-kappaB pathways by diverse genetic abnormalities in multiple myeloma. Cancer Cell 12:115–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baens M, Fevery S, Sagaert X, Noels H, Hagens S, Broeckx V, Billiau AD, De Wolf-Peeters C, Marynen P 2006. Selective expansion of marginal zone B cells in Emicro-API2-MALT1 mice is linked to enhanced IkappaB kinase gamma polyubiquitination. Cancer Res 66:5270–5277 [DOI] [PubMed] [Google Scholar]
- Bajpai UD, Zhang K, Teutsch M, Sen R, Wortis HH 2000. Bruton's tyrosine kinase links the B cell receptor to nuclear factor kappaB activation. J Exp Med 191:1735–1744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin AS 2001. Control of oncogenesis and cancer therapy resistance by the transcription factor NF-kappaB. J Clin Invest 107:241–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, Schinzel AC, Sandy P, Meylan E, Scholl C, et al. 2009. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 462:108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargou RC, Emmerich F, Krappmann D, Bommert K, Mapara MY, Arnold W, Royer HD, Grinstein E, Greiner A, Scheidereit C, et al. 1997. Constitutive nuclear factor-kappaB-RelA activation is required for proliferation and survival of Hodgkin's disease tumor cells. J Clin Invest 100:2961–2969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargou RC, Leng C, Krappmann D, Emmerich F, Mapara MY, Bommert K, Royer HD, Scheidereit C, Dorken B 1996. High-level nuclear NF-kappa B and Oct-2 is a common feature of cultured Hodgkin/Reed-Sternberg cells. Blood 87:4340–4347 [PubMed] [Google Scholar]
- Basso K, Klein U, Niu H, Stolovitzky GA, Tu Y, Califano A, Cattoretti G, Dalla-Favera R 2004. Tracking CD40 signaling during germinal center development. Blood 104:4088–4096 [DOI] [PubMed] [Google Scholar]
- Baud V, Karin M 2009. Is NF-kappaB a good target for cancer therapy? Hopes and pitfalls. Nat Rev Drug Discov 8:33–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Neriah Y 2002. Regulatory functions of ubiquitination in the immune system. Nat Immunol 3:20–26 [DOI] [PubMed] [Google Scholar]
- Bende RJ, Aarts WM, Riedl RG, de Jong D, Pals ST, van Noesel CJ 2005. Among B cell non-Hodgkin's lymphomas, MALT lymphomas express a unique antibody repertoire with frequent rheumatoid factor reactivity. J Exp Med 201:1229–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beraza N, Ludde T, Assmus U, Roskams T, Vander Borght S, Trautwein C 2007. Hepatocyte-specific IKK gamma/NEMO expression determines the degree of liver injury. Gastroenterology 132:2504–2517 [DOI] [PubMed] [Google Scholar]
- Bidere N, Ngo VN, Lee J, Collins C, Zheng L, Wan F, Davis RE, Lenz G, Anderson DE, Arnoult D, et al. 2009. Casein kinase 1alpha governs antigen-receptor-induced NF-kappaB activation and human lymphoma cell survival. Nature 458:92–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blonska M, Lin X 2009. CARMA1-mediated NF-kappaB and JNK activation in lymphocytes. Immunol Rev 228:199–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm JS, Zhao JJ, Yao J, Kim SY, Firestein R, Dunn IF, Sjostrom SK, Garraway LA, Weremowicz S, Richardson AL, et al. 2007. Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell 129:1065–1079 [DOI] [PubMed] [Google Scholar]
- Boone DL, Turer EE, Lee EG, Ahmad RC, Wheeler MT, Tsui C, Hurley P, Chien M, Chai S, Hitotsumatsu O, et al. 2004. The ubiquitin-modifying enzyme A20 is required for termination of Toll-like receptor responses. Nat Immunol 5:1052–1060 [DOI] [PubMed] [Google Scholar]
- Broemer M, Krappmann D, Scheidereit C 2004. Requirement of Hsp90 activity for IkappaB kinase (IKK) biosynthesis and for constitutive and inducible IKK and NF-kappaB activation. Oncogene 23:5378–5386 [DOI] [PubMed] [Google Scholar]
- Brown K, Gerstberger S, Carlson L, Franzoso G, Siebenlist U 1995. Control of I kappa B-alpha proteolysis by site-specific, signal-induced phosphorylation. Science 267:1485–1488 [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Nijman SM, Dirac AM, Bernards R 2003. Loss of the cylindromatosis tumour suppressor inhibits apoptosis by activating NF-kappaB. Nature 424:797–801 [DOI] [PubMed] [Google Scholar]
- Brune V, Tiacci E, Pfeil I, Doring C, Eckerle S, van Noesel CJ, Klapper W, Falini B, von Heydebreck A, Metzler D, et al. 2008. Origin and pathogenesis of nodular lymphocyte-predominant Hodgkin lymphoma as revealed by global gene expression analysis. J Exp Med 205:2251–2268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabannes E, Khan G, Aillet F, Jarrett RF, Hay RT 1999. Mutations in the IkBa gene in Hodgkin's disease suggest a tumour suppressor role for IkappaBalpha. Oncogene 18:3063–3070 [DOI] [PubMed] [Google Scholar]
- Carbone A, Gloghini A, Gruss HJ, Pinto A 1995. CD40 ligand is constitutively expressed in a subset of T cell lymphomas and on the microenvironmental reactive T cells of follicular lymphomas and Hodgkin's disease. Am J Pathol 147:912–922 [PMC free article] [PubMed] [Google Scholar]
- Chan VW, Meng F, Soriano P, DeFranco AL, Lowell CA 1997. Characterization of the B lymphocyte populations in Lyn-deficient mice and the role of Lyn in signal initiation and down-regulation. Immunity 7:69–81 [DOI] [PubMed] [Google Scholar]
- Chen G, Cao P, Goeddel DV 2002. TNF-induced recruitment and activation of the IKK complex require Cdc37 and Hsp90. Mol Cell 9:401–410 [DOI] [PubMed] [Google Scholar]
- Chen ZJ 2005. Ubiquitin signalling in the NF-kappaB pathway. Nat Cell Biol 7:758–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba T, Tanaka K 2004. Cullin-based ubiquitin ligase and its control by NEDD8-conjugating system. Curr Protein Pept Sci 5:177–184 [DOI] [PubMed] [Google Scholar]
- Chien Y, Kim S, Bumeister R, Loo YM, Kwon SW, Johnson CL, Balakireva MG, Romeo Y, Kopelovich L, Gale M Jr., et al. 2006. RalB GTPase-mediated activation of the IkappaB family kinase TBK1 couples innate immune signaling to tumor cell survival. Cell 127:157–170 [DOI] [PubMed] [Google Scholar]
- Claudio E, Brown K, Park S, Wang H, Siebenlist U 2002. BAFF-induced NEMO-independent processing of NF-kappa B2 in maturing B cells. Nat Immunol 3:958–965 [DOI] [PubMed] [Google Scholar]
