Abstract
Understanding the origin of life requires knowledge not only of the origin of biological molecules such as amino acids, nucleotides and their polymers, but also the manner in which those molecules are integrated into the organized systems that characterize cellular life. In this article, we introduce a constructive approach to understand how biological molecules can be arranged to achieve a higher-order biological function: replication of genetic information.
Construction of self-replicating genetic systems in liposomes could reveal how molecules initially assembled to form living systems with higher-order biological functions.
BACKGROUND
What Can We Learn from a Constructive Approach?
In a constructive approach, we aim to reconstitute a biological function, such as genome replication and protein translation, and ultimately fabricate an artificial cell from molecules purified and defined in vitro (Szostak et al. 2001; Deamer 2005). During the process, we can determine what conditions are sufficient to achieve the minimum set of biological functions required for cellular life. For instance, if we can reconstitute a given biological function from a set of defined molecules, we can conclude that the properties of those molecules are sufficient to accomplish that biological function. With regard to the origins of life, this represents a parallel and complementary approach to surveying possible routes from nonliving molecules to extant living systems. If it is difficult for us to reconstruct a biological function, it may have been correspondingly difficult for that function to evolve in a primitive living organism. Knowledge of which functions are difficult to assemble from existing biological molecules, and how such hurdles can be overcome, is expected to provide insights into the origin and evolution of multifunctional extant life.
In the field of synthetic biology, researchers are now constructing artificial networks to understand the “design principles” of biological systems, which is another expression for the “sufficient conditions” concept used here. Most current studies in synthetic biology incorporate modifications of existing cells, but some investigators are constructing artificial networks from defined molecules (Benner and Sismour 2005; Simpson 2006). This is similar to what we refer to here as a constructive approach (Kaneko 2006).
What Should We Construct?
To gain insight into the origins of life, it is important to determine the nonbiological origin of chemical components, such as the amino acids, nucleotides, and lipids discussed in other articles on this topic. These are the small molecules of life that assemble into the proteins, nucleic acids and membranes that are essential for contemporary cells. Primitive versions of such molecules must have given rise to the first forms of cellular life by a process of self-assembly. However, if we observe a mixture of components from disrupted Escherichia coli containing all the molecules originally present in the living cells, no spontaneous regeneration of living cells takes place. It follows that molecules per se are necessary but not sufficient for life. Molecules and their functions must be coordinated in the correct order according to intrinsic chemical and physical rules, as observed by Schrodinger who famously described life as the “orderly and lawful behavior of matter” (Schrodinger 1944).
The primary aim of the constructive approach to protocellular life is to find sufficient conditions under which biological molecules assemble into systems that display higher-order biological functions, such as translation, replication of genetic information, cell growth, division, and nutrient transport. Some of these functions, including membrane growth (Walde 1994; Hanczyc et al. 2003), membrane growth coordinated with internal replication of genetic information (Chen et al. 2004), membrane transport of nutrients (Chakrabarti et al. 1994; Monnard and Deamer 2001; Fischer et al. 2002; Monnard et al. 2007; Mansy et al. 2008), and coupling of translation and nutrient transport (Noireaux and Libchaber 2004) have been reported previously. Here, we focus on the replication of genetic information, which is a fundamental characteristic of life.
Classifying Self-Replication of Genetic Information
One of the characteristics of life is the possession of genetic information, which replicates by using itself as a template, which we will refer to as “self-replication.” Several types of self-replicating systems have been constructed with bioinformational molecules. These include self-replication of DNA by polymerase chain reaction (PCR) (Saiki et al. 1985), self-sustained sequence replication (3SR) (Guatelli et al. 1990), self-replication of RNA by Qβ replicase (Mills et al. 1967; Biebricher et al. 1985; Oberholzer et al. 1995). Other examples include self-replication of peptides (Lee et al. 1996, 1997), low molecular weight compounds (Tjivikua et al. 1990), tetranucleotides (Zielinski and Orgel 1987) and other oligomers (Sievers and von Kiedrowski 1994), and most recently the ligation activity of ribozymes (Lincoln and Joyce 2009).
