Abstract
Electron cryotomography (ECT) is an emerging technology that allows thin samples such as macromolecular complexes and small bacterial cells to be imaged in 3-D in a nearly native state to “molecular” (∼4 nm) resolution. As such, ECT is beginning to deliver long-awaited insight into the positions and structures of cytoskeletal filaments, cell wall elements, motility machines, chemoreceptor arrays, internal compartments, and other ultrastructures. This article describes the technique and summarizes its contributions to bacterial cell biology. For comparable recent reviews, see (Subramaniam 2005; Jensen and Briegel 2007; Murphy and Jensen 2007; Li and Jensen 2009). For reviews on the history, technical details, and broader application of electron tomography in general, see for example (Subramaniam and Milne 2004; Lucić et al. 2005; Leis et al. 2008; Midgley and Dunin-Borkowski 2009).
Electron cryotomography allows 3D imaging of intact bacteria, revealing the organization of cytoskeletal filaments, cell wall components, chemoreceptor arrays, and internal compartments.
INTRODUCTION—THE STORY OF FtsZ
ECT can produce three-dimensional (3-D) reconstructions of intact cells in near-native states to “molecular” resolution (∼4 nm), and has thus begun providing unprecedented views into the ultrastructure of bacterial cells. The exciting potential of ECT as compared to other light and electron microscopy techniques is powerfully illustrated by the history of FtsZ.
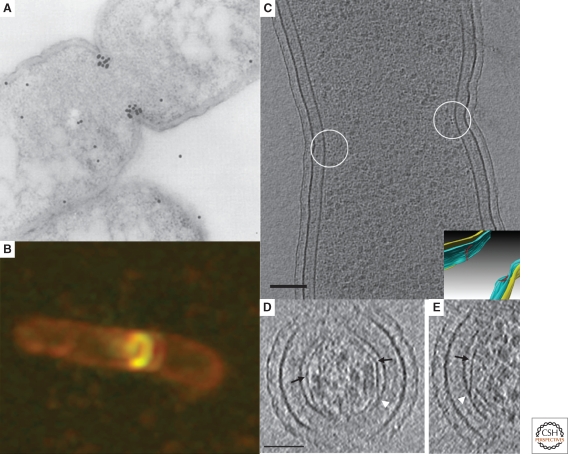
In 1980 Lutkenhaus et al. identified a temperature-sensitive, filament-forming Escherichia coli mutant unable to undergo cell division but unaffected in its ability to replicate and segregate DNA (Lutkenhaus et al. 1980). The gene responsible for the observed phenotype was identified and named ftsZ (for filamentous temperature sensitive Z). A decade later, Bi and Lutkenhaus showed through immuno-electron microscopy that FtsZ protein localized near the constriction site of dividing cells (Bi and Lutkenhaus 1991). This seminal study was performed using what we would now call “traditional” electron microscopy (EM): exponentially growing E. coli cells were chemically fixed (2% glutaraldehyde), pelleted and washed in buffer, dehydrated in a gradient of ethanol (from 30% to 100%), stained with uranyl acetate, resin embedded, cured at 60°C, sectioned, exposed to an FtsZ-specific antibody, and finally reacted with a colloidal-gold-labeled secondary antibody. Although longitudinal cell sections like the one shown in Figure 1A showed that FtsZ localized to the mid-plane of dividing cells, unfortunately no detailed ultrastructure was visible, probably because the fixation, dehydration, and other harsh preparative steps destroyed it. Thus although FtsZ was proposed to form a ringlike structure (the “Z-ring”), no further mechanistic clues could be gained.
Figure 1.
Microscopy of FtsZ. (A) Immunolocalization of FtsZ in a dividing Escherichia coli cell. The location of FtsZ is indicated by the black dots, which are gold-labeled antibodies. Adapted from (Lutkenhaus 1993) with permission from Blackwell Publishing ltd. (B) Fluorescent light microscopy image of E. coli cells expressing low levels of FtsZ-GFP showing the formation of a new FtsZ ring (gold) in a dividing cell (outlined in red). Reprinted from (Margolin 1998), © 1998, with permission from Elsevier Ltd. (C) An 8-nm thick cryo-tomographic slice through a dividing Caulobacter crescentus cell showing FtsZ filaments in cross-section (small dark dots near the center of the circles next to the membrane). Scale bar 100 nm. The inset shows the 3-D segmentation of the same cell. The inner membrane, outer membrane and the FtsZ arclike filaments are colored in blue, yellow and red, respectively. (D) and (E) 6.7-nm thick tomographic slices containing examples of straight and curved (white arrow) segments of FtsZ filaments. Abrupt “kinks” were sometimes seen (black arrows) as well as direct connections of straight filaments to the membrane (white arrowheads). Scale bar 50 nm. Fig. 1C–E adapted from (Li et al. 2007) with permission from Nature Publishing Group.
Just a few years later, the cloning and recombinant expression of the green fluorescent protein (GFP) from Aequorea sp. revolutionized cell biology, allowing specific proteins of interest to be tagged and imaged in living cells by fluorescence light microscopy (fLM) (reviewed by Tsien 1998). In 1996, Ma et al. fused FtsZ with GFP and confirmed that it formed a dynamic ring at the midplane of dividing cells (Ma et al. 1996) (Fig. 1B) (similar results were also obtained by immunofluorescence (Addinall and Lutkenhaus 1996; Levin and Losick 1996)). Later, sophisticated fluorescence recovery after photobleaching (FRAP) experiments showed that FtsZ monomers entered and left the ring with a half-time of ∼9–30 s (Stricker et al. 2002; Anderson et al. 2004). Despite fLM's ability to reveal dynamics and deliver 3-D information (through deconvolution and confocal techniques), the mechanism of constriction remained unclear. The resolution of light microscopy was insufficient to show, for instance, whether FtsZ monomers were actually forming filaments or just localizing to the midcell near the membrane. Crystal structures showing FtsZ's clear homology to eukaryotic tubulin and in vitro polymerization studies argued strongly that it did form filaments (Löwe and Amos 1998). Among many hypotheses, one, for instance, was that in analogy to eukaryotic cytokinesis, perhaps FtsZ formed a complete ring or spiral and an unknown protein drove adjacent protofilaments in opposite directions, generating a constriction force (Bramhill 1997). The FRAP data, however, seemed incompatible with this idea. Moreover, although not a problem in this specific case (of localizing FtsZ), it is worth noting that GFP and its relatives can cause proteins to mislocalize. A recent comprehensive tagging screen showed, for instance, that less than one-third of proteins that show a particular localization pattern when tagged at the amino terminus show the same localization when the tag is moved to the carboxyl terminus (most are delocalized whereas a small percent localize differently) (Werner et al. 2009)! Although immunofluorescence can be used to confirm particular patterns (of untagged proteins), this again introduces the problems of fixation.
It was in this context that the first ECT images of bacterial cell division were obtained. Images of dividing Caulobacter crescentus cells showed that FtsZ forms short, separated, arc-like filaments rather than a complete ring or spiral (Fig. 1C) (Li et al. 2007). In one cell there were, for instance, nine filaments on one side of the cell and none on the other. Individual 4-nm-wide protofilaments were irregularly spaced (rather than bundled) and connected to the inner membrane by additional protein densities. Perhaps most intriguingly, some FtsZ filaments were curved whereas others were straight (Figs. 1D,E). Because like its eukaryotic tubulin homologs, FtsZ appears to favor curved and straight conformations in vitro when bound to GDP and GTP, respectively (Lu et al. 2000), these new structural details strongly supported a model first proposed by Erickson that FtsZ itself generates the force needed to constrict the cell through nucleotide-hydrolysis-driven conformational changes in short, independently acting protofilaments (Erickson 1997).
As illustrated by these imaging studies on FtsZ, the main advantages of ECT over other microscopy techniques are the preservation of the sample in a near-native state (that preserves filaments, for instance) and the resolution to visualize even small details directly (like filament curvature and connectivity), in 3-D. How this is accomplished is described next.
TECHNICAL AND PROCEDURAL DETAILS
Sample Preparation
One of the main challenges in biological electron microscopy is preserving the sample in as native state as possible within the high vacuum of the microscope. As mentioned earlier, this has traditionally been done through chemical fixation, dehydration, and plastic-embedment. To eliminate the artifacts produced by these harsh treatments, Dubochet and his colleagues developed the method of “plunge-freezing” (Dubochet et al. 1983). A few microliters of an aqueous sample such as a cell culture or a solution of purified protein is applied to the surface of a thin, perforated layer of carbon supported by a standard EM grid. This can be done by pipet or by simply dipping the EM grid into the sample. When working with adherent cells, it can be advantageous to add EM grids to the cell culture for hours or days so the cells grow naturally on and attach to the carbon surface (Seybert et al. 2006). In these cases gold grids are usually used in place of the more toxic but cheaper copper grids. To facilitate subsequent image alignment, 5- or 10-nm colloidal gold clusters are also typically added to the sample. These can be either dried onto the grid before the sample is added, mixed into the sample itself, or both.