- Clement JF, Meloche S, Servant MJ 2008. The IKK-related kinases: from innate immunity to oncogenesis. Cell Res 18:889–899 [DOI] [PubMed] [Google Scholar]
- Compagno M, Lim WK, Grunn A, Nandula SV, Brahmachary M, Shen Q, Bertoni F, Ponzoni M, Scandurra M, Califano A, et al. 2009. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature 459:717–721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coope HJ, Atkinson PG, Huhse B, Belich M, Janzen J, Holman MJ, Klaus GG, Johnston LH, Ley SC 2002. CD40 regulates the processing of NF-kappaB2 pp100 to p52. Embo J 21:5375–5385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coornaert B, Baens M, Heyninck K, Bekaert T, Haegman M, Staal J, Sun L, Chen ZJ, Marynen P, Beyaert R 2008. T cell antigen receptor stimulation induces MALT1 paracaspase-mediated cleavage of the NF-kappaB inhibitor A20. Nat Immunol 9:263–271 [DOI] [PubMed] [Google Scholar]
- Copie-Bergman C, Plonquet A, Alonso MA, Boulland ML, Marquet J, Divine M, Moller P, Leroy K, Gaulard P 2002. MAL expression in lymphoid cells: further evidence for MAL as a distinct molecular marker of primary mediastinal large B-cell lymphomas. Mod Pathol 15:1172–1180 [DOI] [PubMed] [Google Scholar]
- Covert MW, Leung TH, Gaston JE, Baltimore D 2005. Achieving stability of lipopolysaccharide-induced NF-kappaB activation. Science 309:1854–1857 [DOI] [PubMed] [Google Scholar]
- Dalton WS 2003. The tumor microenvironment: focus on myeloma. Cancer Treat Rev 29:11–19 [DOI] [PubMed] [Google Scholar]
- Davis RE, Brown KD, Siebenlist U, Staudt LM 2001. Constitutive nuclear factor kappa B activity is required for survival of activated B Cell-like diffuse large B cell lymphoma cells. J Exp Med 194:1861–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis RE, Ngo VN, Lenz G, Tolar P, Young RM, Romesser PB, Kohlhammer H, Lamy L, Zhao H, Yang Y, et al. 2010. Chronic active B-cell-receptor signalling in diffuse large B-cell lymphoma. Nature 463:88–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierlamm J, Baens M, Wlodarska I, Stefanova-Ouzounova M, Hernandez JM, Hossfeld DK, De Wolf-Peeters C, Hagemeijer A, Van den Berghe H, Marynen P 1999. The apoptosis inhibitor gene API2 and a novel 18q gene, MLT, are recurrently rearranged in the t(11;18)(q21;q21)p6ssociated with mucosa- associated lymphoid tissue lymphomas. Blood 93:3601–3609 [PubMed] [Google Scholar]
- Ding BB, Yu JJ, Yu RY, Mendez LM, Shaknovich R, Zhang Y, Cattoretti G, Ye BH 2008. Constitutively activated STAT3 promotes cell proliferation and survival in the activated B-cell subtype of diffuse large B-cell lymphomas. Blood 111:1515–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunleavy K, Pittaluga S, Czuczman MS, Dave SS, Wright G, Grant N, Shovlin M, Jaffe ES, Janik JE, Staudt LM, et al. 2009. Differential efficacy of bortezomib plus chemotherapy within molecular subtypes of diffuse large B-cell lymphoma. Blood 113:6069–6076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eddy SF, Guo S, Demicco EG, Romieu-Mourez R, Landesman-Bollag E, Seldin DC, Sonenshein GE 2005. Inducible IkappaB kinase/IkappaB kinase epsilon expression is induced by CK2 and promotes aberrant nuclear factor-kappaB activation in breast cancer cells. Cancer Res 65:11375–11383 [DOI] [PubMed] [Google Scholar]
- Emmerich F, Meiser M, Hummel M, Demel G, Foss HD, Jundt F, Mathas S, Krappmann D, Scheidereit C, Stein H, et al. 1999. Overexpression of I kappa B alpha without inhibition of NF-kappaB activity and mutations in the I kappa B alpha gene in reed-sternberg cells. Blood 94:3129–3134 [PubMed] [Google Scholar]
- Emmerich F, Theurich S, Hummel M, Haeffker A, Vry MS, Dohner K, Bommert K, Stein H, Dorken B 2003. Inactivating I kappa B epsilon mutations in Hodgkin/Reed-Sternberg cells. J Pathol 201:413–420 [DOI] [PubMed] [Google Scholar]
- Ferch U, Kloo B, Gewies A, Pfander V, Duwel M, Peschel C, Krappmann D, Ruland J 2009. Inhibition of MALT1 protease activity is selectively toxic for activated B cell-like diffuse large B cell lymphoma cells. J Exp Med 206:2313–2320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreri AJ, Zucca E Marginal-zone lymphoma 2007. Crit Rev Oncol Hematol 63:245–256 [DOI] [PubMed] [Google Scholar]
- Feuerhake F, Kutok JL, Monti S, Chen W, LaCasce AS, Cattoretti G, Kurtin P, Pinkus GS, de Leval L, Harris NL, et al. 2005. NFkappaB activity, function, and target-gene signatures in primary mediastinal large B-cell lymphoma and diffuse large B-cell lymphoma subtypes. Blood 106:1392–1399 [DOI] [PubMed] [Google Scholar]
- Funke L, Dakoji S, Bredt DS 2005. Membrane-associated guanylate kinases regulate adhesion and plasticity at cell junctions. Annu Rev Biochem 74:219–245 [DOI] [PubMed] [Google Scholar]
- Garrison JB, Samuel T, Reed JC 2009. TRAF 2-binding BIR 1 domain of c-IAP 2/MALT 1 fusion protein is essential for activation of NF-kappaB. Oncogene 28:1584–1593 [DOI] [PubMed] [Google Scholar]
- Gauld SB, Cambier JC 2004. Src-family kinases in B-cell development and signaling. Oncogene 23:8001–8006 [DOI] [PubMed] [Google Scholar]
- Gazumyan A, Reichlin A, Nussenzweig MC 2006. Ig beta tyrosine residues contribute to the control of B cell receptor signaling by regulating receptor internalization. J Exp Med 203:1785–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler F, Algul H, Paxian S, Schmid RM 2007. Genetic inactivation of RelA/p65 sensitizes adult mouse hepatocytes to TNF-induced apoptosis in vivo and in vitro. Gastroenterology 132:2489–2503 [DOI] [PubMed] [Google Scholar]
- Greten FR, Arkan MC, Bollrath J, Hsu LC, Goode J, Miething C, Goktuna SI, Neuenhahn M, Fierer J, Paxian S, et al. 2007. NF-kappaB is a negative regulator of IL-1beta secretion as revealed by genetic and pharmacological inhibition of IKKbeta. Cell 130:918–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greten FR, Eckmann L, Greten TF, Park JM, Li ZW, Egan LJ, Kagnoff MF, Karin M 2004. IKKbeta links inflammation and tumorigenesis in a mouse model of colitis-associated cancer. Cell 118:285–296 [DOI] [PubMed] [Google Scholar]
- Gruss HJ, Hirschstein D, Wright B, Ulrich D, Caligiuri MA, Barcos M, Strockbine L, Armitage RJ, Dower SK 1994. Expression and function of CD40 on Hodgkin and Reed-Sternberg cells and the possible relevance for Hodgkin's disease. Blood 84:2305–2314 [PubMed] [Google Scholar]