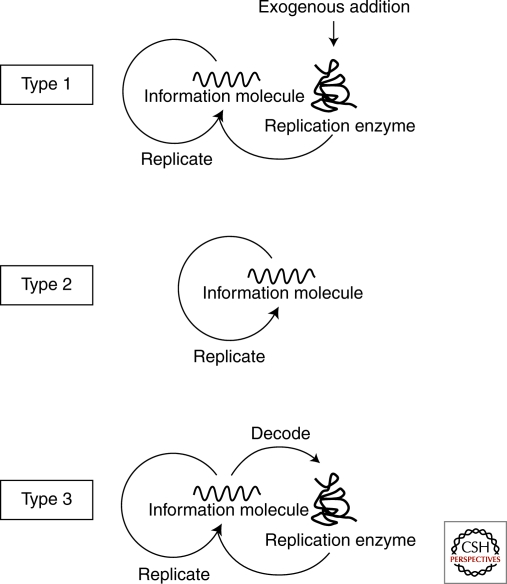
These self-replication reactions can be classified according to the reaction scheme shown in Figure 1. Type 1 is the self-replication reaction in which the information molecule (DNA or RNA) is replicated by an exogenous enzyme and includes PCR (Saiki et al. 1985), 3SR (Guatelli et al. 1990), and RNA replication (Mills et al. 1967; Biebricher et al. 1985; Oberholzer et al. 1995). Type 2 self-replication does not require any replication enzymes and these reactions include self-ligating ribozymes (Lincoln and Joyce 2009), self-replicating peptides (Lee et al. 1996; Lee et al. 1997), and other low molecular weight compounds (Zielinski and Orgel 1987; Tjivikua et al. 1990; Sievers and von Kiedrowski 1994). In this type of reaction, the information molecule replicates itself in a reaction catalyzed by its own activity, and therefore this is the simplest type of self-replication. Type 3 is a modification of type 1 in which the replication enzyme is supplied internally by synthesis rather than being exogenously added. The replication enzyme encoded in the information molecule is first decoded and then catalyzes replication of the original information molecule. The information molecule serves to provide the information required for protein production and the template for replication. The self-replication of the genome in cells or viruses would also be classified as type 3, because the replication enzyme is translated from the genomic DNA or RNA and then catalyzes replication of the genome. Construction of these types of self-replication systems has been reported previously (Ghadessy et al. 2001; Matsuura et al. 2002). In these reactions, however, translation and replication are separated temporally rather than occurring simultaneously. In the next section, we will describe characteristic features of each type of self-replication from the viewpoint of evolution.
Figure 1.
Types of self-replication. Type 1 is the self-replication reaction in which the information molecule (DNA or RNA) is replicated by an exogenous enzyme and includes PCR, 3SR and RNA replication by Qβ replicase. Type 2 self-replication does not require any replication enzymes and these reactions include self-ligating ribozymes, self-replicating peptides. Type 3 is a modification of type 1 in which the replication enzyme is supplied internally by synthesis rather than being exogenously added.
Evolution of Three Types of Self-Replication
If a self-replication process continues for many generations, random mutations can be introduced into the information molecule. This makes it possible for increasingly replicable mutants to appear and dominate the population, as first shown by Bartel and Szostak (1993). There are a number of ways this can occur, including enhancement of template activity (the ability to act as a replication template) and/or catalytic activity that promotes replication. The actual mechanism depends on the type of self-replication. When type 1 self-replication evolves, the template activity is enhanced, as shown previously (Mills et al. 1967). It is notable that although type 1 self-replication requires a replication enzyme, the replication activity does not evolve because the enzyme is not encoded on the information molecule and thus no mutations are introduced to improve the enzyme activity. Therefore, in the evolution of RNA self-replication by Qβ replicase, the template RNA initially encoding three genes were lost during evolution, resulting in a shorter, more rapidly replicating template (Mills et al. 1967).
