Abstract
Forward and reverse genetic experiments have both played important roles in revealing critical aspects of mammalian signal transduction pathways in cell culture experiments. Only recently have we begun to comprehend the depth, breadth, and complexity of these pathways and of their interrelationships. Here, we summarize successful examples in which different forward genetic approaches have led to novel discoveries in NF-κB signaling. We believe that forward genetics will continue to play an irreplaceable role in advancing our understanding of the complexities of the pathways that regulate the functions of this key transcription factor.
Selection systems that identify cells with disrupted NF-κB signaling are revealing novel regulators of this pathway, including proteins such as Act1, RIP1, and FBXL11.
FORWARD GENETICS IN MAMMALIAN CELLS IN TISSUE CULTURE
Genetic approaches to investigate signaling mechanisms in cell culture fall into two broad categories: In forward genetics, one creates random mutations in a population of cells, whereas in reverse genetics, one manipulates a known gene in a single cell clone or in a population of similar clones. Both approaches have played critical roles in revealing the depth and complexity of mammalian signal transduction pathways. It is fair to say that only in recent years have we begun to comprehend the depth, breadth, and complexity of these pathways and of their interrelationships. Many of the intricate networks that provide sophisticated regulation of signaling pathways would have been very difficult to recognize or understand in the absence of powerful genetic techniques. Forward genetics seeks to associate a specific protein with a biological phenotype in a pathway of interest without necessarily relying on any previous knowledge. Typical steps are: (1) Create cell libraries containing millions of random mutations; (2) apply a selective pressure or sorting technique to isolate rare cells in which the targeted phenotype has been altered; (3) identify the mutated gene or gene product; and (4) characterize the function in the pathway of the altered, overexpressed, or missing protein.
A LETHAL SELECTION SYSTEM TO IDENTIFY MUTANTS IN WHICH NF-κB-DEPENDENT SIGNALING IS ALTERED
Starting with HEK293 cells, which express a high level of transfected IL-1 receptor subunits (Cao et al. 1996), we introduced two separate selectable markers, both driven by the NF-κB-dependent E-selectin promoter (Fig. 1). One is a gene whose protein product confers resistance to zeocin (Zeo) and the other is the herpes simplex thymidine kinase (TK) gene (Li et al. 1999). The E-selectin promoter has low basal activity and can be induced strongly by activators of NF-κB, such as IL-1. Clone 293-TK/Zeo survives in ganciclovir (GCV, converted to a toxic metabolite by TK) and dies in GCV plus IL-1; it dies in Zeo and survives in Zeo plus IL-1. This clone has been used extensively, for many different experiments.
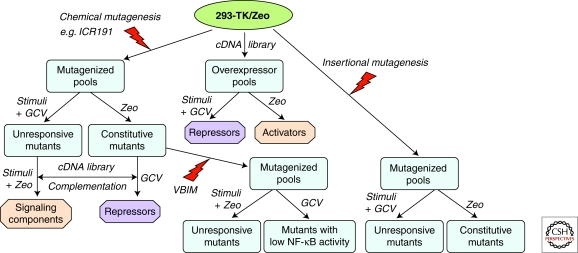
Figure 1.
General scheme for forward genetics analysis of NF-κB-dependent signaling pathway, using lethal selection to identify regulative proteins. 293-TK/Zeo cells, carrying both TK and Zeo selectable markers, can be used in conjunction with chemical mutagenesis (left) or insertional mutagenesis (right) to obtain mutants unresponsive to a stimulus or constitutive mutants, and then to identify either stimulus-specific signaling components or general activators or repressors. A cDNA library (middle) can also be overexpressed to screen directly for activators or repressors. Additionally, constitutive mutants can be further mutated by insertional mutagenesis using VBIM (bottom) to screen for mutants with low NF-κB to identify activators or repressors.
An important feature of the GCV-TK selection is that one can manipulate the concentration of GCV so that cells with a low basal TK expression survive but are killed after induction. There is a difference of 10-fold or more between basal and induced levels of TK in 293-TK/Zeo cells (Li et al. 1999). Moreover, because GCV is a poor substrate for mammalian TK, the GCV selection does not require the use of a TK-null cell line. Therefore, 293-TK/Zeo cells, with their low basal level of NF-κB activity, survive in GCV and die in Zeo. When these cells are treated with a ligand that activates NF-κB or are mutated so that they have high constitutive activation, the expression of both TK and Zeo are induced, so the cells die in GCV and survive in Zeo (Fig. 1). Using this dual selection system, we have used several different forward genetic approaches to randomly alter the expression of several different proteins, thus identifying both positive and negative novel regulators of NF-κB (Fig. 1).
EXAMPLES OF DIFFERENT FORWARD GENETIC APPROACHES TO STUDY NF-κB
The approaches that we have used to dissect NF-κB-dependent signaling pathways, using the 293-TK/Zeo cell system, include: (1) Chemical mutagenesis, followed by identification of the affected gene (Li et al. 1999; Sathe et al. 2004); (2) expression of cDNA libraries (Li et al. 2000); (3) retroviral mutagenesis by promoter insertion (Kandel et al. 2005); (4) transposon-based insertional mutagenesis (Dasgupta et al. 2008); and (5) lentiviral-based insertional mutagenesis (Lu et al. 2009a). Methods three to five include strategies to remove or silence the inserted promoter after a mutant clone has been isolated, to validate the dependence of the mutant phenotype on the function of the promoter.