- Gyrd-Hansen M, Darding M, Miasari M, Santoro MM, Zender L, Xue W, Tenev T, da Fonseca PC, Zvelebil M, Bujnicki JM, et al. 2008. IAPs contain an evolutionarily conserved ubiquitin-binding domain that regulates NF-kappaB as well as cell survival and oncogenesis. Nat Cell Biol 10:1309–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker H, Karin M 2006. Regulation and function of IKK and IKK-related kinases. Sci STKE 2006:pre13. [DOI] [PubMed] [Google Scholar]
- Hailfinger S, Lenz G, Ngo V, Posvitz-Fejfar A, Rebeaud F, Guzzardi M, Penas EM, Dierlamm J, Chan WC, Staudt LM, et al. 2009. Essential role of MALT1 protease activity in activated B cell-like diffuse large B-cell lymphoma. Proc Natl Acad Sci USA 106:19946–19951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S, Hathcock K, Zheng B, Kepler TB, Hodes R, Kelsoe G 1995. Cellular interaction in germinal centers. Roles of CD40 ligand and B7-2 in established germinal centers. J Immunol 155:556–567 [PubMed] [Google Scholar]
- Hantschel O, Rix U, Schmidt U, Burckstummer T, Kneidinger M, Schutze G, Colinge J, Bennett KL, Ellmeier W, Valent P, et al. 2007. The Btk tyrosine kinase is a major target of the Bcr-Abl inhibitor dasatinib. Proc Natl Acad Sci USA 104:13283–13288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hideshima T, Anderson KC 2002. Molecular mechanisms of novel therapeutic approaches for multiple myeloma. Nat Rev Cancer 2:927–937 [DOI] [PubMed] [Google Scholar]
- Hideshima T, Chauhan D, Richardson P, Mitsiades C, Mitsiades N, Hayashi T, Munshi N, Dang L, Castro A, Palombella V, et al. 2002. NF-kappa B as a therapeutic target in multiple myeloma. J Biol Chem 277:16639–16647 [DOI] [PubMed] [Google Scholar]
- Hideshima T, Neri P, Tassone P, Yasui H, Ishitsuka K, Raje N, Chauhan D, Podar K, Mitsiades C, Dang L, et al. 2006. MLN120B, a novel IkappaB kinase beta inhibitor, blocks multiple myeloma cell growth in vitro and in vivo. Clin Cancer Res 12:5887–5894 [DOI] [PubMed] [Google Scholar]
- Hiscott J, Nguyen TL, Arguello M, Nakhaei P, Paz S 2006. Manipulation of the nuclear factor-kappaB pathway and the innate immune response by viruses. Oncogene 25:6844–6867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Du MQ, Park SM, Alcivar A, Qu L, Gupta S, Tang J, Baens M, Ye H, Lee TH, et al. 2006. cIAP2 is a ubiquitin protein ligase for BCL10 and is dysregulated in mucosa-associated lymphoid tissue lymphomas. J Clin Invest 116:174–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel M, Bentink S, Berger H, Klapper W, Wessendorf S, Barth TF, Bernd HW, Cogliatti SB, Dierlamm J, Feller AC, et al. 2006. A biologic definition of Burkitt's lymphoma from transcriptional and genomic profiling. N Engl J Med 354:2419–2430 [DOI] [PubMed] [Google Scholar]
- Hussell T, Isaacson PG, Crabtree JE, Spencer J 1996. Helicobacter pylori-specific tumour-infiltrating T cells provide contact dependent help for the growth of malignant B cells in low-grade gastric lymphoma of mucosa-associated lymphoid tissue. J Pathol 178:122–127 [DOI] [PubMed] [Google Scholar]
- Huynh QK, Boddupalli H, Rouw SA, Koboldt CM, Hall T, Sommers C, Hauser SD, Pierce JL, Combs RG, Reitz BA, et al. 2000. Characterization of the recombinant IKK1/IKK2 heterodimer. Mechanisms regulating kinase activity. J Biol Chem 275:25883–25891 [DOI] [PubMed] [Google Scholar]
- Iqbal J, Greiner TC, Patel K, Dave BJ, Smith L, Ji J, Wright G, Sanger WG, Pickering DL, Jain S, et al. 2007. Distinctive patterns of BCL6 molecular alterations and their functional consequences in different subgroups of diffuse large B-cell lymphoma. Leukemia 21:2332–2343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacson PG, Du MQ 2004. MALT lymphoma: from morphology to molecules. Nat Rev Cancer 4:644–653 [DOI] [PubMed] [Google Scholar]
- Iwakoshi NN, Lee AH, Glimcher LH 2003. The X-box binding protein-1 transcription factor is required for plasma cell differentiation and the unfolded protein response. Immunol Rev 194:29–38 [DOI] [PubMed] [Google Scholar]
- Jun JE, Wilson LE, Vinuesa CG, Lesage S, Blery M, Miosge LA, Cook MC, Kucharska EM, Hara H, Penninger JM, et al. 2003. Identifying the MAGUK protein Carma-1 as a central regulator of humoral immune responses and atopy by genome-wide mouse mutagenesis. Immunity 18:751–762 [DOI] [PubMed] [Google Scholar]
- Jungnickel B, Staratschek-Jox A, Br]auninger A, Spieker T, Wolf J, Diehl V, Hansmann ML, Rajewsky K, R K. 2000. Clonal deleterious mutations in the IkappaBalpha gene in the malignant cells in Hodgkin's lymphoma. J Exp Med 191:395–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalaitzidis D, Davis RE, Rosenwald A, Staudt LM, Gilmore TD 2002. The human B-cell lymphoma cell line RC-K8 has multiple genetic alterations that dysregulate the Rel/NF-kappaB signal transduction pathway. Oncogene 21:8759–8768 [DOI] [PubMed] [Google Scholar]
- Karin M 2010. NF-κB as a critical link between inflammation and cancer. In: Karin M, Staudt LM, eds. NF-κB: A network hub controlling immunity, inflammation, and cancer: Cold Spring Harbor Laboratory Press [Google Scholar]
- Karin M, Lawrence T, Nizet V 2006. Innate immunity gone awry: linking microbial infections to chronic inflammation and cancer. Cell 124:823–835 [DOI] [PubMed] [Google Scholar]
- Karin M, Yamamoto Y, Wang QM 2004. The IKK NF-kappa B system: a treasure trove for drug development. Nat Rev Drug Discov 3:17–26 [DOI] [PubMed] [Google Scholar]
- Karrer U, Althage A, Odermatt B, Hengartner H, Zinkernagel RM 2000. Immunodeficiency of alymphoplasia mice (aly/aly) in vivo: structural defect of secondary lymphoid organs and functional B cell defect. Eur J Immunol 30:2799–2807 [DOI] [PubMed] [Google Scholar]
- Keats JJ, Fonseca R, Chesi M, Schop R, Baker A, Chng WJ, Van Wier S, Tiedemann R, Shi CX, Sebag M, et al. 2007. Promiscuous mutations activate the noncanonical NF-kappaB pathway in multiple myeloma. Cancer Cell 12:131–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein U, Casola S, Cattoretti G, Shen Q, Lia M, Mo T, Ludwig T, Rajewsky K, Dalla-Favera R 2006. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat Immunol 7:773–782 [DOI] [PubMed] [Google Scholar]