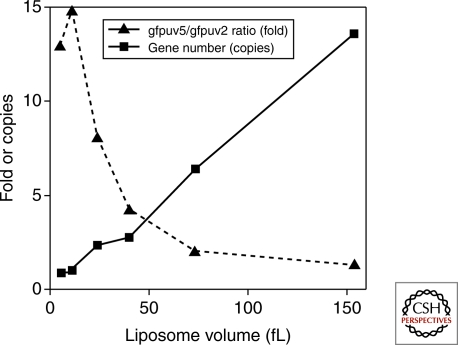
In type 2 self-replication, a single information molecule has template and catalytic functions, thus both activities are able to evolve (Lincoln and Joyce 2009). In type 3 self-replication, even though the two activities (template activity of DNA or RNA and replication activity of replicase) are encoded on different molecules, both can evolve. This is because the replication catalyst is encoded in the information molecule so that mutations will be introduced. To achieve evolution of the replication enzyme in type 3 self-replication, other conditions are required: encapsulation of the reaction in a compartment with a small number of information molecules. These conditions are required to link information molecules with the encoding replicase so that the translated replication enzyme can interact with its origin information molecule. Without compartmentalization of a low number of information molecules, even if a highly active replicase arose because of mutation, the replicase would amplify the wild-type information rather than its own information containing the mutation (Szostak 1999; Szostak et al. 2001, Matsuura et al. 2002). This requirement for a small number of information molecules was recently shown by an experiment (Sunami et al. 2006) where we encapsulated two types of GFP genes, GFPuv5 and GFPuv2 (GFPuv5:GFPuv2= 0.85:0.15), into a liposome population having a diverse size range together with a cell-free transcription-translation system. GFPuv5 shows eightfold higher fluorescence signal than GFPuv2. Following translation, we collected liposomes showing high GFP fluorescence with a fluorescent-activated cell sorter (FACS) and investigated the ratio of the GFPuv5 gene to the GFPuv2 gene in the collected liposome population. We observed that the ratio was dependent on the liposome volume; within liposomes contained in a large volume (150 fL), the total number of genes per liposome was more than ten and the GFPuv5/GFPuv2 gene ratio was low (Fig. 2). In contrast, liposomes in a small volume (5–10 fL) were found to have nearly one gene per liposome and high GFPuv5/GFPuv2 gene ratio. These observations showed that the gene encoding a highly active protein was selected efficiently in the small liposome, which has a small number of genes.
Figure 2.
Relationship between liposome volume and efficiency of selection. Two types of GFP genes, GFPuv5 and GFPuv2 (GFPuv5:GFPuv2=0.85:0.15), were encapsulated into a liposome population having a diverse size range together with a cell-free transcription-translation system. GFPuv5 shows eightfold higher fluorescence signal than GFPuv2. Following translation, liposomes showing high GFP fluorescence were collected with a fluorescent-activated cell sorter, and investigated the copy number of total GFP genes and the ratio of the GFPuv5 gene to the GFPuv2 gene in the collected liposome population.
RECENT EXPERIMENTAL RESULTS
Construction of Type 3 Self-Replication in Liposomes
Primitive life presumably involved type 2 self-replication because of its simplicity. Over time, the replication process would have become more complex by evolutionary selection and approach the type 3 self-replication systems used by extant life. Our purpose was to construct a type 3 self-replication system in which translation and replication occur simultaneously, and to optimize conditions under which type 3 self-replication can function efficiently. The encapsulated system incorporates an information molecule and translation machinery required to decode the information. The information molecule encodes a replication enzyme, which serves to replicate the information molecule.