Use of Chemical Mutagenesis to Help Elucidate the Role of IRAK1 in IL-1-dependent Signaling to NF-κB
The chemical mutagen ICR191, an intercalating agent that causes frame-shift mutations and deletions, has been extremely useful in generating recessive mutants by randomly inactivating both alleles of a specific target gene. We first used this approach to select eight different mutant cell lines defective in responding to interferons, contributing in this way to the discovery of the JAK/STAT pathway and to the elucidation of many details of how cells respond to interferons (Pellegrini et al. 1989; Velazquez et al. 1992; Darnell et al. 1994; Borden et al. 2007). To extend this method to an analysis of NF-κB, 293-TK/Zeo cells were subjected to five rounds of chemical mutagenesis with ICR191, followed by selection in GCV plus IL-1. Four independent mutant cell lines were isolated that failed to respond to IL-1. Attempts to complement these mutant cell lines with cDNA libraries were unsuccessful. Therefore, each mutant was tested with antibodies against known IL-1 signaling components. Mutant I1A lacks the expression of IRAK1 (Li et al. 1999). Two limitations became apparent during the course of this work, for reasons that we still understand incompletely. First, despite repeated attempts, we were unable to obtain mutant cell lines lacking additional proteins already known to be required for IL-1-dependent signaling. Second, we could not functionally complement three mutant cell lines that still express all proteins known to be required for response to IL-1, despite extensive attempts using different cDNA libraries.
Use of the human null cell line I1A helped to position IRAK1 within the IL-1 signaling pathway. IRAK1 is required for the activation of both NF-κB and Jun kinase in response to IL-1, functioning between MyD88 and TRAF6 (Li et al. 1999). Putting the kinase-dead IRAK1 mutant K239A back into I1A cells allowed us to make the surprising discovery that the kinase activity of IRAK is not required for it to function in IL-1-dependent signaling (Li et al. 1999). Later, by expressing a series of IRAK1 deletion constructs in I1A cells, the relationship between the structure of IRAK1 and its function was analyzed in great detail (Li et al. 2001; Jiang et al. 2002). These findings suggested that both the amino-terminal death domain and the carboxy-terminal region of IRAK are required for IL-1-induced NF-κB and JNK activation, whereas the amino-proximal undetermined domain is required for the activation of NF-κB but not JNK. The phosphorylation and ubiquitination of IRAK deletion mutants correlate tightly with their ability to activate NF-κB in response to IL-1, but IRAK can mediate IL-1-induced JNK activation without being phosphorylated. These studies reveal that the IL-1-induced signaling pathways leading to NF-κB and JNK activation diverge either at IRAK or at a point nearer to the receptor.
Chemical Mutagenesis Has Led to the Generation of a Group of Mutant Cells with Constitutive Activation of NF-κB
As shown in Figure 1, a single chemically mutated pool can be selected either in GCV plus activator, for mutants that do not respond to a specific stimulus, or in Zeo in the absence of a stimulus, for mutants with abnormal constitutive activation of NF-κB. Using the latter strategy, we isolated eight mutant cell lines in which NF-κB is constitutively activated (Sathe et al. 2004). These cells have different properties and belong to eight different complementation groups, showing that there must be many more than eight different mutations that can lead to this phenotype. All but one of the mutants is recessive, indicating that a negative regulatory function had been lost because of mutagenesis in these seven cases. The eight mutants represent at least five different biochemical phenotypes, differing in the sets of upstream kinases that were abnormally activated, including IKK, JNK, Akt, p90rsk1, and ERK (Sathe et al. 2004).
The constitutive activation of NF-κB in these mutant cells, and also in many different cancer cell lines, is almost always caused by the constitutive secretion of one or more factors that activate NF-κB in autocrine fashion from outside the cell (Lu et al. 2004a). Interestingly, in the mutant cell line Z12, TGF-β2 was highly expressed and secreted. This observation led to the discovery that TGF-β activates NF-κB in several different tumors and mutant cell lines, and that the basis of increased TGF-β secretion is an increased steady-state level of the corresponding mRNA (Lu et al. 2004a; Lu et al. 2004b; Lu and Stark 2004). In addition, we found that relatively high concentrations of TGF-β (about ten times higher than the concentrations required to activate SMADs) activate NF-κB by recruiting and activating the IL-1 receptor and, conversely, that IL-1 activates SMADs similarly, by recruiting the TGF-β receptor. We propose that this unusual cross talk is especially important in the immediate vicinity of tumors or at sites of inflammation, where the concentrations of TGF-β or IL-1 are likely to be high (Lu et al. 2007).
Use of a cDNA Library to Discover the NF-κB Regulator Act1
A popular approach in forward genetics is to randomly overexpress in a target cell's population proteins, protein fragments, or anti-sense RNAs from libraries constructed in expression vectors, followed by recovery of the causative constructs from mutant clones. Several types of libraries have been used: full-length cDNAs (Miki and Aaronson 1995), anti-sense cDNAs (Deiss and Kimchi 1991), truncated cDNA fragments (genetic suppressor elements or GSEs) (Gudkov et al. 1994), small interfering RNAs (Berns et al. 2004), and hammerhead ribozymes (Wadhwa et al. 2004). These approaches, still actively used in many laboratories, continue to contribute new information to our understanding of complex signaling pathways (Neznanov et al. 2003; Wang et al. 2002). In all of these approaches, the effect of the cloned element on the phenotype of interest can be confirmed by expressing it in naïve cells, not necessarily the same as the ones used for the initial isolation. Significant disadvantages of these approaches include the difficulty of constructing, maintaining, and delivering a comprehensive library of the required complexity.
In our laboratory, a human keratinocyte cDNA library was delivered into 293-TK/Zeo cells, followed by selection in Zeo (Fig. 1). The 2.6 kb cDNA recovered from one of the resistant clones corresponds to a gene of previously unknown function (Li et al. 2000). The predicted 60-kDa polypeptide was named Act1 (NF-κB activator 1). At about the same time, the same protein was discovered by another group and named CIKS (connection to IKK and SAPK/JNK) or TRAF3IP2 (TRAF3 interacting protein 2) (Leonardi et al. 2000).