- Kovalenko A, Chable-Bessia C, Cantarella G, Israel A, Wallach D, Courtois G 2003. The tumour suppressor CYLD negatively regulates NF-kappaB signalling by deubiquitination. Nature 424:801–805 [DOI] [PubMed] [Google Scholar]
- Krappmann D, Emmerich F, Kordes U, Scharschmidt E, Dorken B, Scheidereit C 1999. Molecular mechanisms of constitutive NF-kappaB/Rel activation in Hodgkin/Reed-Sternberg cells. Oncogene 18:943–953 [DOI] [PubMed] [Google Scholar]
- Kraus M, Alimzhanov MB, Rajewsky N, Rajewsky K 2004. Survival of resting mature B lymphocytes depends on BCR signaling via the Igalpha/beta heterodimer. Cell 117:787–800 [DOI] [PubMed] [Google Scholar]
- Kraus M, Saijo K, Torres RM, Rajewsky K 1999. Ig-alpha cytoplasmic truncation renders immature B cells more sensitive to antigen contact. Immunity 11:537–545 [DOI] [PubMed] [Google Scholar]
- Lake A, Shield LA, Cordano P, Chui DT, Osborne J, Crae S, Wilson KS, Tosi S, Knight SJ, Gesk S, et al. 2009. Mutations of NFKBIA, encoding IkappaB alpha, are a recurrent finding in classical Hodgkin lymphoma but are not a unifying feature of non-EBV-associated cases. Int J Cancer 125:1334–1342 [DOI] [PubMed] [Google Scholar]
- Lam KP, Kuhn R, Rajewsky K 1997. In vivo ablation of surface immunoglobulin on mature B cells by inducible gene targeting results in rapid cell death. Cell 90:1073–1083 [DOI] [PubMed] [Google Scholar]
- Lam LT, Davis RE, Ngo VN, Lenz G, Wright G, Xu W, Zhao H, Yu X, Dang L, Staudt LM 2008. Compensatory IKKalpha activation of classical NF-kappaB signaling during IKKbeta inhibition identified by an RNA interference sensitization screen. Proc Natl Acad Sci U S A 105:20798–20803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam LT, Davis RE, Pierce J, Hepperle M, Xu Y, Hottelet M, Nong Y, Wen D, Adams J, Dang L, et al. 2005. Small molecule inhibitors of IkappaB kinase are selectively toxic for subgroups of diffuse large B-cell lymphoma defined by gene expression profiling. Clin Cancer Res 11:28–40 [PubMed] [Google Scholar]
- Lam LT, Wright G, Davis RE, Lenz G, Farinha P, Dang L, Chan JW, Rosenwald A, Gascoyne RD, Staudt LM 2008. Cooperative signaling through the signal transducer and activator of transcription 3 and nuclear factor-{kappa}B pathways in subtypes of diffuse large B-cell lymphoma. Blood 111:3701–3713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitges M, Schmedt C, Guinamard R, Davoust J, Schaal S, Stabel S, Tarakhovsky A 1996. Immunodeficiency in protein kinase cbeta-deficient mice. Science 273:788–791 [DOI] [PubMed] [Google Scholar]
- Lenz G, Davis RE, Ngo VN, Lam L, George TC, Wright GW, Dave SS, Zhao H, Xu W, Rosenwald A, et al. 2008. Oncogenic CARD11 mutations in human diffuse large B cell lymphoma. Science 319:1676–1679 [DOI] [PubMed] [Google Scholar]
- Lenz G, Wright G, Dave SS, Xiao W, Powell J, Zhao H, Xu W, Tan B, Goldschmidt N, Iqbal J, et al. 2008. Stromal gene signatures in large-B-cell lymphomas. N Engl J Med 359:2313–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenz G, Wright GW, Emre NC, Kohlhammer H, Dave SS, Davis RE, Carty S, Lam LT, Shaffer AL, Xiao W, et al. 2008. Molecular subtypes of diffuse large B-cell lymphoma arise by distinct genetic pathways. Proc Natl Acad Sci USA 105:13520–13525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Norris PS, Ni CZ, Havert ML, Chiong EM, Tran BR, Cabezas E, Reed JC, Satterthwait AC, Ware CF, et al. 2003. Structurally distinct recognition motifs in lymphotoxin-beta receptor and CD40 for tumor necrosis factor receptor-associated factor (TRAF)-mediated signaling. J Biol Chem 278:50523–50529 [DOI] [PubMed] [Google Scholar]
- Li Z, Wang H, Xue L, Shin DM, Roopenian D, Xu W, Qi CF, Sangster MY, Orihuela CJ, Tuomanen E, et al. 2009. Emu-BCL10 mice exhibit constitutive activation of both canonical and noncanonical NF-kappaB pathways generating marginal zone (MZ) B-cell expansion as a precursor to splenic MZ lymphoma. Blood 114:4158–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao G, Zhang M, Harhaj EW, Sun SC 2004. Regulation of the NF-kappaB-inducing kinase by tumor necrosis factor receptor-associated factor 3-induced degradation. J Biol Chem 279:26243–26250 [DOI] [PubMed] [Google Scholar]
- Lin SC, Chung JY, Lamothe B, Rajashankar K, Lu M, Lo YC, Lam AY, Darnay BG, Wu H 2008. Molecular basis for the unique deubiquitinating activity of the NF-kappaB inhibitor A20. J Mol Biol 376:526–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Wong K, Calame K 1997. Repression of c-myc transcription by Blimp-1, an inducer of terminal B cell differentiation. Science 276:596–599 [DOI] [PubMed] [Google Scholar]
- Liu H, Ye H, Ruskone-Fourmestraux A, De Jong D, Pileri S, Thiede C, Lavergne A, Boot H, Caletti G, Wundisch T, et al. 2002. T(11;18) is a marker for all stage gastric MALT lymphomas that will not respond to H. pylori eradication. Gastroenterology 122:1286–1294 [DOI] [PubMed] [Google Scholar]
- Lucas PC, Kuffa P, Gu S, Kohrt D, Kim DS, Siu K, Jin X, Swenson J, McAllister-Lucas LM 2007. A dual role for the API2 moiety in API2-MALT1-dependent NF-kappaB activation: heterotypic oligomerization and TRAF2 recruitment. Oncogene 26:5643–5654 [DOI] [PubMed] [Google Scholar]
- Lucas PC, Yonezumi M, Inohara N, McAllister-Lucas LM, Abazeed ME, Chen FF, Yamaoka S, Seto M, Nunez G 2001. Bcl10 and MALT1, independent targets of chromosomal translocation in malt lymphoma, cooperate in a novel NF-kappa B signaling pathway. J Biol Chem 276:19012–19019 [DOI] [PubMed] [Google Scholar]
- Luedde T, Assmus U, Wustefeld T, Meyer zu Vilsendorf A, Roskams T, Schmidt-Supprian M, Rajewsky K, Brenner DA, Manns MP, Pasparakis M, et al. 2005. Deletion of IKK2 in hepatocytes does not sensitize these cells to TNF-induced apoptosis but protects from ischemia/reperfusion injury. J Clin Invest 115:849–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luedde T, Beraza N, Kotsikoris V, van Loo G, Nenci A, De Vos R, Roskams T, Trautwein C, Pasparakis M 2007. Deletion of NEMO/IKKgamma in liver parenchymal cells causes steatohepatitis and hepatocellular carcinoma. Cancer Cell 11:119–132 [DOI] [PubMed] [Google Scholar]