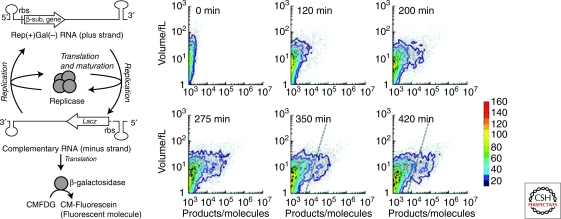
Figure 3A shows a schematic representation of our type 3 self-replication system. The system consists of a template RNA as an information molecule and a reconstituted cell-free translation system (PURE system) as the decoding machinery, all of which were encapsulated in phospholipid vesicles (liposomes) (Kita et al. 2008). The RNA molecule encodes the catalytic subunit of RNA-dependent RNA polymerase (Qβ replicase), derived from an E. coli RNA phage, and has recognition sequences for the Qβ replicase at the termini. During the reaction, the Qβ replicase subunit is first translated from the template RNA and forms active Qβ replicase with EF-Tu and EF-Ts, which are elongation factors for translation and contained in the PURE system. The translated replicase then binds to the original template RNA (plus strand) and synthesizes the complementary RNA strand (minus strand). As the minus strand can also act as a template for the replicase, the RNA strand complementary to the minus strand (i.e., the plus strand) is synthesized in a similar manner. In this way, the information molecule, plus strand RNA, is self-replicated by the self-encoded replication enzyme. Additionally, to monitor self-replication by fluorescence, we introduced the β-galactosidase sequence into the minus strand. The β-galactosidase is translated after minus strand synthesis and catalyzes hydrolyzation of nonfluorescent 5-chloromethylfluorescein di-β-D-galactopyranoside (CMFDG; Invitrogen, USA) to yield green fluorescent 5-chloromethylfluorescein (CM-fluorescein).
Figure 3.
Type 3 self-replication of genetic information in liposomes (A) Schematic representation of the reaction with an additional phenotype that was generated by insertion of the lacZ gene. The Qβ replicase β-subunit was encoded on the plus-strand RNA, and β-galactosidase was encoded on the minus-strand RNA (complement of the plus-strand RNA). Nonfluorescent CMFDG was hydrolyzed by β-galactosidase to yield green fluorescent product, CM-fluorescein. (B) Time course of the reaction analyzed by FACS. The results of 15000 liposomes. The results of FACS analysis of product (horizontal) and internal aqueous volume (vertical) of each liposome are shown. Dots represent the data of individual liposomes. Contour maps are overlaid. The frequency is depicted in color code. At 350 and 420 min, the reacted liposomes were defined as those with a substantial amount of products (right of the dashed lines).
The reaction system consists of 144 gene products (3 rRNAs, 46 tRNAs, 55 ribosomal proteins, 38 proteins), amino acids, other low molecular weight compounds and the template RNA (Table 1). This is a purified reconstituted system in which all of the components and their concentrations are defined. The number of components is amazingly large, yet this is one of the simplest encapsulated systems for carrying out protein translation and RNA replication. With regard to the origin of life, the first living systems would have had functionally identical translation and replication systems, but they must have been simpler and contained machinery for nutrient transport. The complexity of our system implies that extant translation machinery has become highly sophisticated during the evolutionary process.
Table 1.
The structure and composition of a typical functioning liposome
| Size of a vesicle | 1–100 fL (typically 4 fL) * |
|---|---|
| Lipid composition | POPC: cholesterol:DSPE-PEG5000 at a molar ratio of 58:39:3 |
| List of proteins and RNAs in the liposome | Template RNA (70 nM), Ribosome (1.2 µM), translational initiation factors: IF1 (2.5 µM), IF2 (0.21 µM), IF3 (0.95 µM), elongation factors: EF-G (3.2 µM), EF-Tu (12 µM), EF-Ts(8.2 µM), releasing factors: RF1 (0.25 µM), RF2 (0.24 µM), RF3 (0.17 µM), RRF (0.48 µM), aminoacyl-tRNA synthetase: AlaRS (725 nM), ArgRS (31 nM), AsnRS (380 nM), AspRS (127 nM), CysRS (24 nM), GlnRS (60 nM), GluRS (233 nM), GlyRS (87 nM), HisRS (8 nM), IleRS (396 nM), LeuRS (42 nM), LysRS (113 nM), MetRS (27 nM), PheRS (676 nM), ProRS (165 nM), SerRS (39 nM), ThrRS (85 nM), TrpRS (28 nM), TyrRS (7 nM), ValRS (17 nM), methionyl-tRNA transformylase (588 nM), creatine kinase (0.47 µM), myokinase (0.93 µM), nucleoside-diphosphate kinase (1.3 µM), pyrophosphatase (0.62 µM), ribosomal protein S1 (4.6 µM), RNasin Plus RNase Inhibitor (1 U/μL), tRNA mix (48 A260 units) |
| List of substrates in the liposome † | ATP (2 mM), GTP (2 mM), CTP (1 mM), UTP (1 mM), 20 amino acids (0.3 mM each), creatine phosphate (20 mM), 10-formyl-5,6,7,8-tetrahydrofolic acid (10 ng/mL) |
| Other small compounds | Magnesium acetate (13 mM), potassium glutamate (100 mM), spermidine (2 mM), dithiothreitol (1 mM), 2-[4-(2-hydroxyethyl)-1-piperazinyl] ethanesulfonic acid (HEPES) (50 mM, pH 7.6), |
*A liposome of a typical size (4 fL) is expected to carry 17 TyrSR molecules (the minimum among protein components), 28 800 EF-Tu molecules (the maximum among protein components), 72 RNA templates and 2 880 ribosomes. It is worth noting that all components that were required for the reactions were likely to be encapsulated into liposomes (>1 fL).