Next, to reduce the sample to just a thin film (∼0.5-μm thick or less), the grid is blotted by filter-paper pads on either one side or both sides simultaneously for a second or two. The blotted grid is then rapidly plunged into liquid ethane or, more conveniently, into an ethane-propane mixture (Tivol et al. 2008). This freezes the samples so quickly (>106 K/s) (Dubochet et al. 1988) that the water molecules stop diffusing before they can form the energetically favorable hydrogen bond networks of crystalline ice, retaining instead a disordered, liquid-water-like arrangement referred to as “vitreous” ice (Angell 2004). Dissolved macromolecules, their complexes, and even entire cells are thereby preserved in a near-native, “frozen-hydrated” state (sufficiently native in fact to continue living if later thawed (Erk et al. 1998). If the plunging process is not rapid enough, ice crystals form and denature the sample. Fortunately, denatured samples are immediately recognized in the EM, as ice crystals have a pronounced and unambiguous appearance. Automatic plunge-freezers with multiple adjustable parameters have dramatically improved the reproducibility of plunge-freezing; one commonly used, commercially available machine is called the “Vitrobot” (Frederik and Hubert 2005). For a more detailed description of ECT sample preparation with the Vitrobot, see (Iancu et al. 2006a). Once the sample is frozen across a grid, it is ready for imaging in the EM.
Tilt-Series Acquisition and Fundamental Limitations
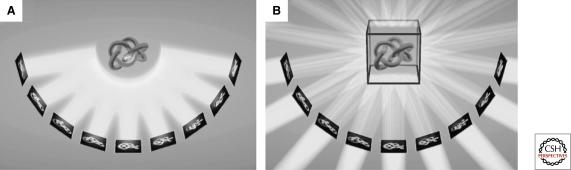
The word “tomography” means imaging by sections or sectioning. The most familiar use of the word is the medical “CT,” or “computed tomography” scan, wherein X-ray projection images through a subject are recorded from a number of directions and then merged to produce a 3-D anatomical model. Similarly, in electron tomography, a “tilt-series” of projection images are recorded of a single object like a bacterial cell as it is incrementally tilted around one, and sometimes two axes, and these images are then merged to produce a 3-D “reconstruction” or “tomogram” (Fig. 2). The basic workflow is that a grid is inserted into the EM, a target is chosen and centered under the electron beam, a projection image is recorded, the sample is rotated (tilted) a degree or two, another projection image is recorded, and the cycle of rotation and imaging is repeated as far as useful images can be obtained (until the sample becomes prohibitively thick or the grid or grid holder begins to block the beam, usually ∼65°). Images of the inverse tilt angles (i.e., 0° to −65°) are recorded similarly, or alternatively, the tilt-series can begin at one extreme tilt angle (like 65°) and proceed through the untilted position to the opposite extreme (i.e., −65°). For a movie showing an example tilt-series of a frozen-hydrated bacterial cell, see Supp. Movie S2 of (Komeili et al. 2006).
Figure 2.
Principles of data acquisition and reconstruction in ECT. (A) A set of 2-D projection images is recorded while tilting the object around one axis. (B) The back-projection method of 3-D reconstruction in ECT: For each projection, a back-projection body is calculated, and the sum of all the back-projection bodies represents the density distribution of the original object. Reprinted, with permission from (Baumeister et al. 1999).
Because the resolution of modern EMs is subatomic (Batson et al. 2002), and there is no limit to how finely the tilt range can be sampled, one might hope that this procedure would deliver atomic-resolution images of cells. Unfortunately, for frozen-hydrated biological materials, radiation damage prohibits this. As the imaging electrons pass through the sample, they can remain unscattered, scatter elastically, scatter inelastically, or suffer multiple scattering events. Although image contrast (the information content) is produced by the interference of the unscattered and the elastically scattered electrons, the inelastically scattered electrons gradually destroy the sample. Inelastic scattering events break covalent bonds, deposit heat, and more rarely even knock atomic nuclei out of place. Because for every useful elastic scattering event there are approximately 3 damaging inelastic scattering events (Henderson 1995), as more and more electrons are used to build up an image, sample damage accumulates. The originally sharp edges of macromolecular structures degrade and eventually “bubbles” of (presumably) radiolytic fragments appear and catastrophically disrupt the structure (Comolli and Downing 2005; Iancu et al. 2006b; Wright et al. 2006). Thus the most fundamentally limiting factor in ECT is the total number of electrons that can be used to record images before the sample is destroyed. Depending on how much damage is tolerated, in practice total doses of ∼1–200 electrons/Å2 are typically used, spread across the tilt-series. As a result, individual atoms or even protein secondary structures are not resolved, but the position of domains and the rough morphology of complexes can be, as was described for FtsZ earlier.
After damaging the sample, inelastically scattered electrons emerge with lower energies (and thus longer wavelengths), and therefore focus differently as they pass through the electron lenses. To prevent inelastically scattered electrons from blurring the image, energy filters remove them. Because the mean free path (i.e., the average distance between scattering events) in aqueous materials of the 300 kV electrons that are typically used in ECT is only ∼350 nm (Grimm et al. 1996), about half of the electrons sent through even a very slender bacterium are inelastically scattered and then removed by the energy filter. As the sample is tilted, the path length the electrons must traverse through the sample increases as the secant of the tilt angle (becoming twice as thick, for instance, at 60° tilt), and more and more electrons are lost to inelastic scattering. To build up meaningful images, higher doses can be used, but at the cost of increased radiation damage. Moreover, in addition to requiring higher doses per image, thicker samples in general also require more images to achieve the same resolution because the tilt range has to be sampled more finely (Derosier and Klug 1968). Thus a second fundamental limitation in ECT is that the interpretability (clarity and resolution) of reconstructions degrades with sample thickness, with 0.5 µm being a useful practical limit.
Traditionally, thicker samples such as cells and tissues have always been sectioned. This is straightforward for plastic-embedded materials, but methods to “cryo-section” frozen-hydrated samples have only recently met success (Al-Amoudi et al. 2004). Larger samples (such as suspensions of large bacterial cells, biofilms, or even tissues) are first frozen under high pressure (2100 atmospheres) to prevent ice crystal formation (McDonald and Auer 2006), and then the frozen blocks are cryo-sectioned. Although the procedure is technically challenging and fraught with artifacts such as knife-marks, crevasses, and compression, nevertheless it is beginning to provide much-needed images of larger samples in near-native states, as illustrated in some of the examples presented later.
Because with even the thinnest samples, useful images at tilt angles higher than ∼65–70° cannot usually be collected, there is a “wedge” of information (the tilt angles surrounding 90°) that remains unmeasured. As a result, the resolution of the 3-D reconstruction in the direction parallel to the electron beam is significantly worse than the resolution perpendicular. In simple visual terms, this causes spherical objects to appear somewhat ellipsoidal (smeared in the direction of the beam), and continuous objects such as filaments and membranes are more visible in some orientations than in others. This is why in “xz” or “yz” tomographic slices (such as Fig. 1D), the membranes do not appear to connect around the “top” and “bottom” of the cell. Although the missing wedge may be reduced to a missing “pyramid” by rotating the grid 90° and collecting a second, orthogonal tilt-series (a so-called “dual-axis” data set), this procedure is more than twice as time consuming, the dose that can be used per image is halved, and alignment errors between the tilt-series can erode the benefit (Nickell et al. 2003; Iancu et al. 2005). Thus a third fundamental limitation in ECT is the anisotropic resolution caused by tilt limitations (the “missing wedge”).
In practice, the acquisition of even one tilt-series is complicated. Once the sample is plunge-frozen, it must be kept at temperatures below the vitrification point (∼120 K) throughout storage, loading, and data collection to prevent the water from crystallizing (which would denature the specimen). For this purpose, special cryo-stages have been developed for transmission electron microscopes that are cooled by liquid nitrogen or helium. Unfortunately, no stage is perfectly eucentric and these cryo-stages are particularly susceptible to thermal drift. As a result, as the sample is rotated, it is displaced laterally and vertically within the column. Thus to kept the target centered under the beam and at a constant focus, before each image is taken the beam must be electronically shifted, the image has to be correspondingly unshifted to center it back on the CCD camera, and the strength of the objective lens has to be adjusted. To find the magnitude and direction of the shifts and focus changes that should be applied before each image is taken, without exposing the target to any unnecessary dose, the movements can either be “predicted” by modeling trends from previous images in the tilt-series (Zheng et al. 2004) or measured indirectly by deflecting the beam to an off-target, “tracking” position and recording reference images. Fortunately there are several software packages that perform these tedious and intricate operations automatically now, including SerialEM (Mastronarde 2005), Leginon (Suloway et al. 2009), UCSF Tomo (Zheng et al. 2007), the TOM toolbox (Nickell et al. 2005), and others.
The ideal instrument for ECT is a fully automatable intermediate-voltage (∼300 kV) EM with a highly stable and eucentric cryo-stage, an energy filter, and a large format and low-noise CCD. Unfortunately because of their extreme cost, there are still only a few such instruments being used for ECT in the world today. The natural contrast of unstained biological macromolecules suspended in vitreous ice in such microscopes is extremely low, because it arises from the small difference in their densities. Because the ultimate resolution limitation is radiation damage, imaging parameters such as defocus, magnification, and tilt increment are therefore carefully chosen to maximize contrast and reduce noise without becoming even more limiting (than radiation damage). The defocus is set high enough (∼4–16 µm) to generate strong phase contrast, but not so high that the highest spatial frequencies of interest are inverted by the contrast transfer function (Fernández et al. 2006). Magnifications are set high enough that the pixel size of the reconstruction (typically 0.5–1.0 nm) is one-third or less of the desired resolution, but not further, so that the dose per pixel and field of view can remain as large as possible. The tilt increment is set fine enough to capture the highest spatial frequency of interest, but no smaller so the total dose per image can be maximized.