Since its discovery, Act1 has been found to play major roles in NF-κB-dependent signaling, with about 300 publications describing its functions. Act1 turns out to be an important regulator of CD40-dependent signaling (Qian et al. 2004). Genetic deficiency in Act1 results in a dramatic increase in peripheral B cells, which culminates in lymphadenopathy and splenomegaly, hyper-γ-globulinemia, and autoantibodies. Although the B-cell specific Act1 knockout mice displayed a similar phenotype with less severity, the pathology of the Act1-deficient mice was mostly blocked in CD40-Act1 and BAFF-Act1 double knockout mice. CD40- and BAFF-mediated survival is significantly increased in Act1-deficent B cells, with stronger IκB phosphorylation, processing of NF-κB2 (p100/p52), and activation of JNK, ERK, and p38 pathways, indicating that Act1 negatively regulates CD40- and BAFF-mediated signaling events. These findings demonstrate that Act1 plays an important role in the homeostasis of B cells by attenuating CD40 and BAFFR signaling.
Recently, Act1 has been linked to IL-17-dependent signaling, where it acts as an essential adaptor in this newly discovered pathway that is associated with autoimmune and inflammatory diseases when misregulated (Hunter 2007; Qian et al. 2007). T-helper cells that produce IL-17 are associated with inflammation and the control of certain bacteria. After stimulation with IL-17, recruitment of Act1 to IL-17R required the IL-17R conserved cytoplasmic “SEFIR” domain, followed by recruitment of the kinase TAK1 and the E3 ubiquitin ligase TRAF6, which mediate downstream activation of NF-κB. IL-17-induced expression of inflammation-related genes was abolished in Act1-deficient primary astroglial and gut epithelial cells. This reduction was associated with much less inflammatory disease in vivo in both autoimmune encephalomyelitis and dextran sodium-sulfate-induced colitis (Qian et al. 2007).
Thus, the adapter molecule Act1 regulates autoimmunity through its impact on both T- and B-cell mediated immune responses. Whereas Act1 is an important negative regulator for B-cell-mediated humoral immune responses through its function in CD40L and BAFF signaling, it is also a key positive signaling component for IL-17 signaling pathway, critical for T(H)17-mediated autoimmune and inflammatory responses. The dual, seemingly opposite, functions of Act1 in CD40- BAFFR- and IL-17R-dependent signaling are orchestrated by different domains. Whereas Act1 interacts with the IL-17R through the carboxy-terminal SEFIR domain, it is recruited to CD40 and BAFFR indirectly, mediated by TRAF3 through the TRAF binding site in Act1. Such delicate regulatory mechanisms may provide a common means to promote balance between host defense to pathogens and tolerance to self (Qian et al. 2004, 2007 and 2008).
Insertional Mutagenesis
In this group of forward genetic methods, a defined DNA fragment is inserted approximately randomly throughout the genome. Thus, these methods have the potential to target any gene and can be applied in many different experimental systems. If the inserted DNA includes a strong promoter, dominant mutants can be obtained by driving transcription into an adjacent gene, leading to the overexpression of an mRNA encoding a full-length or truncated protein, or an antisense RNA, depending on the position and orientation of the inserted promoter. We have developed three types of insertional mutagenesis vectors. All use physical removal or silencing of the inserted promoter to prove that the mutant phenotype depends on its function (“validation”).
Retroviral Insertional Mutagenesis Has Led to New Information about p65 and TAB3
We used this technique (Fig. 2A) to search for proteins that regulate NF-κB-dependent signaling in 293-TK/Zeo cells. Two reversible mutants were characterized and the affected genes were identified as relA, encoding the NF-κB p65 subunit, and act1 (Kandel et al. 2005). Conditioned medium from the mutant overexpressing p65 activates NF-κB, and the ability to secrete factors that activate NF-κB was reduced sharply in medium from cells in which the phenotype was reversed by introducing Cre, revealing that the overexpression of p65 could activate NF-κB and that the secretion of NF-κB-activating factors occurs in a p65-dependent manner (Kandel et al. 2005).
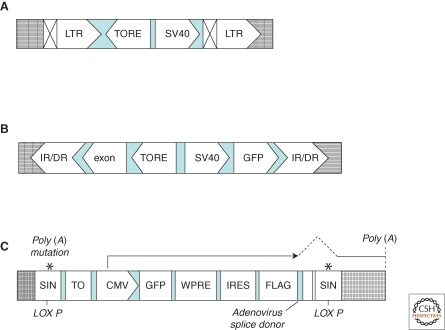
Figure 2.