†Our system does not have nutrient transport machinery such as α-hemolysin (Noireaux and Libchaber 2004). It works by consuming internal substrates that have been encapsulated.
The self-replication system was encapsulated in lipid vesicles prepared by the freeze-dried empty liposome method (Sato et al. 2005) using the phospholipid mixture, 1-Palmitoyl-2-oleoyl-sn-phosphatidylcholine (POPC):cholesterol: distearoyl phosphatidylethanolamine-polyethylene glycol 5000 (DSPE-PEG5000) at a molar ratio of 58:39:3). The liposomes were multilamellar and the internal volume was found to range from 1 to 100 fL with the most frequent volume about 4 fL. The internal volume was estimated from the fluorescence intensity of the red fluorescent protein, R-phycoerythrin (R-PE), encapsulated as a volume marker and measured by a FACS (Sato et al. 2006; Sunami et al. 2006).
The encapsulated self-replication reaction produces the minus strand, from which β-galactosidase was translated. The translated β-galactosidase hydrolyses the fluorogenic substrate to produce a green fluorescent product. Hence we could monitor the progress of the self-replication reaction by measuring green fluorescence. FACS analysis showed that the number of liposomes showing green fluorescence increased over time (Fig. 3). We defined the liposomes harboring a substantial amount of products as “reacted liposomes” (liposomes with green fluorescence larger than the dotted line in Fig. 3B). The frequency of the reacted liposome depended on the liposome volume. Statistical analysis showed that the frequency of the reaction occurring per unit volume was constant (0.013 per femtoliter) indicating that the frequency of self-replication was only 5.2% in the case of a typical 4 fL liposome. This implies low efficiency of self-replication in the liposome. The low efficiency is not because of the lack of components because even the smallest liposome at 1 fL is considered to contain all of the components and substrates in our system (Table 1). There are many possible reasons for the low efficiency including degradation of RNAs, inactivation of enzymes, accumulation of inhibitory products and competition between translation and replication. The most plausible possibility for type 3 self-replication is the last one: competition between translation and replication, which could lower reaction efficiency. In the next section, we describe evaluation of the effects of competition for our self-replication system.
Competition between Translation and Replication
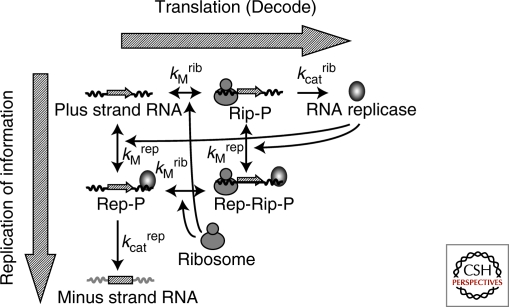
Type 3 self-replication is characterized by the dual roles of the information molecule: the information for protein production and template for replication. If the two roles compete, the efficiencies of these reactions in the self-replication reaction become lower than what we expect for the individual activities of translation or replication. Occurrence of this effect arises from the nature of the ribosomes and replication enzyme. In this case, the ribosomes and Qβ replicase were presumed to compete because it was shown that if either is bound to an RNA molecule, the other cannot use the bound RNA as a template (Kolakofsky and Weissmann 1971). The existence of such competition implies that there is an optimum balance between translation and replication. That is, translation of replicase is required for minus strand synthesis, but excess translation by too many ribosomes inhibits minus strand synthesis. To evaluate this competition effect quantitatively, we used a kinetic model to describe part of the self-replication reaction (Fig. 4) (Ichihashi et al. 2008).