3-D Reconstruction and Interpretation
As mentioned earlier, because no goniometer is perfect, specimens move laterally and vertically within the column throughout the tilt-series. The images must therefore be precisely aligned before a 3-D reconstruction can be calculated. As an additional challenge, because of the physics of electron optics, changes in height/focus within the column cause images to rotate and show subtly different magnifications. Further, although the tilt angle of each image is approximately known, the actual angles reached must be determined more accurately. Sophisticated software has therefore been written to refine estimates of the translations, rotation, magnification, tilt axis, and tilt angle of each image in the tilt-series (Mastronarde 2008). The colloidal gold beads typically added to the samples provide precise fiducial markers to facilitate this process.
Once the images are aligned, 3-D reconstructions can be calculated with a variety of algorithms. The most intuitive is “back-projection,” in which a reconstruction is built up by “smearing” the densities in each image back through space in the opposite direction they were projected (Fig. 2B) (Crowther et al. 1970b). To understand this reconstruction in Fourier space, the key principle is that the 2-D Fourier transform of a projection image is a central slice of the 3-D Fourier transform of the object (the “Projection theorem”) (Crowther et al. 1970a). Thus the 3-D Fourier transform of the sample can be “filled” with the transforms of the 2-D images, and then re-sampled onto a regular (for instance Cartesian) coordinate system and inverse transformed to produce a real-space reconstruction (Lee et al. 2008). Various software packages have been written to perform these calculations, including IMOD (Mastronarde 2008), the TOM toolbox (Nickell et al. 2005), and RAPTOR (Amat et al. 2008). Once reconstructed, tomograms can be “denoised” to improve image contrast and enhance interpretability (Frangakis and Hegerl 2001; Narasimha et al. 2008) and/or “segmented” to allow specific features to be visualized in isolation or as surfaces (Pruggnaller et al. 2008).
Tomograms are rich in detail. The layers of the cell envelope and large ultrastructures such as flagella, magnetosomes, storage granules, carboxysomes, and gas vesicles are readily recognizable through their unique and known characteristics, but more sophisticated methods must be used to identify others within the crowded environment of the cell (Grunewald et al. 2003). In the case of chemoreceptor arrays, for instance, bundles of parallel filament-like densities extending into the cytoplasm from the inner membrane were immediately suspected to be chemoreceptor arrays because they matched expectations for their gross architecture and dimensions. To confirm this assignment, however, Weis et al. chemically fixed cells and immunolabeled components of the chemoreceptor arrays (Weis et al. 2003). This worked well because the distinctive “zippered” and “micellar” structures of overexpressed receptors persisted through the required chemical fixation, and so could still be recognized in the immunolabeled sections. Unfortunately this is not always true, as described earlier in the case of FtsZ, where the labels revealed the general location of FtsZ but the filaments themselves were no longer visible.
A potentially more general strategy for identifying structures is correlated fLM and ECT. The basic idea is to immobilize cells expressing a fluorescent fusion protein on an EM grid, record fLM images marking the localization of the fusion protein in each cell, then move the sample into an EM and record high-resolution cryo-tomograms of the same cells. For this purpose light microscope cryo-stages have recently been developed (Sartori et al. 2007; Schwartz et al. 2007), but because the sample is kept frozen, oil immersion lenses cannot be used and unfortunately the resolution is too poor to localize fusion proteins within small bacterial cells (unpublished observations). Briegel et al. developed an alternative protocol, immobilizing cells on an EM grid with poly-l-lysine before inspection at room temperature in an oil-immersion, high-resolution fLM (Briegel et al. 2008). These cells remained in place when they were removed from the fLM and plunge-frozen, so matching cryo-tomograms were obtainable. The perfect correlation between the location of (mCherry-fused) chemoreceptor foci in the fLM images and the presence of the putative chemoreceptor array structure in the cryo-tomograms confirmed their identity. Unfortunately, to preserve the cells' integrity and stop internal rearrangements, they had to be lightly fixed before they were adhered to the grid surface with poly-l-lysine, so this method has not worked for other structures like certain cytoskeletal filaments that are destroyed by light fixation. Ideally, methods to label proteins with genetically fusable tags that are directly visible in cryo-tomograms of native cells are needed. To this end, several groups are trying to develop heavy metal cluster tags based on metallothionein or other metal-binding proteins (Nishino et al. 2007; Diestra et al. 2008).
In the absence of specific tags, another strategy that has been employed to identify objects in tomograms is manipulating their abundance. In the case of FtsZ, for example, the filaments at the constriction site were identified by comparing tomograms of wild-type cells with cells overexpressing FtsZ, where there were many more filaments showing the same localization patterns and structural characteristics, and cells overexpressing a hyper-stable mutant of FtsZ, in which hundreds of again similar but much longer filaments were seen. No similar filaments were seen in cells depleted for FtsZ.
Objects in tomograms can also sometimes be identified by their structural “signatures.” ParM filaments within cryo-sections of E. coli cells were identified this way, for instance (Salje et al. 2008). Power spectra of the filaments showing their longitudinal repeat and interfilament packing distances matched spacings known from crystal structures and images of ParM filament bundles in vitro. Sophisticated software has been written to identify particular isolated complexes by their morphological similarity to a known template (Bohm et al. 2000). In this way the positions and orientations of the ribosomes in Spiroplasma melliferum, a very slender bacterium (∼0.3 µm diameter) were estimated (Ortiz et al. 2006). When a large number of identical or nearly identical objects are present, higher signal-to-noise ratios can be obtained by averaging subtomograms (Schmid and Booth 2008).
In summary, ECT produces 3-D images of intact cells in near-native states to “molecular resolution,” but the sample must be thin (< 0.5 µm), the interpretability of the resulting tomograms is limited by radiation damage, the resolution is anisotropic because of tilt limitations, the procedure is complicated and requires expensive electron cryo-microscopes, and identifying structures of interest in the tomograms can be challenging. Nevertheless in just the last few years since it has become practical, ECT has already provided a wealth of new details about bacterial ultrastructures like the cytoskeleton, cell envelope, surface appendages, chemoreceptor arrays, and other large structures. A number of examples are summarized later.
FIRST CONTRIBUTIONS OF ECT TO BACTERIAL CELL BIOLOGY
The Bacterial Cytoskeleton
For decades it was thought that bacteria lacked cytoskeletons. Homolog of all three major eukaryotic cytoskeletal protein families (actin, tubulin, and intermediate filaments) were then identified in bacteria by their sequence, structural, and biochemical similarities (Michie and Löwe 2006), and were further found by fLM to localize in elongated patterns consistent with a filamentous structure (Pogliano 2008). As illustrated by the FtsZ example in the Introduction, ECT has now both confirmed by direct visualization that at least some of these do form filaments in vivo and provided vital new clues to their structures and functions.
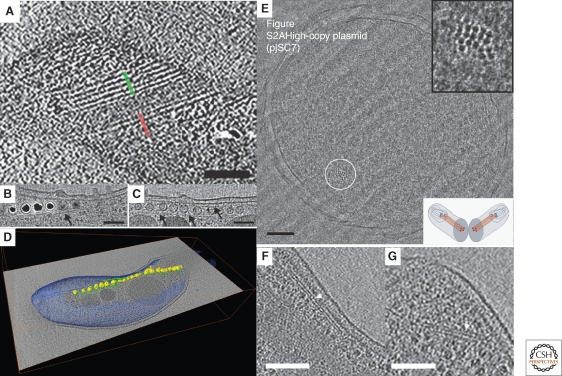
The first published report of the direct visualization of a cytoskeletal filament in a bacterium by ECT described the species Spiroplasma melliferum (Fig. 3A) (Kürner et al. 2005). Two ribbons of parallel 11-nm filaments were seen underneath the cytoplasmic membrane flanking a central ribbon of 4-nm filaments. Although the peripheral filaments were identified as the Fib protein, the central ribbon was speculated to be MreB. This “three-ribbon” model corresponds to a total ribbon width of 146 nm. More recently, however, results by Trachtenberg et al. reported that the filament bundle was continuous, uniform, and only 70 nm wide and proposed a “single-ribbon” model (Trachtenberg et al. 2008). The cause of these differences is unclear.
Figure 3.