Structures of different promoter insertion vectors. (A) Retroviral vectors. Retroviral vectors optimized for insertional mutagenesis were generated by using the MMLV (Moloney murine leukemia virus) backbone of pBabe vectors. The recognition sequence for Cre recombinase was placed inside the 3′ LTR so that, on reverse transcription, this motif would be copied into the 5′-LTR (Kandel et al. 2005). Abbreviations: TORE, tetracycline-regulated promoter; SV40, promoter and enhancer from the SV40 virus; LTR, long terminal repeat. Symbols: ⊠, the LTR modification, including a LoxP site for cre-mediated recombination, leading to deletion of the promoter; grid lines, host genomic DNA. (B) Sleeping Beauty transposon-based vectors. This series of mutagenesis vectors contain a coding region for GFP controlled by TORE, which is placed in the vicinity of a divergent SV40 promoter, followed by a mini-exon, which ends with an unpaired adenoviral splice site (Kandel et al. 2005). These vectors can be cotransfected with the Sleeping Beauty transposase to randomly generate mutations. Because the promoter is tet-regulated, mutant phenotypes caused by the inserted promoter can be reverted on promoter shutdown on treatment with the tetracycline analog doxycycline (Dasgupta et al. 2008). Abbreviations: IR/DR, terminal repeat from Sleeping Beauty; TORE, modified tetracycline-regulated promoter; GFP, coding region of enhanced GFP. Symbol: grid lines, host genomic DNA. (C) VBIM vectors. The VBIM vectors use a lentiviral backbone with polyadenylation mutations in both 5′ and 3′ LTRs and a lox P site in the 3′ LTR. This design allows excision of all but 238 bp of inert proviral DNA, lacking both promoter activity and polyadenylation signals, following cleavage by Cre, a critical feature for complete and consistent phenotypic reversion. The polyadenylation mutations also permit the mutagenic promoter to be placed in the same direction as transcription from the 5′ LTR during virus packaging, resulting in high virus titers that are comparable to those obtained with standard lentiviral vectors, eliminating promoter conflicts that occur with alternative designs. Furthermore, eliminating promoter interference also permits the use of a strong full-length CMV promoter rather than a minimal tetracycline-regulated promoter, which requires the tet activator protein tTA to be present in the target cells (Kandel et al. 2005). Thus, primary and even differentiated or senescent cells can be mutated without prior manipulation to express tTA or an ecotropic receptor, as previously required (Kandel et al. 2005). Besides these, a tetracycline-binding element upstream of the full-length CMV promoter is introduced to allow tetracycline-regulated control of the mutagenic promoter after the mutant has been created, by using a TR-KRAB fusion protein. Detailed description of this series of vectors has been reported recently (Lu et al. 2009a). Abbreviations: CMV, cytomegalovirus promoter; GFP, green fluorescent protein; IRES, internal ribosome entry sequence; LoxP, site for cre-mediated recombination; SIN, self-inactivating LTR; TO, tetracycline operon; WPRE, woodchuck hepatitis virus post-transcriptional regulatory element. Symbol: grid lines, host genomic DNA.
Jin et al. (2004) used a similar stratagem to identify TAB3 as an NF-κB-activating protein when overexpressed in human cells. The activation of NF-κB by TAB3 could be blocked by the NF-κB inhibitor SN50 and by the expression of dominant–negative forms of TRAF6 (TNFα-associated factor 6) and TAK1 (TGF-β-activated kinase 1), suggesting that TAB3 is a component of an NF-κB-dependent signaling pathway functioning upstream of TRAF6/TAK1. Furthermore, overexpression of TAB3 in NIH3T3 cells resulted in transformation; TAB3 overexpression was also found in some cancer tissues, suggesting a causative link between elevated TAB3 expression, constitutive NF-κB activation, and oncogenesis (Jin et al. 2004).
Use of Transposon-mediated Insertional Mutagenesis to Discover that Short RIP is an Activator of NF-κB
The benefits of transposons include a DNA-only life cycle, avoiding some constraints imposed by retroviral insertional vectors, and lack of known endogenous homolog in mammalian cells. Because the integration biases of transposons and various retroviruses differ (Yant et al. 2005), the two approaches may be complementary in attempts to achieve full-genome coverage. The cell-autonomous nature of transposition is reflected in in vivo mutagenesis. Transposons have been successfully used to search for oncogenes in mice using insertion of transposon constitutive promoters (Collier et al. 2005; Dupuy et al. 2005).
We used a vector based on the Sleeping Beauty transposon to search for constitutive activators of NF-κB in cultured cells. Dominant mutations were produced by random insertion of the tetracycline-regulated promoter TORE (Fig. 2B), which provided robust and exceptionally well-regulated expression of downstream genes. The ability to regulate the mutant phenotype was used to attribute it to the insertional event. In one such mutant, the promoter was inserted in the middle of the gene encoding receptor-interacting protein kinase 1 (RIP1). The protein encoded by the hybrid transcript lacks the putative kinase domain of RIP1, but potently stimulates NF-κB activity (Dasgupta et al. 2008). Similar to TNFα treatment, the expression of short RIP1 is toxic to cells that fail to up-regulate NF-κB. The effect of short RIP1 did not require endogenous RIP1 or cytokine treatment. More importantly, a similar short RIP1 is produced naturally from the ripk1 locus. Elevated expression of short RIP1 resulted in a loss of full length RIP1 from cells, indicating a novel mechanism through which the abundance of RIP1 and the related signals could be regulated (Dasgupta et al. 2008).
The previous results demonstrate the successful use of promoter insertion to isolate mutants in which constitutive activators are overexpressed. However, because many different activators of NF-κB are already known, the probability that novel activators will be found against such a high background is not great.
Use of the VBIM Technique to Discover FBXL11 as a Negative Regulator of NF-κB
To improve the features of reversible promoter insertional technique to facilitate the creation of dominant mutants in which a strong promoter, inserted into the genome approximately randomly, drives high-level expression of downstream genes, we designed a set of lentiviral validation-based insertional mutagenesis (VBIM) vectors (Fig. 2C) that extend the application of reversible promoter insertion previously described by our laboratory (Fig. 2A) to nearly any type of mammalian cell, even cells that are not dividing (Lu et al. 2009a). The VBIM lentiviruses are designed to increase the expression of downstream genomic sequences that encode full-length proteins, truncated proteins, or antisense RNAs, and potentially even microRNAs. These dominant mutations can thus identify either positive or negative regulators from the same genetic screen. The VBIM vectors also allow the mutant phenotype to be reversed, either by removing the inserted promoter with Cre recombinase or by silencing it with the transcriptional repressor Kruppel-associated box (KRAB) domain of the human Kox1 zinc finger protein. One can then readily associate a mutant phenotype with a specific target gene by cloning sequences flanking the insertion site. In several different screens, we obtained validated mutant clones with frequencies of the order of 10−6 to 10−5, a high yield that allows the selection of multiple mutants in experiments of very reasonable scale. Therefore, the VBIM technique is a powerful tool for gene discovery that has broad applications in many different systems.