Figure 4.
Kinetic model of a part of the type 3 self-replication The kinetic model contains four components: Plus strand RNA, minus strand RNA, RNA replicase (Qβ replicase) and ribosome. It carries out six reactions encompassing binding and dissociation of the plus strand RNA with RNA replicase and ribosome, translation, and minus strand synthesis. The binding and dissociation reactions are assumed to be in equilibrium. The forward reactions favor translation of RNA replicase (decoding processes). The downward reactions tend toward minus strand synthesis (replication processes). The ternary complex of the plus strand with ribosome and replicase (Rep-Rib-P) is incapable of translation and replication. The kinetic model has four measurable parameters: dissociation constants for ribosome (KMrib) and replicase (KMrep), and catalytic constants for translation (kcatrib) and replication (kcatrep).
The kinetic model contains four components: plus strand RNA, minus strand RNA, RNA replicase (Qβ replicase) and ribosome. It carries out six reactions encompassing binding and dissociation of the plus strand RNA with RNA replicase and ribosome, translation, and minus strand synthesis. The binding and dissociation reactions are assumed to be in equilibrium. The forward reactions favor translation of RNA replicase (decoding processes). The downward reactions tend toward minus strand synthesis (replication processes). The competition effect is represented as the ternary complex of the plus strand with ribosome and replicase (Rep-Rib-P), which is incapable of translation and replication.
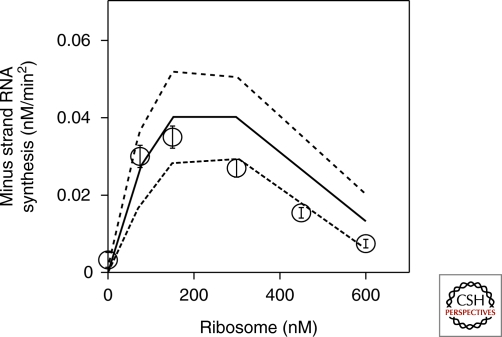
The kinetic model has four measurable parameters: dissociation constants for ribosome and replicase, and catalytic constants for translation and replication. Taking advantage of the reconstituted system, we varied the concentration of each component to estimate all four parameters. From the above model and parameters, we could predict an optimum ribosome concentration for minus strand synthesis because of the competition effect (Fig. 5). To examine this prediction experimentally, we varied ribosome concentration in a cell-free translation system, and measured the amount of synthesized minus strand by quantitative PCR after reverse transcription. The experimental results yielded a bell-shaped curve (Fig. 5) and the optimum ribosome concentration was close to the predicted value, indicating the validity of the kinetic model.
Figure 5.
Effects of ribosome on minus strand RNA synthesis Ribosome concentration in a cell-free translation system was varied, and the amount of synthesized minus strand was measured by quantitative PCR after reverse transcription (open circle). Theoretical prediction from the kinetic model and experimentally estimated parameters were shown (solid line) with standard deviation (dotted lines).
To summarize, we found that our self-replication system showed competition between translation and replication, and we were able to evaluate the effect quantitatively. Such a competition effect in type 3 self-replication is inevitable as long as the dual roles (template and information) are inherent in the information molecule which causes the mutual inhibition of the translation and replication machineries. However, optimizing the ribosome concentration minimizes the inhibitory effect, indicating that the balance between translation and replication is important for efficient type 3 self-replication.
The kinetic analysis provides another implication, with respect to possible size of an evolvable artificial cell. The kinetic analysis revealed that the dissociation constant of the RNA with the replicase was about 20 nM (Ichihashi et al. 2008), indicating that the concentration of RNA should be at more than nanomolar levels for efficient reaction. This requirement limits the possible size of an evolvable artificial cell, considering that the information molecule (RNA in this case) exists in low number for evolution as described above. For example, one information molecule corresponds to approximately 1 nM in a 1 µm cell, but 1 pM in 10 µm cell. Therefore, the size of an evolvable artificial cell including the self-replication system should be 1 µm order for efficient internal RNA replication. This notion implies that the possible size of an evolvable cell would be limited by the affinity of internal components.