Examples of bacterial cytoskeletal filaments. (A) Tomographic slice through a Spiroplasma melliferum cell. The two outer ribbons are composed of five thick filaments with a spacing of ∼11 nm (highlighted in green and red). Adapted from (Kürner et al. 2005) with permission from the American Association for the Advancement of Science. (B) Tomographic slice showing the magnetosome chains in Magnetospirillum magneticum sp. AMB-1 flanked by long MamK filaments (black arrows). The magnetite crystals appear as dense black balls. (C) Tomographic view of a magnetosome chain grown in the absence of iron. (D) 3-D segmentation of a M. magneticum cell showing the magnetosomes (yellow) and their associated filaments (green) with respect to the whole cell (blue), superimposed on a tomographic slice through the same cell (gray level image). Fig. 3B–D adapted from (Komeili et al. 2006) with permission from the American Association for the Advancement of Science. (E) Projection image through a vitreous cryo-section of an E. coli cell carrying the high-copy ParMRC plasmid pJSC7. Small bundles of filaments appear as black dots (circled). Upper inset: enlarged view of bundle, lower inset: Schematic representation of the plane of imaging which is perpendicular to the filaments to increase contrast and reveal their packing. Adapted from (Salje et al. 2008) with permission from the American Association for the Advancement of Science. (F) and (G) Tomographic slices through Bdellovibrio bacteriovorus cells showing cytoskeletal filaments (white arrowhead) oriented parallel to the plane of the plasma membrane. Adapted from (Borgnia et al. 2008) with permission from the American Society for Microbiology. Scale bar for Fig. 3E–G is 100 nm.
ECT led to the discovery of a novel bacterial actin homolog, MamK, in magnetotactic bacteria. Magnetotactic bacteria can navigate in geomagnetic fields with the help of unique organelle-like compartments called magnetosomes. Magnetosomes are magnetite (Fe3O4) or gregite (Fe3S4) crystals partially enclosed by a lipid bilayer, and are found in linear chains of 15 or more within cells (Bazylinski and Frankel 2004). Because traditional thin-section EM images often showed the magnetosomes near but not connected to the cytoplasmic membrane, it was thought that they were free-standing, fully-membrane-enclosed bacterial organelles. When frozen-hydrated Magnetospirillum magneticum sp. AMB-1 cells were imaged in 3-D by ECT, however, it became clear that the magnetosomes were instead just invaginations of the inner membrane (Komeili et al. 2006). A network of cytoskeletal filaments was seen flanking the magnetosome chain (Fig. 3B–D). One gene, mamK, from the mam operon involved in magnetosome genesis and function was identified as a homolog of the bacterial actin MreB. ECT reconstructions of cells with an in-frame deletion of mamK showed no filaments, and the magnetosome chains were disordered. When the mutant cells were complemented with a plasmid containing the mamK gene, the filaments reappeared and the magnetosomes were once again aligned. Concurrently, Scheffel et al. imaged the strain Magnetospirillum gryphiswaldense sp. MSR-1 and revealed similar filamentous structures associated with magnetosome chains (Scheffel et al. 2006). Surprisingly, in cells missing the mamJ gene, which encodes a small acidic protein, filaments were still present but the magnetosomes did not appear to be connected to them and no longer showed a linear arrangement. Together, these studies led to the model that MamJ links individual magnetosomes to MamK filaments. Importantly, these studies also showed that one of the functions of bacterial cytoskeletal filaments is to organize internal organelle-like compartments, just as they do in eukaryotic cells.
As mentioned briefly earlier, ParM has also been visualized in vivo by ECT. ParM is a bacterial actin homolog involved in low-copy R1 plasmid segregation. Because E. coli cells are too thick for direct imaging after plunge-freezing, Salje et al. cryo-sectioned cells overexpressing ParM and saw filament bundles (Fig. 3E) (Salje et al. 2008). The bundles were identified as ParM by noting that their computed diffraction patterns matched the diffraction patterns of ParM filaments assembled in vitro. In a wild-type (low copy R1 plasmid number) strain, the authors observed bundles of three to five intracellular ParM filaments localized within the periphery of the nucleoid, strongly supporting the model that one filament is needed to separate each R1 plasmid pair.
A number of other filaments have been described that still need to be identified. Four different types of cytoskeletal bundles, for instance, were observed in Caulobacter crescentus (Briegel et al. 2006). A different filament bundle was seen in Bdellovibrio bacteriovorus (Fig. 3F,G) (Borgnia et al. 2008). Taken together these examples show that filamentous proteins are both general and abundant in bacteria. Not surprisingly, the list of candidate filamentous proteins is rapidly expanding and now includes, for instance, actin homologs MreB, ParM, FtsA, MamK, AlfA, and others; tubulin homologs FtsZ, BtubA/B, TubZ-Ba, TubZ-Bt, and others; intermediate filaments CreS, CfpA, Scc, and AglZ, and the recently identified ParA/MinD superfamily (Pogliano 2008). ECT will likely be used to study the structure and function of all of these and others yet to be identified.
The Bacterial Cell Envelope
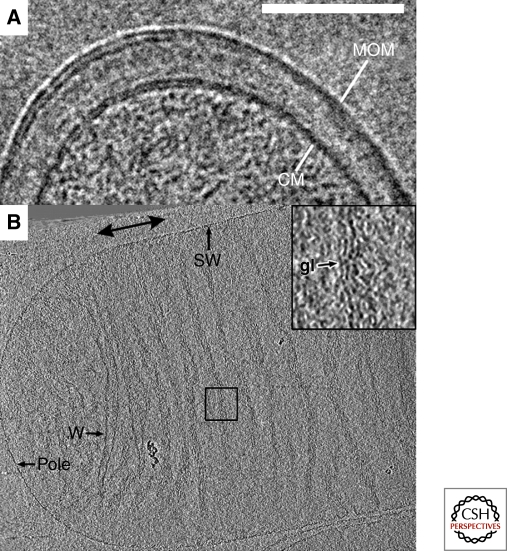
Although traditional thin-section EM images revealed the basic architecture of Gram-positive and negative bacterial envelopes, ECT and simple projection imaging of vitreous cryo-sections has now revealed key new details free of the potential complications of fixation artifacts. The mycobacterial cell wall has been intensively studied for decades, for instance, because its unique composition makes Mycobacterium tuberculosis remarkably resistant to antibiotics (Dover et al. 2004). Because the cell wall is comprised of an unusual peptidoglycan-arabinogalactan polymer with covalently bound mycolic acids and pore-forming proteins, it was thought to form an asymmetrical bilayer. In two independent studies, Hoffmann et al. and Zuber et al. applied ECT to vitreous cryo-sections of Mycobacterium smegmatis, Mycobacterium bovis BCG, and Corynebacterium glutamicum (Hoffmann et al. 2008; Zuber et al. 2008). The images showed that the outermost layer of the envelope is indeed a symmetric lipid bilayer (Fig. 4A). By imaging mycolic acid-deficient C. glutamicum, both groups also showed that mycolic acids are constituents of the outer membrane. Their models differed, however, in whether the meromycolic chains were folded or unfolded.
Figure 4.
The bacterial cell envelope. (A) Projection image from a cryo-section of Mycobacterium smegmatis. The cytoplasmic and mycobacterial outer membranes are marked as CM and MOM, respectively and the bilayer structure of both is discernible. Scale bar 100 nm. Adapted from (Hoffmann et al. 2008) with permission. © 2008 National Academy of Sciences, USA. (B) 10-nm thick tomographic slice through an E. coli strain XL-10 sacculus. Abbreviations: gl, glycan strand; SW, side wall; w, wrinkle. The double-headed arrow denotes the saccular polar axis. Inset shows four-fold enlarged view of the boxed region. Adapted from (Gan et al. 2008) with permission. © 2008 National Academy of Sciences, USA.
The mechanical strength of the bacterial cell envelope arises from a mesh of peptide-crosslinked glycan strands. Although the chemical composition and subunit structure of this peptidoglycan layer has been known for decades, its higher-order architecture had remained unclear. Two fundamentally different models had been suggested, namely the “layered” and “scaffold” models (Vollmer and Holtje 2004). By imaging intact sacculi purified from two Gram-negative bacteria, E. coli (Fig. 4B) and C. crescentus, Gan et al. showed that the peptidoglycan is just a single layer thick and that individual glycan strands are oriented in the plane of the sacculus perpendicular to the long axis of the cell approximately 5–8 nm apart (Gan et al. 2008). This observation ruled out the scaffold model and instead established a “disordered, circumferential, layered” model.
Motility and Surface Appendages
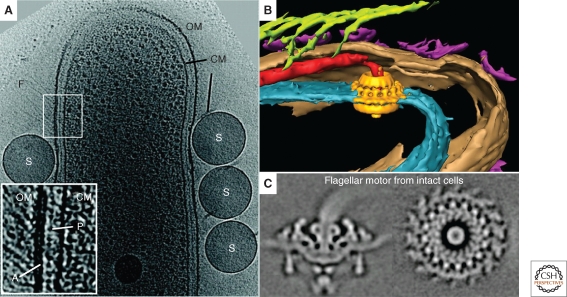
ECT has also provided insight into several different forms of bacterial motility. The cryo-tomograms of S. melliferum mentioned earlier led to a model of its motility based on differential contraction of the filament ribbons (Kürner et al. 2005). The gliding bacterium Flavobacterium johnsoniae was shown to possess tufts of ∼5-nm-wide external filaments emanating from the inner surface of the outer membrane (Fig. 5A) (Liu et al. 2007). These filaments were absent in a nonmotile gldF mutant cell but were restored in the same mutant complemented with plasmid-encoded GldF, a component of a putative ATP-binding cassette transporter. Cryo-tomograms of Mycoplasma pneumoniae revealed the molecular architecture of its “attachment organelle” and suggested that it was in fact a multijointed, flexible motility machine (Henderson and Jensen 2006; Seybert et al. 2006). ECT of the pathogenic spirochete Treponema denticola revealed cytoplasmic filaments and platelike structures possibly anchoring those filaments to the inner membrane (Izard et al. 2008). The cytoplasmic filaments, earlier suggested to be composed of CfpA (a unique protein to spirochetes) (Izard et al. 2001), were visible directly beneath the flagella in the cytoplasm, suggesting a role in motility. Cryo-tomograms of the spirochete Treponema primitia revealed novel structures such as bowls, arches, fibrils, and two layers of peptidoglycan (Fig. 5B) (Murphy et al. 2008), strongly supporting the “rolling cylinder” model of motility originally proposed by Berg (Berg 1976). Charon et al. imaged the Lyme disease spirochete, Borrelia burgdorferi, and revealed that the periplasmic flagella adopt a flat-ribbon configuration (Charon et al. 2008) instead of the previously reported stacked bundle observed by traditional-EM (Motaleb et al. 2000).