To use VBIM to screen for negative regulators of NF-κB, we started with mutant Z3 cells, in which constitutive NF-κB activation had been generated by chemical mutagenesis (Sathe et al. 2004). These cells survive in Zeo and die in GCV (Fig. 1). Constitutive overexpression of a protein in Z3 cells that causes the constitutive NF-κB activity to shut down can be selected for by requiring the cells to survive in GCV and die in Zeo (Fig. 1).
Z3 cells infected with VBIM viruses were selected in GCV for suppression of NF-κB activity (Fig. 1). Three reversible mutants were obtained, at a frequency of ca. 10−6. The mutant SD1-11 was characterized in detail, and characterization of the other two mutants is still in progress. SD1-11 cells were infected with a vector encoding Cre recombinase to verify that the phenotype is reversible. A variety of approaches (including drug selections, electrophoretic mobility gel shift assays, luciferase assays, and Western and Northern assays) were used to confirm that the phenotype of SD1-11 is reversed by Cre. A Southern experiment showed that there is only one insertion in this mutant, and inverse PCR was used to clone the flanking genomic sequences. The gene up-regulated by the inserted promoter is F-box leucine repeat rich protein 11 (FBXL11), encoding a known histone H3 lysine 36 (H3K36) demethylase (Tsukada et al. 2006). The insertion in the second intron of FBXL11 produces a Flag-tagged fusion protein lacking only 14 of 1162 amino acid residues at its amino terminus, leaving all the functional domains intact (Lu et al. 2009a).
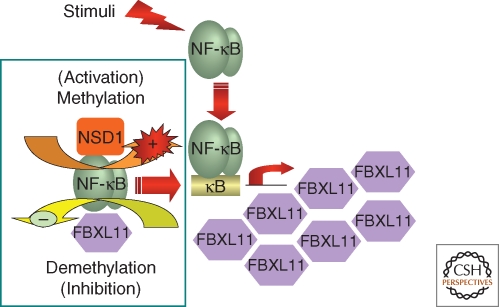
Mechanistic studies show that a point mutation that knocks out the demethylase activity of full-length FBXL11 also abolishes its ability to inhibit NF-κB activation. Knocking down the expression of FBXL11 activates NF-κB, as does overexpressing the histone H3K36 methylase NSD1 (the nuclear receptor-binding SET domain-containing protein 1). In cells with constitutively active NF-κB, or in cytokine-treated cells, the p65 subunit binds to both FBXL11 and NSD1 and significant mono-methylation of K218 and di-methylation of K221 of p65 occurs. Single K-A mutations of K218 or K221 reveal that these two residues are targets of FBXL11, and the K-A double mutants are much less active than wild-type p65. Importantly, the FBXL11 gene is transcribed in response to NF-κB activation and thus, similarly to the well known inhibitor IκB, FBXL11 participates in an autoregulatory negative-feedback loop (Fig. 3) (Lu et al. 2009b). In summary, we have uncovered a novel regulatory pathway for NF-κB that is driven by cycles of lysine methylation and demethylation of its p65 subunit. The discovery of reversible lysine methylation of NF-κB represents an important new contribution to our understanding of how fine control of this key transcription factor is achieved, and the discovery of FBXL11 as a negative regulator of NF-κB by utilizing the VBIM technique vividly demonstrates its power.
Figure 3.
Regulation of NF-κB by lysine methylation. NF-κB is regulated through reversible methylation of p65, catalyzed by the NSD1–FBXL11 enzyme pair. The FBXL11 gene is activated by NF-κB, forming a negative-feedback loop that down-regulates NF-κB.
Recently, Huang et al. (2007) showed that p53 is demethylated on K370 by LSD1, a histone H3K4 or H3K9 demethylase. The dual function of LSD1 in regulating both histone H3 and p53 is strikingly similar to our finding that FBXL11 regulates both histone H3 and NF-κB. These two examples suggest that the regulation of transcription factors by histone methylases and demethylases may be a general phenomenon in mammalian cells, and probably in other biological systems as well.