- Ma H, Yankee TM, Hu J, Asai DJ, Harrison ML, Geahlen RL 2001. Visualization of Syk-antigen receptor interactions using green fluorescent protein: differential roles for Syk and Lyn in the regulation of receptor capping and internalization. J Immunol 166:1507–1516 [DOI] [PubMed] [Google Scholar]
- Malinin NL, Boldin MP, Kovalenko AV, Wallach D 1997. MAP3K-related kinase involved in NF-kappaB induction by TNF, CD95 and IL-1. Nature 385:540–544 [DOI] [PubMed] [Google Scholar]
- Marsters SA, Yan M, Pitti RM, Haas PE, Dixit VM, Ashkenazi A 2000. Interaction of the TNF homologues BLyS and APRIL with the TNF receptor homologues BCMA and TACI. Curr Biol 10:785–788 [DOI] [PubMed] [Google Scholar]
- Massoumi R, Chmielarska K, Hennecke K, Pfeifer A, Fassler R 2006. Cyld inhibits tumor cell proliferation by blocking Bcl-3-dependent NF-kappaB signaling. Cell 125:665–677 [DOI] [PubMed] [Google Scholar]
- Matsumoto R, Wang D, Blonska M, Li H, Kobayashi M, Pappu B, Chen Y, Lin X 2005. Phosphorylation of CARMA1 plays a critical role in T Cell receptor-mediated NF-kappaB activation. Immunity 23:575–585 [DOI] [PubMed] [Google Scholar]
- Matsuzawa A, Tseng PH, Vallabhapurapu S, Luo JL, Zhang W, Wang H, Vignali DA, Gallagher E, Karin M 2008. Essential cytoplasmic translocation of a cytokine receptor-assembled signaling complex. Science 321:663–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauro C, Pacifico F, Lavorgna A, Mellone S, Iannetti A, Acquaviva R, Formisano S, Vito P, Leonardi A 2006. ABIN-1 binds to NEMO/IKKgamma and co-operates with A20 in inhibiting NF-kappaB. J Biol Chem 281:18482–18488 [DOI] [PubMed] [Google Scholar]
- McCully RR, Pomerantz JL 2008. The protein kinase C-responsive inhibitory domain of CARD11 functions in NF-kappaB activation to regulate the association of multiple signaling cofactors that differentially depend on Bcl10 and MALT1 for association. Mol Cell Biol 28:5668–5686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meylan E, Dooley AL, Feldser DM, Shen L, Turk E, Ouyang C, Jacks T 2009. Requirement for NF-kappaB signalling in a mouse model of lung adenocarcinoma. Nature 462:104–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migliazza A, Lombardi L, Rocchi M, Trecca D, Chang CC, Antonacci R, Fracchiolla NS, Ciana P, Maiolo AT, Neri A 1994. Heterogeneous chromosomal aberrations generate 3' truncations of the NFKB2/lyt-10 gene in lymphoid malignancies. Blood 84:3850–3860 [PubMed] [Google Scholar]
- Miyawaki S, Nakamura Y, Suzuka H, Koba M, Yasumizu R, Ikehara S, Shibata Y 1994. A new mutation, aly, that induces a generalized lack of lymph nodes accompanied by immunodeficiency in mice. Eur J Immunol 24:429–434 [DOI] [PubMed] [Google Scholar]
- Molin D, Edstrom A, Glimelius I, Glimelius B, Nilsson G, Sundstrom C, Enblad G 2002. Mast cell infiltration correlates with poor prognosis in Hodgkin's lymphoma. Br J Haematol 119:122–124 [DOI] [PubMed] [Google Scholar]
- Monroe JG 2006. ITAM-mediated tonic signalling through pre-BCR and BCR complexes. Nat Rev Immunol 6:283–294 [DOI] [PubMed] [Google Scholar]
- Monti S, Savage KJ, Kutok JL, Feuerhake F, Kurtin P, Mihm M, Wu B, Pasqualucci L, Neuberg D, Aguiar RC, et al. 2005. Molecular profiling of diffuse large B-cell lymphoma identifies robust subtypes including one characterized by host inflammatory response. Blood 105:1851–1861 [DOI] [PubMed] [Google Scholar]
- Moreaux J, Cremer FW, Reme T, Raab M, Mahtouk K, Kaukel P, Pantesco V, De Vos J, Jourdan E, Jauch A, et al. 2005. The level of TACI gene expression in myeloma cells is associated with a signature of microenvironment dependence versus a plasmablastic signature. Blood 106:1021–1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JA, Yin Y, Borowsky AD, Kuo F, Nourmand N, Koontz JI, Reynolds C, Soreng L, Griffin CA, Graeme-Cook F, et al. 1999. Breakpoints of the t(11;18)(q21;q21) in mucosa-associated lymphoid tissue (MALT) lymphoma lie within or near the previously undescribed gene MALT1 in chromosome 18. Cancer Res 59:6205–6213 [PubMed] [Google Scholar]
- Nagashima K, Sasseville VG, Wen D, Bielecki A, Yang H, Simpson C, Grant E, Hepperle M, Harriman G, Jaffee B, et al. 2006. Rapid TNFR1-dependent lymphocyte depletion in vivo with a selective chemical inhibitor of IKKbeta. Blood 107:4266–4273 [DOI] [PubMed] [Google Scholar]
- Ngo VN, Davis RE, Lamy L, Yu X, Zhao H, Lenz G, Lam LT, Dave S, Yang L, Powell J, et al. 2006. A loss-of-function RNA interference screen for molecular targets in cancer. Nature 441:106–110 [DOI] [PubMed] [Google Scholar]
- Ni CZ, Oganesyan G, Welsh K, Zhu X, Reed JC, Satterthwait AC, Cheng G, Ely KR 2004. Key molecular contacts promote recognition of the BAFF receptor by TNF receptor-associated factor 3: implications for intracellular signaling regulation. J Immunol 173:7394–7400 [DOI] [PubMed] [Google Scholar]
- Ni CZ, Welsh K, Leo E, Chiou CK, Wu H, Reed JC, Ely KR 2000. Molecular basis for CD40 signaling mediated by TRAF3. Proc Natl Acad Sci U S A 97:10395–10399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niiro H, Allam A, Stoddart A, Brodsky FM, Marshall AJ, Clark EA 2004. The B lymphocyte adaptor molecule of 32 kilodaltons (Bam32) regulates B cell antigen receptor internalization. J Immunol 173:5601–5609 [DOI] [PubMed] [Google Scholar]
- Noels H, van Loo G, Hagens S, Broeckx V, Beyaert R, Marynen P, Baens M 2007. A Novel TRAF6 binding site in MALT1 defines distinct mechanisms of NF-kappaB activation by API2middle dotMALT1 fusions. J Biol Chem 282:10180–10189 [DOI] [PubMed] [Google Scholar]
- O'Connor BP, Raman VS, Erickson LD, Cook WJ, Weaver LK, Ahonen C, Lin LL, Mantchev GT, Bram RJ, Noelle RJ 2004. BCMA is essential for the survival of long-lived bone marrow plasma cells. J Exp Med 199:91–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Mahony A, Lin X, Geleziunas R, Greene WC 2000. Activation of the heterodimeric IkappaB kinase alpha (IKKalpha)-IKKbeta complex is directional: IKKalpha regulates IKKbeta under both basal and stimulated conditions. Mol Cell Biol 20:1170–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oeckinghaus A, Wegener E, Welteke V, Ferch U, Arslan SC, Ruland J, Scheidereit C, Krappmann D 2007. Malt1 ubiquitination triggers NF-kappaB signaling upon T-cell activation. Embo J 26:4634–4645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Z, Scheerens H, Li SJ, Schultz BE, Sprengeler PA, Burrill LC, Mendonca RV, Sweeney MD, Scott KC, Grothaus PG, et al. 2007. Discovery of selective irreversible inhibitors for Bruton's tyrosine kinase. ChemMedChem 2:58–61 [DOI] [PubMed] [Google Scholar]