Figure 5.
Motility and surface appendages. (A) 3-nm thick tomographic slice through a wild-type Flavobacterium johnsoniae cell. Features arising from the cytoplasmic membrane (CM), outer membrane (OM), peptidoglycan (P), cell surface filaments (F), and added latex spheres (S) are labeled. The inset shows an expanded view of the periplasmic region at a location where filaments are observed. The densities arising from the outer membrane, cytoplasmic membrane, peptidoglycan layer, and patch (A) at the base of the outer membrane can be seen. Scale bar 300 nm. Adapted from (Liu et al. 2007) with permission from the American Society for Microbiology. (B) Manually segmented 3-D view of a Treponema primitia cell shows surface hooks (green), bowls (purple), inner membrane (blue), outer membrane (brown), and the averaged flagellar motor (yellow) embedded in the position of the motor. Adapted from (Murphy and Jensen 2007). © 2007 BioTechniques. Used by Permission. (C) Side (left) and top (right) views of the averaged flagellar motor from Borrelia burgdorferi, without rotational symmetry imposed. Adapted from (Liu et al. 2009) with permission from the American Society for Microbiology.
At the heart of swimming motility is the flagellar motor. This quintessential molecular machine is a structural biological marvel, but because the torque-generating stator is embedded in the inner membrane and binds to the peptidoglycan, isolating intact motors for structure determination has not been possible. The first structure of a complete flagellar motor, at ∼7 nm resolution, was therefore obtained by averaging 20 individual motors from cryo-tomograms of T. primitia (Fig. 5B) (Murphy et al. 2006). The stator showed 16-fold symmetry and possessed direct contacts to the rotor, C ring, and a novel P-ring-like structure. This work illustrated how some important macromolecular complexes may not be purifiable, and will therefore have to be studied in situ. ECT is a technology that can do that, albeit at low resolution. More recently Liu et al. reached ∼3.5 nm resolution by averaging over a thousand flagellar motors from cryo-tomograms of B. burgdorferi (Fig. 5C) (Liu et al. 2009). By not imposing rotational symmetry, asymmetric aspects of the stator structure and rotor-stator interactions were revealed including curvature in the stator itself and flexibility in the rotor-stator interaction. It was proposed that flexibility in the motor structure may be needed to maintain connectivity among the flagellar proteins during rotation.
Chemotaxis
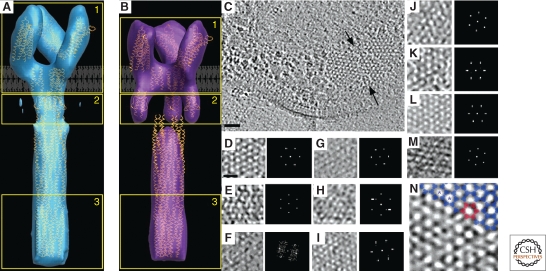
The chemotactic system in bacteria senses attractants and repellants in the environment. The main structural units of the system (the methyl-accepting chemotaxis proteins, or MCPs) are arranged in arrays and are usually located at the cell pole. After numerous biochemical, structural, and modeling studies (Kim et al. 1999; Shimizu et al. 2000; Studdert and Parkinson 2005), averaging cryo-tomograms of the serine Tsr receptor overexpressed in E. coli gave rise to the first full-length 3-D structure of an MCP (Fig. 6A,B) (Khursigara et al. 2008b). The result fit well with the known atomic models of the various domains. Structures were obtained both in the presence and absence of attractant, revealing conformational changes in the HAMP domain of the receptor likely responsible for signal conversion. The averaged structures of the two states showed a “trimer-of-dimers” interaction, confirming the functional unit suggested by previous crystallographic studies (Kim et al. 2002).
Figure 6.
Chemotaxis. (A) Compact and (B) Expanded averaged conformations of the TsrQEQE receptor. The structural coordinates corresponding to chemoreceptor models are loosely fit to a single trimer of receptor dimers. Adapted from (Khursigara et al. 2008b) with permission. © 2008 National Academy of Sciences, USA. (C) Hexagonal arrangement of receptors in different bacteria. Top view of a chemoreceptor array (between black arrows) in Thermotoga maritima. A subregion of the hexagonally ordered lattice and its corresponding power spectrum showing the ∼12 nm periodicity are enlarged in the inset. Scale bar 50 nm. (D–M) Top views of receptor arrays in other organisms. (D) T. maritima; (E) A. longum; (F) C. jejuni; (G) H. hepaticus; (H) M. magneticum; (I) H. neapolitanus; (J) R. sphaeroides; (K) E. coli; (L) V. cholerae; (M) T. primitia. Scale bar 25 nm, power spectra enlarged. (N) Trimer of dimers (blue) fit into the vertices of the hexagonal lattice in a chemoreceptor array. Six trimers of dimers (red) enclose one hexagon. The spacing from the center of one hexagon to another (blue asterisks) is 12 nm. Adapted from (Briegel et al. 2009) with permission. © 2009 National Academy of Sciences, USA.
The extraordinary sensitivity, gain, and signal integration showed by chemoreceptor systems is thought to be made possible by the grouping of chemoreceptors into organized arrays (Hazelbauer et al. 2008). Several competing models of these interactions have been proposed over the years based on crystal structures (Kim et al. 2002), pulsed electron spin relaxation (Park et al. 2006), cross-linking (Studdert and Parkinson 2005), cryo-EM reconstructions (Wolanin et al. 2006), and tomograms of cells overexpressing chemoreceptors (Zhang et al. 2004). ECT of C. crescentus cells by two groups then showed that chemoreceptors are arranged in a hexagonal array with a center-to-center spacing of 12 nm (Briegel et al. 2008; Khursigara et al. 2008a). Docking crystal structures of chemoreceptor “trimer-of-dimers” at the vertices of this hexagonal pattern yielded a compelling fit. ECT of 13 different species has now shown that this same 12-nm hexagonal architecture is conserved across a broad array of bacterial phyla, and is thus probably universally conserved (Fig. 6C–G) (Briegel et al. 2009).
Metabolism
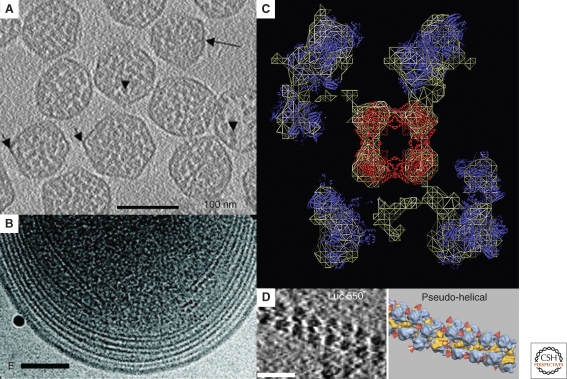
Bacteria were once thought to be simple bags of enzymes. It is now understood, however, that many reactions are spatially organized and localized (Norris et al. 2007). Because ECT can produce molecular resolution 3-D images of unique assemblies of macromolecules, it will be an essential tool to understand such spatial regulation. One of the most stunning examples of spatial organization is the carboxysome, a proteinaceous microcompartment that sequesters the enzyme ribulose 1,5-bisphosphate carboxylase/oxygenase (RuBisCO) (Yeates et al. 2008). In cyanobacteria and many chemoautotrophic bacteria, the encapsulated RuBisCO catalyzes the first step of carbon fixation in the Calvin-Benson-Bassham cycle. It is thought that by sequestering RuBisCO near carbonic anhydrases, carboxysomes increase the local concentration of substrate and enzyme and inhibit side reactions. Schmid et al. first reported the structure of carboxysomes by ECT from Halothiobacillus neapolitanus and showed that they are regular icosahedra of different sizes (Fig. 7A) (Schmid et al. 2006). The authors averaged subclasses of particles and suggested that the different sizes might be a result of different packing arrangements of the shell proteins. Iancu et al. imaged carboxysomes from Synechococcus strain WH8102, showing that they too were regular icosahedra, but suggested that the different sizes arose from different “T-numbers” (Iancu et al. 2007). These authors further showed through simulation that simple packing forces could explain the concentric shells of RuBisCO seen in both studies.
Figure 7.