PERSPECTIVES
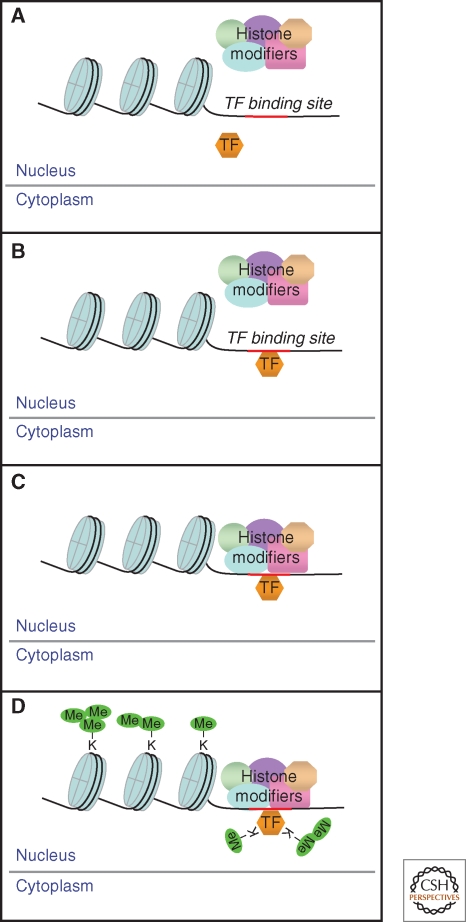
A crucial question follows from the mechanism represented in Figure 3: What controls the activating methylation of NF-κB in response to a ligand such as IL-1? A strong clue is given by the observations that NF-κB (our study) and p53 (Huang and Berger 2008; Huang et al. 2007) are methylated by enzymes that also modify histones in the context of chromatin. A likely possibility is that NF-κB and NSD1 are in different compartments of untreated cells and that activation by the H3K36 methylase NSD1 occurs only after NF-κB enters the nucleus and binds to target promoters (Fig. 4). In support of this possibility, we have recent evidence that the signal transducer and activator of transcription 3 (STAT3) is methylated only when it is bound to DNA (unpubl. data). In response to IL-6, lysine 140 of STAT3 is reversibly di-methylated, reducing its ability to bind to the promoters of a subset of IL-6-induced genes. Methylation of K140 does not occur on variant STAT3 proteins that are tyrosine-phosphorylated normally but that are prevented by point mutations from entering the nucleus or binding to DNA. Consistently, wild-type di-methyl STAT3 remains in the nucleus and is not observed in the cytoplasm, even following long exposure of cells to IL-6. Furthermore, modifications other than methylation may also occur only following the binding of transcription factors to promoters. The well known IL-6-induced phosphorylation of STAT3 on S727 occurs only when tyrosine-phosphorylated STAT3 can enter the nucleus and bind to DNA (unpubl. data), similar to an earlier observation concerning the serine phosphorylation of STAT1 (Sadzak et al. 2008).
Figure 4.
Model for transcription factor methylation by histone-modifying enzymes. (A) The activated transcription factor (TF) enters the nucleus and (B) binds to a promoter. (C) The histone-modifying complex is recruited. (D) This complex then methylates both the histones and the promoter-bound TF. Multiple sites are mono-, di-, and tri-methylated, profoundly affecting the functions of both the histones and the TF.
It is well known that activated inducible transcription factors such as NF-κB drive chromatin remodeling (Natoli 2009). Genes induced in response to NF-κB without new protein synthesis (primary response genes) fall into “fast” and “slow” subclasses. NF-κB binds to the promoters of fast genes rapidly, because the chromatin landscape is typical of genes “poised for immediate activation, including high levels of histone H3/H4 acetylation and trimethylation of histone H3K4, a histone modification specifically enriched at active or poised transcription start sites.” “Conversely, slow genes in unstimulated cells are associated with hypo-acetylated histones and are negative for H3K4me3. In response to activation, both acetylation and H3K4 trimethylation progressively increase, with a kinetics that apparently precedes NF-κB recruitment.” (Natoli 2009).
Taken together, the previous observations lead to the following working hypotheses: (1) Reversible methylation and serine phosphorylation (and probably other modifications) of NF-κB and STAT3 occur, in concert with histone modifications, only when these transcription factor targets bind to specific promoters and thus become available as substrates for the local chromatin remodeling machinery (Fig. 4); (2) these modifications profoundly affect NF-κB and STAT3 functions at these promoters, thus affecting the strength and duration of inducible expression; (3) these effects may be gene-specific, leading to differential effects on gene expression that give plasticity to the dependent biological responses. For example, (1) interpretation of a single extracellular signal might be different for different genes in the same cell; (2) the downstream effects of different mechanisms of activating NF-κB (for example, in response to IL-1 vs. TNF, or vs. toll-like receptor [TLR] ligands) might be interpreted differently in the same cell; (3) a given gene might be regulated differentially in response to a given signal in different cell types.
We have discussed several different applications of forward genetics to some of the pathways of NF-κB activation. In the future, these methods could also be applied to uncover novel positive and negative regulators of additional pathways in which NF-κB plays a role, for example the TLRs, major mediators of innate immunity whose regulation and function are under intense investigation in many laboratories. Upon stimulation by double-stranded RNA (dsRNA), NF-κB and the interferon regulatory factor 3 (IRF3) are activated, both through the membrane-bound receptor TLR3 and through the internal cytoplasmic retinoic acid-inducible gene I (RIG-I) pathway. The expression of TLR3 in 293-TK/Zeo cells should allow the use of insertional mutagenesis to reveal additional details of the pathway through which dsRNA activates NF-κB.
The VBIM technique generates dominant mutants in which RNAs encoded downstream of the inserted promoter are overexpressed. Most of the “positive” phenotypes described here and in additional unpublished studies result from the overexpression of mRNAs encoding either full-length proteins or truncated proteins that function similarly to their full-length counterparts. Overexpressed truncated proteins might also have dominant–negative functions or unregulated positive functions. Additionally, in one case (unpubl. data), we observed the overexpression of a dominant–negative RNA, resulting from the intragenic insertion of the promoter in the antisense direction. It would be extremely valuable to have a strategy in which recessive mutants, in which the expression of a protein required for signaling has been ablated, are obtained from a validation-based insertional mutagenesis approach. To do this, one might use a gene-trap vector that inactivates the expression of one allele of a gene when inserted into any intron or exon, and promote the loss of the remaining unmutated allele, for example by stimulating chromosome nondisjunction so that the entire homologous chromosome is lost, by stimulating mitotic recombination, or by performing experiments in hypo-diploid cells (Guo et al. 2004; Nobukuni et al. 2005; Banks and Bradley 2007; Kim et al. 2007). It is certain that more powerful and user-friendly methods of using forward genetics in mammalian cells will continue to be developed, leading to many additional exciting discoveries.