- Pasparakis M, Schmidt-Supprian M, Rajewsky K 2002. IkappaB kinase signaling is essential for maintenance of mature B cells. J Exp Med 196:743–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasqualucci L, Compagno M, Houldsworth J, Monti S, Grunn A, Nandula SV, Aster JC, Murty VV, Shipp MA, Dalla-Favera R 2006. Inactivation of the PRDM1/BLIMP1 gene in diffuse large B cell lymphoma. J Exp Med 203:311–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters RT, Liao SM, Maniatis T 2000. IKKepsilon is part of a novel PMA-inducible IkappaB kinase complex. Mol Cell 5:513–522 [DOI] [PubMed] [Google Scholar]
- Petro JB, Khan WN 2001. Phospholipase C-gamma 2 couples Bruton's tyrosine kinase to the NF-kappaB signaling pathway in B lymphocytes. J Biol Chem 276:1715–1719 [DOI] [PubMed] [Google Scholar]
- Petro JB, Rahman SM, Ballard DW, Khan WN 2000. Bruton's tyrosine kinase is required for activation of IkappaB kinase and nuclear factor kappaB in response to B cell receptor engagement. J Exp Med 191:1745–1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A, Reis e Sousa C 2007. Innate recognition of viruses. Immunity 27:370–383 [DOI] [PubMed] [Google Scholar]
- Pinto A, Aldinucci D, Gloghini A, Zagonel V, Degan M, Improta S, Juzbasic S, Todesco M, Perin V, Gattei V, et al. 1996. Human eosinophils express functional CD30 ligand and stimulate proliferation of a Hodgkin's disease cell line. Blood 88:3299–3305 [PubMed] [Google Scholar]
- Podust VN, Brownell JE, Gladysheva TB, Luo RS, Wang C, Coggins MB, Pierce JW, Lightcap ES, Chau V 2000. A Nedd 8 conjugation pathway is essential for proteolytic targeting of p27Kip1 by ubiquitination. Proc Natl Acad Sci U S A 97:4579–4584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomerantz JL, Baltimore D 1999. NF-kappaB activation by a signaling complex containing TRAF2, TANK and TBK1, a novel IKK-related kinase. EMBO J 18:6694–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan P, Wang W, Wallach D 2004. Receptor-specific signaling for both the alternative and the canonical NF-kappaB activation pathways by NF-kappaB-inducing kinase. Immunity 21:477–89 [DOI] [PubMed] [Google Scholar]
- Rawlings DJ, Sommer K, Moreno-Garcia ME 2006. The CARMA 1 signalosome links the signalling machinery of adaptive and innate immunity in lymphocytes. Nat Rev Immunol 6:799–812 [DOI] [PubMed] [Google Scholar]
- Read MA, Brownell JE, Gladysheva TB, Hottelet M, Parent LA, Coggins MB, Pierce JW, Podust VN, Luo RS, Chau V, et al. 2000. Nedd8 modification of cul-1 activates SCF(beta(TrCP))-dependent ubiquitination of IkappaBalpha. Mol Cell Biol 20:2326–2333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebeaud F, Hailfinger S, Posevitz-Fejfar A, Tapernoux M, Moser R, Rueda D, Gaide O, Guzzardi M, Iancu EM, Rufer N, et al. 2008. The proteolytic activity of the paracaspase MALT1 is key in T cell activation. Nat Immunol 9:272–281 [DOI] [PubMed] [Google Scholar]
- Regamey A, Hohl D, Liu JW, Roger T, Kogerman P, Toftgard R, Huber M 2003. The tumor suppressor CYLD interacts with TRIP and regulates negatively nuclear factor kappaB activation by tumor necrosis factor. J Exp Med 198:1959–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimold AM, Iwakoshi NN, Manis J, Vallabhajosyula P, Szomolanyi-Tsuda E, Gravallese EM, Friend D, Grusby MJ, Alt F, Glimcher LH 2001. Plasma cell differentiation requires the transcription factor XBP-1. Nature 412:300–307 [DOI] [PubMed] [Google Scholar]
- Reth M 1989. Antigen receptor tail clue. Nature 338:383–384 [PubMed] [Google Scholar]
- Richardson PG, Sonneveld P, Schuster MW, Irwin D, Stadtmauer EA, Facon T, Harousseau JL, Ben-Yehuda D, Lonial S, Goldschmidt H, et al. 2005. Bortezomib or high-dose dexamethasone for relapsed multiple myeloma. N Engl J Med 352:2487–2498 [DOI] [PubMed] [Google Scholar]
- Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, Gascoyne RD, Muller-Hermelink HK, Smeland EB, Giltnane JM, et al. 2002. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med 346:1937–1947 [DOI] [PubMed] [Google Scholar]
- Rosenwald A, Wright G, Leroy K, Yu X, Gaulard P, Gascoyne RD, Chan WC, Zhao T, Haioun C, Greiner TC, et al. 2003. Molecular diagnosis of primary mediastinal B cell lymphoma identifies a clinically favorable subgroup of diffuse large B cell lymphoma related to Hodgkin lymphoma. J Exp Med 198:851–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruland J, Duncan GS, Wakeham A, Mak TW 2003. Differential requirement for Malt1 in T and B cell antigen receptor signaling. Immunity 19:749–758 [DOI] [PubMed] [Google Scholar]
- Saijo K, Mecklenbrauker I, Santana A, Leitger M, Schmedt C, Tarakhovsky A 2002. Protein kinase C beta controls nuclear factor kappaB activation in B cells through selective regulation of the IkappaB kinase alpha. J Exp Med 195:1647–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samuel T, Welsh K, Lober T, Togo SH, Zapata JM, Reed JC 2006. Distinct BIR domains of cIAP1 mediate binding to and ubiquitination of tumor necrosis factor receptor-associated factor 2 and second mitochondrial activator of caspases. J Biol Chem 281:1080–1090 [DOI] [PubMed] [Google Scholar]
- Sanchez-Izquierdo D, Buchonnet G, Siebert R, Gascoyne RD, Climent J, Karran L, Marin M, Blesa D, Horsman D, Rosenwald A, et al. 2003. MALT1 is deregulated by both chromosomal translocation and amplification in B-cell non-Hodgkin lymphoma. Blood 101:4539–4546 [DOI] [PubMed] [Google Scholar]
- Satterthwaite AB, Witte ON 2000. The role of Bruton's tyrosine kinase in B-cell development and function: a genetic perspective. Immunol Rev 175:120–127 [PubMed] [Google Scholar]