Metabolic macromolecular complexes. (A) 4-nm tomographic slice through a field of isolated carboxysomes from H. neapolitanus. Long arrow: thin shell surrounding the carboxysome. Shorter arrows: fivefold vertices of several particles. Arrowheads: RuBisCO molecules. Scale bar 100 nm. Adapted from (Schmid et al. 2006) with permission from Elsevier Ltd. (B) Projection image of the intracytoplasmic membranes at the pole of a Prochlorococcus MIT9313 cell. The black particles are colloidal gold. Scale bar 100 nm. Adapted from (Ting et al. 2007) with permission from the American Society for Microbiology. (C) Tomographic slice through the reconstruction of a multimeric PDMC complex. The wire surface encloses the part of the volume of the complete complex that appears in this slice. Crystal structures of the central E2 core (red) and the peripheral E1 or E3 domains (blue) are shown as one of several potential fits. Note that although the crystal structure of the symmetrically arranged E2 core could be unambiguously docked, the resolution was insufficient to distinguish the E1 from E3 subunits or their precise orientations. Adapted from (Murphy and Jensen 2005) with permission from Elsevier Ltd. (D) Tomographic slice (left) through a translationally stalled polysome in vitro. Isosurfaces (right) are represented in color (large ribosomal subunit in blue and small subunit in yellow). Red cones point to the peptide exit tunnel on each 50S subunit. Scale bar 50 nm. Adapted from (Brandt et al. 2009) with permission from Elsevier Ltd.
Because the presumed function of carboxysomes is to sequester and organize specific reactions within the cell, however, their positions and relationships to the rest of the cell must also be understood. ECT has been used by two groups now to image carboxysomes within their cellular context (Ting et al. 2007; Iancu et al. 2010). ECT of intact H. neapolitanus cells by Iancu et al. showed that carboxysomes in vivo are clustered around and have extensive proteinaceous contacts with polyphosphate storage granules. This work also showed that carboxysomes in vivo are much more structurally heterogeneous than purified preparations and frequently contain internal storage granules that are lost during purification. Finally, ECT of cells provided images of assembling carboxysomes, giving insight into processes and sorting phenomena that are unlikely to ever be reproduced in vitro.
ECT has also been used to elucidate the organization of bacterial photosynthetic membranes (Fig. 7B). Ting et al. applied ECT to frozen-hydrated cells and vitreous sections and showed that closely related strains of cyanobacteria from the genus Prochlorococcus differ in their cell size, cell-wall structure, carboxysomes size, and photosynthetic lamellae structure (Ting et al. 2007). Studies on another photosynthetic bacterium, Rhodopseudomonas viridis (now Blastochloris viridis), revealed tunnel-like structures connecting the tightly packed photosynthetic membranes to the inner membrane (Konorty et al. 2008). A tightly packed two-dimensional hexagonal lattice of proteins within the membranes prompted questions about whether electron carriers such as quinones could diffuse freely. To address that issue, Konorty et al. then used traditional-EM, ECT and spectroscopy to identify structural and functional differences between cells grown in the presence or absence of light. The authors show that increased molecular order is a general trend in cells characterized by low metabolic activity likely as a result of slower quinone diffusion and fewer bc1 complexes (Konorty et al. 2009).
Although individual enzyme structures can be obtained by X-ray crystallography, NMR spectroscopy, electron crystallography, and/or cryo-EM-based single-particle analysis, all of these techniques require large numbers of identical copies of the biological specimen. Because many enzymes function as parts of heterogeneous complexes that are too dynamic to present a single uniform conformation, techniques like ECT that can reveal the structure of unique objects such as these, albeit at much lower resolution, are essential. ECT was used, for example, to determine the quaternary structures of the E. coli pyruvate and 2-oxoglutarate dehydrogenases (PDHC and OGDHC, respectively) to ∼5.5 nm resolution (Fig. 7C) (Murphy and Jensen 2005). PDHC and OGDHC are multimeric enzymes composed of three protein subunits (E1, E2, and E3). Atomic models of all three subunits (de Kok et al. 1998) and single particle reconstructions of the E1E2 complex (Milne et al. 2002) were available, but the architecture of the intact complex remained unclear. Murphy et al. showed that the E1 and E3 subunits are flexibly tethered to the cubic E2 core. Protein complexes as small as 80 kDa were clearly visible in the reconstructions, suggesting that ECT might prove helpful in elucidating the quaternary structure and conformational flexibility of many large complexes. Concurrent tomographic study of the E2E3 complex in vitro further supported the notion that the distribution of E1 and E3 molecules around the E2 core is random (Milne et al. 2006).
Brandt et al. used a template-matching approach to identify 70S ribosomes in actively translating E. coli cell lysates (Brandt et al. 2009). Even though various crystal structures of ribosomal complexes have been solved, little is known about the spatial arrangement of ribosomes in the cell during protein synthesis. Ribosomes in lysates were found to be densely packed around an mRNA template and showed preferred orientations (either staggered or helical) (Fig. 7D). Although the mRNA template was found to be on the inside of the ribosome cluster, the tRNAs and newly synthesized proteins were entering and exiting from the outside, minimizing interaction between newly synthesized proteins and lowering the probability of aggregate formation.
CONCLUSION AND FUTURE DIRECTIONS
As can be seen from these examples, ECT has become a powerful tool to investigate bacterial ultrastructure. Improvements in both the quality and number of cryo-tomograms being produced should be expected. Beginning with the sample, further improvements in methods for cryo-sectioning will make ECT of serial cryo-sections routine for larger cells and even biofilms. The development of genetically encodable, electron-dense tags (Mercogliano and DeRosier 2007; Nishino et al. 2007; Diestra et al. 2008), improvements in the technologies for correlating fluorescence and cryo-EM images (Lucić et al. 2007; Schwartz et al. 2007; Briegel et al. 2008), and increasingly sophisticated computational tools will allow specific molecules to be located and identified. Instrumentally, the development of direct electron detectors, phase plates, and aberration correctors are expected to dramatically improve image quality in just the next few years. As additional institutions acquire the expensive microscopes needed and more practitioners are trained, the number of ECT studies will also obviously increase. Lastly, automated data collection (Suloway et al. 2009) and processing (Amat et al. 2008), coupled with the development of databases for the management and distribution of the large datasets produced by ECT (Martone et al. 2003) (Ding, J.H. and Jensen, G.J., in preparation), will make it possible to collect and compare thousands of tomograms. Together with complementary advances in genetics, biochemistry, structural genomics, light microscopy, and computational modeling, it therefore seems that elucidating the structure, function, and location of every macromolecule in key model bacterial cells is a plausible goal for our generation (Morris and Jensen 2008).
Footnotes
Editors: Lucy Shapiro and Richard Losick
Additional Perspectives on Cell Biology of Bacteria available at www.cshperspectives.org
REFERENCES
- Addinall SG, Lutkenhaus J 1996. FtsA is localized to the septum in an FtsZ-dependent manner. J Bacteriol 178:7167–7172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Amoudi A, Chang JJ, Leforestier A, McDowall A, Salamin LM, Norlén LP, Richter K, Blanc NS, Studer D, Dubochet J 2004. Cryo-electron microscopy of vitreous sections. EMBO J 23:3583–3588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amat F, Moussavi F, Comolli LR, Elidan G, Downing KH, Horowitz M 2008. Markov random field based automatic image alignment for electron tomography. J Struct Biol 161:260–275 [DOI] [PubMed] [Google Scholar]
- Anderson DE, Gueiros-Filho FJ, Erickson HP 2004. Assembly dynamics of FtsZ rings in Bacillus subtilis and Escherichia coli and effects of FtsZ-regulating proteins. J Bacteriol 186:5775–5781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angell CA 2004. Amorphous water. Annu Rev Phys Chem 55:559–583 [DOI] [PubMed] [Google Scholar]
- Batson PE, Dellby N, Krivanek OL 2002. Sub-angstrom resolution using aberration corrected electron optics. Nature 418:617–620 [DOI] [PubMed] [Google Scholar]