Footnotes
Editors: Louis M. Staudt and Michael Karin
Additional Perspectives on NF-κB available at www.cshperspectives.org
REFERENCES
- Banks DJ, Bradley KA 2007. SILENCE: A new forward genetic technology. Nat Methods 4:51–53 [DOI] [PubMed] [Google Scholar]
- Berns K, Hijmans EM, Mullenders J, Brummelkamp TR, Velds A, Heimerikx M, Kerkhoven RM, Madiredjo M, Nijkamp W, Weigelt B, et al. 2004. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature 428:431–437 [DOI] [PubMed] [Google Scholar]
- Borden EC, Sen GC, Uze G, Silverman RH, Ransohoff RM, Foster GR, Stark GR 2007. Interferons at age 50: Past, current and future impact on biomedicine. Nat Rev Drug Discov 6:975–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Z, Henzel WJ, Gao X 1996. IRAK: A kinase associated with the interleukin-1 receptor. Science 271:1128–1131 [DOI] [PubMed] [Google Scholar]
- Collier LS, Carlson CM, Ravimohan S, Dupuy AJ, Largaespada DA 2005. Cancer gene discovery in solid tumours using transposon-based somatic mutagenesis in the mouse. Nature 436:272–276 [DOI] [PubMed] [Google Scholar]
- Darnell JE Jr, Kerr IM, Stark GR 1994. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science 264:1415–1421 [DOI] [PubMed] [Google Scholar]
- Dasgupta M, Agarwal MK, Varley P, Lu T, Stark GR, Kandel ES 2008. Transposon-based mutagenesis identifies short RIP1 as an activator of NF-κB. Cell Cycle 7:2249–2256 [DOI] [PubMed] [Google Scholar]
- Deiss LP, Kimchi A 1991. A genetic tool used to identify thioredoxin as a mediator of a growth inhibitory signal. Science 252:117–120 [DOI] [PubMed] [Google Scholar]
- Dupuy AJ, Akagi K, Largaespada DA, Copeland NG, Jenkins NA 2005. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature 436:221–226 [DOI] [PubMed] [Google Scholar]
- Gudkov AV, Kazarov AR, Thimmapaya R, Axenovich SA, Mazo IA, Roninson IB 1994. Cloning mammalian genes by expression selection of genetic suppressor elements: Association of kinesin with drug resistance and cell immortalization. Proc Natl Acad Sci 91:3744–3748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo G, Wang W, Bradley A 2004. Mismatch repair genes identified using genetic screens in Blm-deficient embryonic stem cells. Nature 429:891–895 [DOI] [PubMed] [Google Scholar]
- Huang J, Berger SL 2008. The emerging field of dynamic lysine methylation of non-histone proteins. Curr Opin Genet Dev 18:152–158 [DOI] [PubMed] [Google Scholar]
- Huang J, Sengupta R, Espejo AB, Lee MG, Dorsey JA, Richter M, Opravil S, Shiekhattar R, Bedford MT, Jenuwein T, et al. 2007. p53 is regulated by the lysine demethylase LSD1. Nature 449:105–108 [DOI] [PubMed] [Google Scholar]
- Hunter CA 2007. Act1-ivating IL-17 inflammation. Nat Immunol 8:232–234 [DOI] [PubMed] [Google Scholar]
- Jiang Z, Ninomiya-Tsuji J, Qian Y, Matsumoto K, Li X 2002. Interleukin-1 (IL-1) receptor-associated kinase-dependent IL-1-induced signaling complexes phosphorylate TAK1 and TAB2 at the plasma membrane and activate TAK1 in the cytosol. Mol Cell Biol 22:7158–7167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin G, Klika A, Callahan M, Faga B, Danzig J, Jiang Z, Li X, Stark GR, Harrington J, Sherf B 2004. Identification of a human NF-κB-activating protein, TAB3. Proc Natl Acad Sci 101:2028–2033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandel ES, Lu T, Wan Y, Agarwal MK, Jackson MW, Stark GR 2005. Mutagenesis by reversible promoter insertion to study the activation of NF-κB. Proc Natl Acad Sci 102:6425–6430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SO, Ha SD, Lee S, Stanton S, Beutler B, Han J 2007. Mutagenesis by retroviral insertion in chemical mutagen-generated quasi-haploid mammalian cells. Biotechniques 42:493–501 [DOI] [PubMed] [Google Scholar]
- Leonardi A, Chariot A, Claudio E, Cunningham K, Siebenlist U 2000. CIKS, a connection to IκB kinase and stress-activated protein kinase. Proc Natl Acad Sci 97:10494–10499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Commane M, Burns C, Vithalani K, Cao Z, Stark GR 1999. Mutant cells that do not respond to interleukin-1 (IL-1) reveal a novel role for IL-1 receptor-associated kinase. Mol Cell Biol 19:4643–4652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Commane M, Nie H, Hua X, Chatterjee-Kishore M, Wald D, Haag M, Stark GR 2000. Act1, an NFκB-activating protein. Proc Natl Acad Sci 97:10489–10493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Commane M, Jiang Z, Stark GR 2001. IL-1-induced NF-κB and c-Jun N-terminal kinase (JNK) activation diverge at IL-1 receptor-associated kinase (IRAK). Proc Natl Acad Sci 98:4461–4465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Yang X, Yang D, Zhou S 2000. Genetic screens in mammalian cells by enhanced retroviral mutagens. Oncogene 19:5964–5972 [DOI] [PubMed] [Google Scholar]
- Lu T, Stark GR 2004. Cytokine overexpression and constitutive NF-κB in cancer. Cell Cycle 3:1114–1117 [PubMed] [Google Scholar]