- Savage KJ, Monti S, Kutok JL, Cattoretti G, Neuberg D, De Leval L, Kurtin P, Dal Cin P, Ladd C, Feuerhake F, et al. 2003. The molecular signature of mediastinal large B-cell lymphoma differs from that of other diffuse large B-cell lymphomas and shares features with classical Hodgkin lymphoma. Blood 102:3871–3879 [DOI] [PubMed] [Google Scholar]
- Schmitz R, Hansmann ML, Bohle V, Martin-Subero JI, Hartmann S, Mechtersheimer G, Klapper W, Vater I, Giefing M, Gesk S, et al. 2009. TNFAIP3 (A20) is a tumor suppressor gene in Hodgkin lymphoma and primary mediastinal B cell lymphoma. J Exp Med 206:981–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz R, Stanelle J, Hansmann ML, Kuppers R 2009. Pathogenesis of classical and lymphocyte-predominant Hodgkin lymphoma. Annual review of pathology 4:151–174 [DOI] [PubMed] [Google Scholar]
- Schwab U, Stein H, Gerdes J, Lemke H, Kirchner H, Schaadt M, Diehl V 1982. Production of a monoclonal antibody specific for Hodgkin and Sternberg-Reed cells of Hodgkin's disease and a subset of normal lymphoid cells. Nature 299:65–67 [DOI] [PubMed] [Google Scholar]
- Schwabe RF, Brenner DA Nuclear factor-kappaB in the liver: friend or foe? 2007. Gastroenterology 132:2601–2604 [DOI] [PubMed] [Google Scholar]
- Sciammas R, Shaffer AL, Schatz JH, Zhao H, Staudt LM, Singh H 2006. Graded expression of interferon regulatory factor-4 coordinates isotype switching with plasma cell differentiation. Immunity 25:225–236 [DOI] [PubMed] [Google Scholar]
- Senftleben U, Cao Y, Xiao G, Greten FR, Krahn G, Bonizzi G, Chen Y, Hu Y, Fong A, Sun SC, et al. 2001. Activation by IKKalpha of a second, evolutionary conserved, NF-kappa B signaling pathway. Science 293:1495–1499 [DOI] [PubMed] [Google Scholar]
- Shaffer AL, Emre NC, Lamy L, Ngo VN, Wright G, Xiao W, Powell J, Dave S, Yu X, Zhao H, et al. 2008. IRF4 addiction in multiple myeloma. Nature 454:226–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer AL, Emre NC, Romesser PB, Staudt LM 2009. IRF4: Immunity. Malignancy! Therapy? Clin Cancer Res 15:2954–2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer AL, Lin KI, Kuo TC, Yu X, Hurt EM, Rosenwald A, Giltnane JM, Yang L, Zhao H, Calame K, et al. 2002. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity 17:51–62 [DOI] [PubMed] [Google Scholar]
- Shaffer AL, Rosenwald A, Hurt EM, Giltnane JM, Lam LT, Pickeral OK, Staudt LM 2001. Signatures of the immune response. Immunity 15:375–385 [DOI] [PubMed] [Google Scholar]
- Shaffer AL, Shapiro-Shelef M, Iwakoshi NN, Lee A-H, Qian S-B, Zhao H, Yu X, Yang L, Tan BK, Rosenwald A, et al. 2004. XBP1, Downstream of Blimp-1, Expands the Secretory Apparatus and Other Organelles, and Increases Protein Synthesis in Plasma Cell Differentiation. Immunity 21:81–93 [DOI] [PubMed] [Google Scholar]
- Shaffer AL, Yu X, He Y, Boldrick J, Chan EP, Staudt LM 2000. BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity 13:199–212 [DOI] [PubMed] [Google Scholar]
- Shapiro-Shelef M, Calame K 2005. Regulation of plasma-cell development. Nat Rev Immunol 5:230–242 [DOI] [PubMed] [Google Scholar]
- Shapiro-Shelef M, Lin KI, McHeyzer-Williams LJ, Liao J, McHeyzer-Williams MG, Calame K 2003. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity 19:607–620 [DOI] [PubMed] [Google Scholar]
- Shapiro-Shelef M, Lin KI, Savitsky D, Liao J, Calame K 2005. Blimp-1 is required for maintenance of long-lived plasma cells in the bone marrow. J Exp Med 202:1471–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada T, Kawai T, Takeda K, Matsumoto M, Inoue J, Tatsumi Y, Kanamaru A, Akira S 1999. IKK-i, a novel lipopolysaccharide-inducible kinase that is related to IkappaB kinases. Int Immunol 11:1357–1362 [DOI] [PubMed] [Google Scholar]
- Shinkura R, Kitada K, Matsuda F, Tashiro K, Ikuta K, Suzuki M, Kogishi K, Serikawa T, Honjo T 1999. Alymphoplasia is caused by a point mutation in the mouse gene encoding Nf-kappa b-inducing kinase. Nat Genet 22:74–77 [DOI] [PubMed] [Google Scholar]
- Sommer K, Guo B, Pomerantz JL, Bandaranayake AD, Moreno-Garcia ME, Ovechkina YL, Rawlings DJ 2005. Phosphorylation of the CARMA1 linker controls NF-kappaB activation. Immunity 23:561–574 [DOI] [PubMed] [Google Scholar]
- Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, et al. 2009. An inhibitor of NEDD8-activating enzyme as a new approach to treat cancer. Nature 458:732–736 [DOI] [PubMed] [Google Scholar]
- Srinivasan L, Sasaki Y, Calado DP, Zhang B, Paik JH, DePinho RA, Kutok JL, Kearney JF, Otipoby KL, Rajewsky K 2009. PI3 kinase signals BCR-dependent mature B cell survival. Cell 139:573–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudt LM, Dave S 2005. The biology of human lymphoid malignancies revealed by gene expression profiling. Adv Immunol 87:163–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Deng L, Ea CK, Xia ZP, Chen ZJ 2004. The TRAF 6 ubiquitin ligase and TAK 1 kinase mediate IKK activation by BCL 10 and MALT 1 in T lymphocytes. Mol Cell 14:289–301 [DOI] [PubMed] [Google Scholar]
- Sun SC 2009. CYLD: a tumor suppressor deubiquitinase regulating NF-kappaB activation and diverse biological processes. Cell death and differentiation 17:25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam W, Gomez M, Chadburn A, Lee JW, Chan WC, Knowles DM 2006. Mutational analysis of PRDM1 indicates a tumor-suppressor role in diffuse large B-cell lymphomas. Blood 107:4090–4100 [DOI] [PubMed] [Google Scholar]
- Tan JE, Wong SC, Gan SK, Xu S, Lam KP 2001. The adaptor protein BLNK is required for B cell antigen receptor- induced activation of NFk-B and cell cycle entry and Survival of B lymphocytes. J Biol Chem 23:23. [DOI] [PubMed] [Google Scholar]
- Tanner MJ, Hanel W, Gaffen SL, Lin X 2007. CARMA1 coiled-coil domain is involved in the oligomerization and subcellular localization of CARMA1 and is required for T cell receptor-induced NF-kappaB activation. J Biol Chem 282:17141–17147 [DOI] [PubMed] [Google Scholar]
- Thome M 2004. CARMA1, BCL-10 and MALT 1 in lymphocyte development and activation. Nat Rev Immunol 4:348–359 [DOI] [PubMed] [Google Scholar]
- Tojima Y, Fujimoto A, Delhase M, Chen Y, Hatakeyama S, Nakayama K, Kaneko Y, Nimura Y, Motoyama N, Ikeda K, et al. 2000. NAK is an IkappaB kinase-activating kinase. Nature 404:778–782 [DOI] [PubMed] [Google Scholar]