- Baumeister W, Grimm R, Walz J 1999. Electron tomography of molecules and cells. Trends Cell Biol 9:81–85 [DOI] [PubMed] [Google Scholar]
- Bazylinski DA, Frankel RB 2004. Magnetosome formation in prokaryotes. Nat Rev Microbiol 2:217–230 [DOI] [PubMed] [Google Scholar]
- Berg HC 1976. How spirochetes may swim. J Theor Biol 56:269–273 [DOI] [PubMed] [Google Scholar]
- Bi EF, Lutkenhaus J 1991. FtsZ ring structure associated with division in Escherichia coli. Nature 354:161–164 [DOI] [PubMed] [Google Scholar]
- Bohm J, Frangakis AS, Hegerl R, Nickell S, Typke D, Baumeister W 2000. Toward detecting and identifying macromolecules in a cellular context: Template matching applied to electron tomograms. Proc Natl Acad Sci 97:14245–14250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgnia MJ, Subramaniam S, Milne JL 2008. Three-dimensional imaging of the highly bent architecture of Bdellovibrio bacteriovorus by using cryo-electron tomography. J Bacteriol 190:2588–2596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramhill D 1997. Bacterial cell division. Annu Rev Cell Dev Biol 13:395–424 [DOI] [PubMed] [Google Scholar]
- Brandt F, Etchells SA, Ortiz JO, Elcock AH, Hartl FU, Baumeister W 2009. The native 3D organization of bacterial polysomes. Cell 136:261–271 [DOI] [PubMed] [Google Scholar]
- Briegel A, Dias DP, Li Z, Jensen RB, Frangakis AS, Jensen GJ 2006. Multiple large filament bundles observed in Caulobacter crescentus by electron cryotomography. Mol Microbiol 62:5–14 [DOI] [PubMed] [Google Scholar]
- Briegel A, Ding HJ, Li Z, Werner J, Gitai Z, Dias DP, Jensen RB, Jensen GJ 2008. Location and architecture of the Caulobacter crescentus chemoreceptor array. Mol Microbiol 69:30–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briegel A, Ortega DR, Tocheva EI, Wuichet K, Li Z, Chen S, Muller A, Iancu CV, Murphy GE, Dobro MJ et al. 2009. Universal architecture of bacterial chemoreceptor arrays. Proc Natl Acad Sci 106:17181–17186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charon NW, Goldstein SF, Marko M, Hsieh C, Gebhardt LL, Motaleb MA, Wolgemuth CW, Limberger RJ, Rowe N 2008. The flat ribbon configuration of the periplasmic flagella of Borrelia burgdorferi and its relationship to motility and morphology. J Bacteriol 191:600–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comolli LR, Downing KH 2005. Dose tolerance at helium and nitrogen temperatures for whole cell electron tomography. J Struct Biol 152:149–156 [DOI] [PubMed] [Google Scholar]
- Crowther RA, DeRosier DJ, Klug A 1970b. The reconstruction of a three-dimensional structure from projections and its application to electron microscopy. Proc Roy Soc Lond A 317:319–340 [DOI] [PubMed] [Google Scholar]
- Crowther RA, Amos LA, Finch JT, De Rosier DJ, Klug A 1970a. Three dimensional reconstructions of spherical viruses by fourier synthesis from electron micrographs. Nature 226:421–425 [DOI] [PubMed] [Google Scholar]
- de Kok A, Hengeveld AF, Martin A, Westphal AH 1998. The pyruvate dehydrogenase multi-enzyme complex from Gram-negative bacteria. Biochim Biophys Acta 1385:353–366 [DOI] [PubMed] [Google Scholar]
- Derosier DJ, Klug A 1968. Reconstruction of 3 dimensional structures from electron micrographs. Nature 217:130. [DOI] [PubMed] [Google Scholar]
- Diestra E, Fontana J, Guichard P, Marco S, Risco C 2008. Visualization of proteins in intact cells with a clonable tag for electron microscopy. J Struct Biol 165:157–168 [DOI] [PubMed] [Google Scholar]
- Dover LG, Cerdeño-Tárrage AM, Pallen MJ, Parkhill J, Besra GS 2004. Comparative cell wall core biosynthesis in the mycolated pathogens, Mycobacterium tuberculosis and Corynebacterium diphtheriae. FEMS Microbiol Rev 28:225–250 [DOI] [PubMed] [Google Scholar]
- Dubochet J, Adrian M, Chang JJ, Homo JC, Lepault J, McDowall AW, Schultz P 1988. Cryo-electron microscopy of vitrified specimens. Q Rev Biophys 21:129–228 [DOI] [PubMed] [Google Scholar]
- Dubochet J, McDowall AW, Menge B, Schmid EN, Lickfeld KG 1983. Electron microscopy of frozen-hydrated bacteria. J Bacteriol 155:381–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson HP 1997. FtsZ, a tubulin homologue in prokaryote cell division. Trends Cell Biol 7:362–367 [DOI] [PubMed] [Google Scholar]
- Erk I, Nicolas G, Caroff A, Lepault J 1998. Electron microscopy of frozen biological objects: A study using cryosectioning and cryosubstitution. J Microsc 189:236–248 [DOI] [PubMed] [Google Scholar]
- Fernández JJ, Li S, Crowther RA 2006. CTF determination and correction in electron cryotomography. Ultramicroscopy 106:587–596 [DOI] [PubMed] [Google Scholar]
- Frangakis AS, Hegerl R 2001. Noise reduction in electron tomographic reconstructions using nonlinear anisotropic diffusion. J Struct Biol 135:239–250 [DOI] [PubMed] [Google Scholar]
- Frederik PM, Hubert DH 2005. Cryoelectron microscopy of liposomes. Methods Enzymol 391:431–448 [DOI] [PubMed] [Google Scholar]
- Gan L, Chen S, Jensen GJ 2008. Molecular organization of Gram-negative peptidoglycan. Proc Natl Acad Sci 105:18953–18957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm R, Typke D, Barmann M, Baumeister W 1996. Determination of the inelastic mean free path in ice by examination of tilted vesicles and automated most probable loss imaging. Ultramicroscopy 63:169–179 [DOI] [PubMed] [Google Scholar]
- Grunewald K, Medalia O, Gross A, Steven AC, Baumeister W 2003. Prospects of electron cryotomography to visualize macromolecular complexes inside cellular compartments: Implications of crowding. Biophys Chem 100:577–591 [DOI] [PubMed] [Google Scholar]
- Hazelbauer GL, Falke JJ, Parkinson JS 2008. Bacterial chemoreceptors: High-performance signaling in networked arrays. Trends Biochem Sci 33:9–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson R 1995. The potential and limitations of neutrons, electrons and X-rays for atomic resolution microscopy of unstained biological molecules. Q Rev Biophys 28:171–193 [DOI] [PubMed] [Google Scholar]
- Henderson GP, Jensen GJ 2006. Three-dimensional structure of Mycoplasma pneumoniae's attachment organelle and a model for its role in gliding motility. Mol Microbiol 60:376–385 [DOI] [PubMed] [Google Scholar]
- Hoffmann C, Leis A, Niederweis M, Plitzko JM, Engelhardt H 2008. Disclosure of the mycobacterial outer membrane: Cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Proc Natl Acad Sci 105:3963–3967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iancu CV, Ding HJ, Morris DM, Dias DP, Gonzales AD, Martino A, Jensen GJ 2007. The structure of isolated Synechococcus strain WH8102 carboxysomes as revealed by electron cryotomography. J Mol Biol 372:764–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iancu CV, Morris DM, Dou Z, Heinhorst S, Cannon GC, Jensen GJ 2010. Organization, Structure, and Assembly of alpha-Carboxysomes Determined by Electron Cryotomography of Intact Cells. J Mol Biol 396:105–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iancu CV, Tivol WF, Schooler JB, Dias DP, Henderson GP, Murphy GE, Wright ER, Li Z, Yu Z, Briegel A et al. 2006a. Electron cryotomography sample preparation using the Vitrobot. Nat Protocols 1:2813–2819 [DOI] [PubMed] [Google Scholar]
- Iancu CV, Wright ER, Benjamin J, Tivol WF, Dias DP, Murphy GE, Morrison RC, Heymann JB, Jensen GJ 2005. A “flip-flop” rotation stage for routine dual-axis electron cryotomography. J Struct Biol 151:288–297 [DOI] [PubMed] [Google Scholar]
- Iancu CV, Wright ER, Heymann JB, Jensen GJ 2006b. A comparison of liquid nitrogen and liquid helium as cryogens for electron cryotomography. J Struct Biol 153:231–240 [DOI] [PubMed] [Google Scholar]
- Izard J, Samsonoff WA, Limberger RJ 2001. Cytoplasmic filament-deficient mutant of Treponema denticola has pleiotropic defects. J Bacteriol 183:1078–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izard J, Hsieh CE, Limberger RJ, Mannella CA, Marko M 2008. Native cellular architecture of Treponema denticola revealed by cryo-electron tomography. J Strct Biol 163:10–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen GJ, Briegel A 2007. How electron cryotomography is opening a new window onto prokaryotic ultrastructure. Curr Opin Struct Biol 17:260–267 [DOI] [PubMed] [Google Scholar]
- Khursigara CM, Wu X, Subramaniam S 2008a. Chemoreceptors in Caulobacter crescentus: Trimers of receptor dimers in a partially ordered hexagonally packed array. J Bacteriol 190:6805–6810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khursigara CM, Wu X, Zhang P, Lefman J, Subramaniam S 2008b. Role of HAMP domains in chemotaxis signaling by bacterial chemoreceptors. Proc Natl Acad Sci 105:16555–16560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KK, Yokota H, Kim SH 1999. Four-helical-bundle structure of the cytoplasmic domain of a serine chemotaxis receptor. Nature 400:787–792 [DOI] [PubMed] [Google Scholar]