- Lu T, Burdelya LG, Swiatkowski SM, Boiko AD, Howe PH, Stark GR, Gudkov AV 2004a. Secreted transforming growth factor β2 activates NF-κB, blocks apoptosis, and is essential for the survival of some tumor cells. Proc Natl Acad Sci 101:7112–7117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T, Sathe SS, Swiatkowski SM, Hampole CV, Stark GR 2004b. Secretion of cytokines and growth factors as a general cause of constitutive NF-κB activation in cancer. Oncogene 23:2138–2145 [DOI] [PubMed] [Google Scholar]
- Lu T, Tian L, Han Y, Vogelbaum M, Stark GR 2007. Dose-dependent cross-talk between the transforming growth factor β and interleukin-1 signaling pathways. Proc Natl Acad Sci 104:4365–4370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T, Jackson MW, Singhi AD, Kandel ES, Yang M, Zhang Y, Gudkov AV, Stark GR 2009a. Validation-based insertional mutagenesis identifies lysine demethylase FBXL11 as a negative regulator of NF-κB. Proc Natl Acad Sci 106:16339–16344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu T, Jackson MW, Wang B, Yang M, Chance MR, Miyagi M, Gudkov AV, Stark GR 2009b. Regulation of NF-B by NSD1/FBXL11-dependent reversible lysine methylation of p65. Proc Natl Acad Sci (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miki T, Aaronson SA 1995. Isolation of oncogenes by expression cDNA cloning. Methods Enzymol 254:196–206 [DOI] [PubMed] [Google Scholar]
- Natoli G 2009. Control of NF-κB-dependent transcriptional responses by chromatin organization. Cold Spring Harb Perspect Biol 1:a000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neznanov N, Neznanova L, Kondratov RV, Burdelya L, Kandel ES, O’Rourke DM, Ullrich A, Gudkov AV 2003. Dominant negative form of signal-regulatory protein-α (SIRPα /SHPS-1) inhibits tumor necrosis factor-mediated apoptosis by activation of NF-κB. J Biol Chem 278:3809–3815 [DOI] [PubMed] [Google Scholar]
- Nobukuni Y, Kohno K, Miyagawa K 2005. Gene trap mutagenesis-based forward genetic approach reveals that the tumor suppressor OVCA1 is a component of the biosynthetic pathway of diphthamide on elongation factor 2. J Biol Chem 280:10572–10577 [DOI] [PubMed] [Google Scholar]
- Pellegrini S, John J, Shearer M, Kerr IM, Stark GR 1989. Use of a selectable marker regulated by α interferon to obtain mutations in the signaling pathway. Mol Cell Biol 9:4605–4612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian Y, Qin J, Cui G, Naramura M, Snow EC, Ware CF, Fairchild RL, Omori SA, Rickert RC, Scott M, et al. 2004. Act1, a negative regulator in CD40- and BAFF-mediated B cell survival. Immunity 21:575–587 [DOI] [PubMed] [Google Scholar]
- Qian Y, Liu C, Hartupee J, Altuntas CZ, Gulen MF, Jane-Wit D, Xiao J, Lu Y, Giltiay N, Liu J, et al. 2007. The adaptor Act1 is required for interleukin 17-dependent signaling associated with autoimmune and inflammatory disease. Nat Immunol 8:247–256 [DOI] [PubMed] [Google Scholar]
- Qian Y, Giltiay N, Xiao J, Wang Y, Tian J, Han S, Scott M, Carter R, Jorgensen TN, Li X 2008. Deficiency of Act1, a critical modulator of B cell function, leads to development of Sjögren’s syndrome. Eur J Immunol 38:2219–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadzak I, Schiff M, Gattermeier I, Glinitzer R, Sauer I, Saalmüller A, Yang E, Schaljo B, Kovarik P 2008. Recruitment of Stat1 to chromatin is required for interferon-induced serine phosphorylation of Stat1 transactivation domain. Proc Natl Acad Sci 105:8944–8999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathe SS, Sizemore N, Li X, Vithalani K, Commane M, Swiatkowski SM, Stark GR 2004. Mutant human cells with constitutive activation of NFκB. Proc Natl Acad Sci U S A 101:192–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y 2006. Histone demethylation by a family of JmjC domain-containing proteins. Nature 439:811–816 [DOI] [PubMed] [Google Scholar]
- Velazquez L, Fellous M, Stark GR, Pellegrini S 1992. A protein tyrosine kinase in the interferon α/β signaling pathway. Cell 70:313–322 [DOI] [PubMed] [Google Scholar]
- Wadhwa R, Yaguchi T, Kaur K, Suyama E, Kawasaki H, Taira K, Kaul SC 2004. Use of a randomized hybrid ribozyme library for identification of genes involved in muscle differentiation. J Biol Chem 279:51622–51629 [DOI] [PubMed] [Google Scholar]
- Wang D, You Y, Case SM, McAllister-Lucas LM, Wang L, DiStefano PS, Nuñez G, Bertin J, Lin X 2002. A requirement for CARMA1 in TCR-induced NF-κB activation. Nat Immun 3:830–835 [DOI] [PubMed] [Google Scholar]
- Yant SR, Ehrhardt A, Mikkelsen JG, Meuse L, Pham T, Kay MA 2002. Transposition from a gutless adeno-transposon vector stabilizes transgene expression in vivo. Nat Biotechnol 20:999–1005 [DOI] [PubMed] [Google Scholar]
- Yant SR, Wu X, Huang Y, Garrison B, Burgess SM, Kay MA 2005. High-resolution genome-wide mapping of transposon integration in mammals. Mol Cell Biol 25:2085–2094 [DOI] [PMC free article] [PubMed] [Google Scholar]