- Tolar P, Hanna J, Krueger PD, Pierce SK 2009. The constant region of the membrane immunoglobulin mediates B cell-receptor clustering and signaling in response to membrane antigens. Immunity 30:44–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres RM, Hafen K 1999. A negative regulatory role for Ig-alpha during B cell development. Immunity 11:527–536 [DOI] [PubMed] [Google Scholar]
- Trompouki E, Hatzivassiliou E, Tsichritzis T, Farmer H, Ashworth A, Mosialos G 2003. CYLD is a deubiquitinating enzyme that negatively regulates NF-kappaB activation by TNFR family members. Nature 424:793–796 [DOI] [PubMed] [Google Scholar]
- Turer EE, Tavares RM, Mortier E, Hitotsumatsu O, Advincula R, Lee B, Shifrin N, Malynn BA, Ma A 2008. Homeostatic MyD 8 8-dependent signals cause lethal inflamMation in the absence of A 20. J Exp Med 205:451–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner CA Jr., Mack DH, Davis MM 1994. Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell 77:297–306 [DOI] [PubMed] [Google Scholar]
- Uren AG, O'Rourke K, Aravind LA, Pisabarro MT, Seshagiri S, Koonin EV, Dixit VM 2000. Identification of paracaspases and metacaspases: two ancient families of caspase-like proteins, one of which plays a key role in MALT lymphoma. Mol Cell 6:961–967 [DOI] [PubMed] [Google Scholar]
- Vallabhapurapu S, Karin M 2009. Regulation and function of NF-kappaB transcription factors in the immune system. Annu Rev Immunol 27:693–733 [DOI] [PubMed] [Google Scholar]
- Vallabhapurapu S, Matsuzawa A, Zhang W, Tseng PH, Keats JJ, Wang H, Vignali DA, Bergsagel PL, Karin M 2008. Nonredundant and complementary functions of TRAF2 and TRAF3 in a ubiquitination cascade that activates NIK-dependent alternative NF-kappaB signaling. Nat Immunol 9:1364–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CY, Mayo MW, Korneluk RG, Goeddel DV, Baldwin AS 1998. NF-kappaB antiapoptosis: induction of TRAF1 and TRAF2 and c-IAP1 and c-IAP2 to suppress caspase-8 activation. Science 281:1680–1683 [DOI] [PubMed] [Google Scholar]
- Werner SL, Barken D, Hoffmann A 2005. Stimulus specificity of gene expression programs determined by temporal control of IKK activity. Science 309:1857–1861 [DOI] [PubMed] [Google Scholar]
- Wertz IE, O'Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, Wu P, Wiesmann C, Baker R, Boone DL, et al. 2004. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature 430:694–699 [DOI] [PubMed] [Google Scholar]
- Willis TG, Jadayel DM, Du MQ, Peng H, Perry AR, Abdul-Rauf M, Price H, Karran L, Majekodunmi O, Wlodarska I, et al. 1999. Bcl10 is involved in t(1;14)(p22;q32) of MALT B cell lymphoma and mutated in multiple tumor types. Cell 96:35–45 [DOI] [PubMed] [Google Scholar]
- Workman P, Burrows F, Neckers L, Rosen N 2007. Drugging the cancer chaperone HSP90: combinatorial therapeutic exploitation of oncogene addiction and tumor stress. Ann N Y Acad Sci 1113:202–216 [DOI] [PubMed] [Google Scholar]
- Woronicz JD, Gao X, Cao Z, Rothe M, Goeddel DV 1997. IkappaB kinase-beta: NF-kappaB activation and complex formation with IkappaB kinase-alpha and NIK. Science 278:866–869 [DOI] [PubMed] [Google Scholar]
- Wotherspoon AC, Doglioni C, Diss TC, Pan L, Moschini A, de Boni M, Isaacson PG 1993. Regression of primary low-grade B-cell gastric lymphoma of mucosa- associated lymphoid tissue type after eradication of Helicobacter pylori. Lancet 342:575–577 [DOI] [PubMed] [Google Scholar]
- Wright CW, Duckett CS 2005. Reawakening the cellular death program in neoplasia through the therapeutic blockade of IAP function. J Clin Invest 115:2673–2678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Harder KW, Huntington ND, Hibbs ML, Tarlinton DM 2005. Lyn tyrosine kinase: accentuating the positive and the negative. Immunity 22:9–18 [DOI] [PubMed] [Google Scholar]
- Yaccoby S, Pennisi A, Li X, Dillon SR, Zhan F, Barlogie B, Shaughnessy JD Jr. 2008. Atacicept (TACI-Ig) inhibits growth of TACI (high) primary myeloma cells in SCID-hu mice and in coculture with osteoclasts. Leukemia 22:406–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Liao X, Agarwal MK, Barnes L, Auron PE, Stark GR 2007. Unphosphorylated STAT 3 accumulates in response to IL- 6 and activates transcription by binding to NFkappaB. Genes Dev 21:1396–1408 Epub 2007 May 17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Chang HY, Baltimore D 1998. Autoproteolytic activation of pro-caspases by oligomerization. Mol Cell 1:319–325 [DOI] [PubMed] [Google Scholar]
- Yaron A, Hatzubai A, Davis M, Lavon I, Amit S, Manning AM, Andersen JS, Mann M, Mercurio F, Ben-Neriah Y 1998. Identification of the receptor component of the IkappaBalpha-ubiquitin ligase. Nature 396:590–594 [DOI] [PubMed] [Google Scholar]
- Yin L, Wu L, Wesche H, Arthur CD, White JM, Goeddel DV, Schreiber RD 2001. Defective lymphotoxin-beta receptor-induced NF-kappaB transcriptional activity in NIK-deficient mice. Science 291:2162–2165 [DOI] [PubMed] [Google Scholar]
- Zarnegar B, Yamazaki S, He JQ, Cheng G 2008. Control of canonical NF-kappaB activation through the NIK-IKK complex pathway. Proc Natl Acad Sci U S A 105:3503–3508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarnegar BJ, Wang Y, Mahoney DJ, Dempsey PW, Cheung HH, He J, Shiba T, Yang X, Yeh WC, Mak TW, et al. 2008. Noncanonical NF-kappaB activation requires coordinated assembly of a regulatory complex of the adaptors cIAP1, cIAP2, TRAF2 and TRAF3 and the kinase NIK. Nat Immunol 9:1371–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Siebert R, Yan M, Hinzmann B, Cui X, Xue L, Rakestraw KM, Naeve CW, Beckmann G, Weisenburger DD, et al. 1999. Inactivating mutations and overexpression of BCL10, a caspase recruitment domain-containing gene, in MALT lymphoma with t(1;14)(p22;q32). Nat Genet 22:63–68 [DOI] [PubMed] [Google Scholar]
- Zhou H, Du MQ, Dixit VM 2005. Constitutive NF-kappaB activation by the t(11;18)(q21;q21) product in MALT lymphoma is linked to deregulated ubiquitin ligase activity. Cancer Cell 7:425–431 [DOI] [PubMed] [Google Scholar]
- Zhou H, Wertz I, O'Rourke K, Ultsch M, Seshagiri S, Eby M, Xiao W, Dixit VM 2004. Bcl10 activates the NF-kappaB pathway through ubiquitination of NEMO. Nature 427:167–171 [DOI] [PubMed] [Google Scholar]