CHALLENGES AND FUTURE RESEARCH DIRECTIONS
In this article, we present our recent investigation of a type 3 self-replication system and show the importance of the translation/replication balance. However, with the optimum ribosome concentration, self-replication in liposomes is still inefficient. That is because the system has more than a hundred components and each molecule in the system does not function efficiently, probably because of unexpected interactions such as competition. Further studies are required to determine the conditions under which all components function in a coordinated fashion to achieve efficient self-replication (Pohorille and Deamer 2002).
How do we find these coordinated conditions? One approach is to adopt an evolutionary strategy, mutating the RNA of the self-replication system and selecting mutants showing greater replication. For instance, the selective process could be enabled by selecting for liposomes with higher levels of fluorescence by FACS. This type of evolutionary strategy is also likely to have been adopted by primitive cells, which would need to acquire new functions to replicate efficiently in different environments. The new function or functions acquired by an ancient/primitive cell could sometimes cause a conflict with pre-existing functions. To resolve this conflict, a mutant would evolve such that it would be able to coordinate new functions with pre-existing ones. This conflict resolution process is the same evolutionary strategy that we aim to emulate. Therefore, the construction and improvement of model self-replication systems by evolutionary strategies will provide a deeper understanding of the origin of coordinated biological systems.
Footnotes
Editors: David Deamer and Jack W. Szostak
Additional Perspectives on The Origins of Life available at www.cshperspectives.org
REFERENCES
- Bartel DP, Szostak JW 1993. Isolation of new ribozymes from a large pool of random sequences. Science 261:1411–1418 [DOI] [PubMed] [Google Scholar]
- Benner SA, Sismour AM 2005. Synthetic biology. Nat Rev Genet 6:533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biebricher CK, Eigen M, Gardiner WC Jr 1985. Kinetics of RNA replication: competition and selection among self-replicating RNA species. Biochemistry 24:6550–6560 [DOI] [PubMed] [Google Scholar]
- Chakrabarti AC, Breaker RR, Joyce GF, Deamer DW 1994. Production of RNA by a polymerase protein encapsulated within phospholipid vesicles. J Mol Evol 39:555–559 [DOI] [PubMed] [Google Scholar]
- Chen IA, Roberts RW, Szostak JW 2004. The emergence of competition between model protocells. Science 305:1474–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deamer D 2005. A giant step towards artificial life? Trends Biotechnol 23:336–338 [DOI] [PubMed] [Google Scholar]
- Fischer A, Franco A, Oberholzer T 2002. Giant vesicles as microreactors for enzymatic mRNA synthesis. Chembiochem 3:409–417 [DOI] [PubMed] [Google Scholar]
- Ghadessy FJ, Ong JL, Holliger P 2001. Directed evolution of polymerase function by compartmentalized self-replication. Proc Natl Acad Sci 98:4552–4557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guatelli JC, Whitfield KM, Kwoh DY, Barringer KJ, Richman DD, Gingeras TR 1990. Isothermal, in vitro amplification of nucleic acids by a multienzyme reaction modeled after retroviral replication. Proc Natl Acad Sci 87:1874–1878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanczyc MM, Fujikawa SM, Szostak JW 2003. Experimental models of primitive cellular compartments: Encapsulation, growth, and division. Science 302:618–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichihashi N, Matsuura T, Kita H, Hosoda K, Sunami T, Tsukada K, Yomo T 2008. Importance of translation-replication balance for efficient replication by the self-encoded replicase. Chembiochem 9:3023–3028 [DOI] [PubMed] [Google Scholar]
- Kaneko K 2006. Life: An introduction to complex systems biology Springer, Berlin, Germany [Google Scholar]
- Kita H, Matsuura T, Sunami T, Hosoda K, Ichihashi N, Tsukada K, Urabe I, Yomo T 2008. Replication of genetic information with self-encoded replicase in liposomes. Chembiochem 9:2403–2410 [DOI] [PubMed] [Google Scholar]
- Kolakofsky D, Weissmann C 1971. Possible mechanism for transition of viral RNA from polysome to replication complex. Nat New Biol 231:42–46 [DOI] [PubMed] [Google Scholar]
- Lee DH, Granja JR, Martinez JA, Severin K, Ghadri MR 1996. A self-replicating peptide. Nature 382:525–528 [DOI] [PubMed] [Google Scholar]
- Lee DH, Severin K, Yokobayashi Y, Ghadiri MR 1997. Emergence of symbiosis in peptide self-replication through a hypercyclic network. Nature 390:591–594 [DOI] [PubMed] [Google Scholar]
- Lincoln TA, Joyce GF 2009. Self-sustained replication of an RNA enzyme. Science 323:1229–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noireaux V, Libchaber A 2004. A vesicle bioreactor as a step toward an artificial cell assembly. Proc Natl Acad Sci 101:17669–17674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansy SS, Schrum JP, Krishnamurthy M, Tobe S, Treco DA, Szostak JW 2008. Template-directed synthesis of a genetic polymer in a model protocell. Nature 454:122–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuura T, Yamaguchi M, Ko-Mitamura EP, Shima Y, Urabe I, Yomo T 2002. Importance of compartment formation for a self-encoding system. Proc Natl Acad Sci 99:7514–7517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills DR, Peterson RL, Spiegelman S 1967. An extracellular Darwinian experiment with a self-duplicating nucleic acid molecule. Proc Natl Acad Sci 58:217–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnard PA, Deamer DW 2001. Nutrient uptake by protocells: A liposome model system. Orig Life Evol Biosph 31:147–155 [DOI] [PubMed] [Google Scholar]
- Monnard PA, Luptak A, Deamer DW 2007. Models of primitive cellular life: Polymerases and templates in liposomes. Philos Trans R Soc Lond B Biol Sci 362:1741–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberholzer T, Wick R, Luisi PL, Biebricher CK 1995. Enzymatic RNA replication in self-reproducing vesicles: an approach to a minimal cell. Biochem Biophys Res Commun 207:250–257 [DOI] [PubMed] [Google Scholar]
- Walde P, Wick R, Fresta M, Mangone A, Luisi PL 1994. Autopoietic self-reproduction of fatty acid vesicles. J Am Chem Soc 116:11649–11654 [Google Scholar]
- Pohorille A, Deamer D 2002. Artificial cells: Prospects for biotechnology. Trends Biotechnol 20:123–128 [DOI] [PubMed] [Google Scholar]
- Saiki RK, Scharf S, Faloona F, Mullis KB, Horn GT, Erlich HA, Arnheim N 1985. Enzymatic amplification of β-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science 230:1350–1354 [DOI] [PubMed] [Google Scholar]
- Sato K, Obinata K, Sugawara T, Urabe I, Yomo T 2006. Quantification of structural properties of cell-sized individual liposomes by flow cytometry. J Biosci Bioeng 102:171–178 [DOI] [PubMed] [Google Scholar]
- Schrodinger E 1944. What is life? Cambridge University Press, Cambridge, UK [Google Scholar]
- Sievers D, von Kiedrowski G 1994. Self-replication of complementary nucleotide-based oligomers. Nature 369:221–224 [DOI] [PubMed] [Google Scholar]
- Simpson ML 2006. Cell-free synthetic biology: A bottom-up approach to discovery by design. Mol Syst Biol 2:69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunami T, Sato K, Matsuura T, Tsukada K, Urabe I, Yomo T 2006. Femtoliter compartment in liposomes for in vitro selection of proteins. Anal Biochem 357:128–136 [DOI] [PubMed] [Google Scholar]
- Szostak JW 1999. Constraints on the sizes of the earliest cells. In Size limits of very small microorganisms, pp. 120–125 National Academy Press [Google Scholar]
- Szostak JW, Bartel DP, Luisi PL 2001. Synthesizing life. Nature 409:387–390 [DOI] [PubMed] [Google Scholar]
- Tjivikua T, Ballester P, Rebek J 1990. A self-replicating system. J Am Chem Soc 112:1249–1250 [Google Scholar]
- Zielinski WS, Orgel LE 1987. Autocatalytic synthesis of a tetranucleotide analogue. Nature 327:346–347 [DOI] [PubMed] [Google Scholar]