- Kim SH, Wang W, Kim KK 2002. Dynamic and clustering model of bacterial chemotaxis receptors: Structural basis for signaling and high sensitivity. Proc Natl Acad Sci 99:11611–11615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komeili A, Li Z, Newman DK, Jensen GJ 2006. Magnetosomes are cell membrane invaginations organized by the actin-like protein MamK. Science 311:242–245 [DOI] [PubMed] [Google Scholar]
- Konorty M, Brumfeld V, Vermeglio A, Kahana N, Medalia O, Minsky A 2009. Photosynthetic system in Blastochloris viridis revisited. Biochemistry 48:4753–4761 [DOI] [PubMed] [Google Scholar]
- Konorty M, Kahana N, Linaroudis A, Minsky A, Medalia O 2008. Structural analysis of photosynthetic membranes by cryo-electron tomography of intact Rhodopseudomonas viridis cells. J Struct Biol 161:393–400 [DOI] [PubMed] [Google Scholar]
- Kürner J, Frangakis A, Baumeister W 2005. Cryo-electron tomography reveals the cytoskeletal structure of Spiroplasma melliferum. Science 307:436–438 [DOI] [PubMed] [Google Scholar]
- Lee E, Fahimian BP, Iancu CV, Suloway C, Murphy GE, Wright ER, Castano-Diez D, Jensen GJ, Miao J 2008. Radiation dose reduction and image enhancement in biological imaging through equally-sloped tomography. J Struct Biol 164:221–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leis A, Rockel B, Andrees L, Baumeister W 2008. Visualizing cells at the nanoscale. Trends Biochem Sci 34:60–70 [DOI] [PubMed] [Google Scholar]
- Levin PA, Losick R 1996. Transcription factor Spo0A switches the localization of the cell division protein FtsZ from a medial to a bipolar pattern in Bacillus subtilis. Genes Development 10:478–488 [DOI] [PubMed] [Google Scholar]
- Li Z, Jensen GJ 2009. Electron cryotomography: A new view into microbial ultrastructure. Curr Opin Microbiol 12:333–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Trimble MJ, Brun YV, Jensen GJ 2007. The structure of FtsZ filaments in vivo suggests a force-generating role in cell division. EMBO J 26:4694–4708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Lin T, Botkin DJ, McCrum E, Winkler H, Norris SJ 2009. Intact flagellar motor of Borrelia burgdorferi revealed by cryo-electron tomography: Evidence for stator ring curvature and rotor/C-ring assembly flexion. J Bacteriol 191:5026–5036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, McBride MJ, Subramaniam S 2007. Cell surface filaments of the gliding bacterium Flavobacterium johnsoniae revealed by cryo-electron tomography. J Bacteriol 189:7503–7506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löwe J, Amos LA 1998. Crystal structure of the bacterial cell-division protein FtsZ. Nature 391:203–206 [DOI] [PubMed] [Google Scholar]
- Lu C, Reedy M, Erickson HP 2000. Straight and curved conformations of FtsZ are regulated by GTP hydrolysis. J Bacteriol 182:164–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucić V, Förster F, Baumeister W 2005. Structural studies by electron tomography: from cells to molecules. Annu Rev Biochem 74:833–865 [DOI] [PubMed] [Google Scholar]
- Lucić V, Kossel AH, Yang T, Bonhoeffer T, Baumeister W, Sartori A 2007. Multiscale imaging of neurons grown in culture: from light microscopy to cryo-electron tomography. J Struct Biol 160:146–156 [DOI] [PubMed] [Google Scholar]
- Lutkenhaus J 1993. FtsZ ring in bacterial cytokinesis. Mol Microbiol 9:403–409 [DOI] [PubMed] [Google Scholar]
- Lutkenhaus JF, Wolf-Watz H, Donachie WD 1980. Organization of genes in the ftsA-envA region of the Escherichia coli genetic map and identification of a new fts locus (ftsZ). J Bacteriol 142:615–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Ehrhardt DW, Margolin W 1996. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc Natl Acad Sci 93:12998–13003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin W 1998. A green light for the bacterial cytoskeleton. Trends Microbiol 6:233–238 [DOI] [PubMed] [Google Scholar]
- Martone ME, Zhang S, Gupta A, Qian X, He H, Price DL, Wong M, Santini S, Ellisman MH 2003. The cell-centered database: A database for multiscale structural and protein localization data from light and electron microscopy. Neuroinformatics 1:379–395 [DOI] [PubMed] [Google Scholar]
- Mastronarde DN 2005. Automated electron microscope tomography using robust prediction of specimen movements. J Struct Biol 152:36–51 [DOI] [PubMed] [Google Scholar]
- Mastronarde DN 2008. Correction for non-perpendicularity of beam and tilt axis in tomographic reconstructions with the IMOD package. J Microsc 230:212–217 [DOI] [PubMed] [Google Scholar]
- McDonald KL, Auer M 2006. High-pressure freezing, cellular tomography, and structural cell biology. BioTechniques 41:137–143 [DOI] [PubMed] [Google Scholar]
- Mercogliano C, DeRosier D 2007. Concatenated metallothionein as a clonable gold label for electron microscopy. J Struct Biol 160:70–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michie KA, Löwe J 2006. Dynamic filaments of the bacterial cytoskeleton. Annu Rev Biochem 75:467–492 [DOI] [PubMed] [Google Scholar]
- Midgley PA, Dunin-Borkowski RE 2009. Electron tomography and holography in materials science. Nat Mater 8:271–280 [DOI] [PubMed] [Google Scholar]
- Milne JL, Shi D, Rosenthal PB, Sunshine JS, Domingo GJ, Wu X, Brooks BR, Perham RN, Henderson R, Subramaniam S 2002. Molecular architecture and mechanism of an icosahedral pyruvate dehydrogenase complex: A multifunctional catalytic machine. EMBO J 21:5587–5598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne JL, Wu X, Borgnia MJ, Lengyel JS, Brooks BR, Shi D, Perham RN, Subramaniam S 2006. Molecular structure of a 9-MDa icosahedral pyruvate dehydrogenase subcomplex containing the E2 and E3 enzymes using cryoelectron microscopy. J Biol Chem 281:4364–4370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris DM, Jensen GJ 2008. Toward a biomechanical understanding of whole bacterial cells. Annu Rev Biochem 77:583–613 [DOI] [PubMed] [Google Scholar]
- Motaleb MA, Corum L, Bono JL, Elias AF, Rosa P, Samuels DS, Charon NW 2000. Borrelia burgdorferi periplasmic flagella have both skeletal and motility functions. Proc Natl Acad Sci 97:10899–10904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy GE, Jensen GJ 2005. Electron cryotomography of the E. coli pyruvate and 2-oxoglutarate dehydrogenase complexes. Structure 13:1765–1773 [DOI] [PubMed] [Google Scholar]
- Murphy GE, Jensen GJ 2007. Electron cryotomography. BioTechniques 43:413–421 [DOI] [PubMed] [Google Scholar]
- Murphy GE, Leadbetter JR, Jensen GJ 2006. In situ structure of the complete Treponema primitia flagellar motor. Nature 442:1062–1064 [DOI] [PubMed] [Google Scholar]
- Murphy GE, Matson EG, Leadbetter JR, Berg HC, Jensen GJ 2008. Novel ultrastructures of Treponema primitia and their implications for motility. Mol Microbiol 67:1184–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narasimha R, Aganj I, Bennett AE, Borgnia MJ, Zabransky D, Sapiro G, McLaughlin SW, Milne JL, Subramaniam S 2008. Evaluation of denoising algorithms for biological electron tomography. J Struct Biol 164:7–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickell S, Forster F, Linaroudis A, Net WD, Beck F, Hegerl R, Baumeister W, Plitzko JM 2005. TOM software toolbox: Acquisition and analysis for electron tomography. J Struct Biol 149:227–234 [DOI] [PubMed] [Google Scholar]
- Nickell S, Hegerl R, Baumeister W, Rachel R 2003. Pyrodictium cannulae enter the periplasmic space but do not enter the cytoplasm, as revealed by cryo-electron tomography. J Struct Biol 141:34–42 [DOI] [PubMed] [Google Scholar]
- Nishino Y, Yasunaga T, Miyazawa A 2007. A genetically encoded metallothionein tag enabling efficient protein detection by electron microscopy. J Electron Microsc 56:93–101 [DOI] [PubMed] [Google Scholar]
- Norris V, Den Blaauwen T, Cabin-Flaman A, Doi R, Harshey R, Janniere L, Jimenez-Sanchez A, Jin D, Levin P, Mileykovskaya E et al. 2007. Functional Taxonomy of Bacterial Hyperstructures. Microbiol Mol Biol R 71:230–253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortiz JO, Förster F, Kürner J, Linaroudis AA, Baumeister W 2006. Mapping 70S ribosomes in intact cells by cryoelectron tomography and pattern recognition. J Struct Biol 156:334–341 [DOI] [PubMed] [Google Scholar]
- Park SY, Borbat PP, Gonzalez-Bonet G, Bhatnagar J, Pollard AM, Freed JH, Bilwes AM, Crane BR 2006. Reconstruction of the chemotaxis receptor-kinase assembly. Nat Struct Mol Biol 13:400–407 [DOI] [PubMed] [Google Scholar]
- Pogliano J 2008. The bacterial cytoskeleton. Curr Opin Cell Biol 20:19–27 [DOI] [PubMed] [Google Scholar]
- Pruggnaller S, Mayr M, Frangakis A 2008. A visualization and segmentation toolbox for electron microscopy. J Struct Biol 164:161–165 [DOI] [PubMed] [Google Scholar]
- Salje J, Zuber B, Löwe J 2008. Electron Cryomicroscopy of E. coli Reveals Filament Bundles Involved in Plasmid DNA Segregation. Science 323:509–512 [DOI] [PubMed] [Google Scholar]
- Sartori A, Gatz R, Beck F, Rigort A, Baumeister W, Plitzko JM 2007. Correlative microscopy: Bridging the gap between fluorescence light microscopy and cryo-electron tomography. J Struct Biol 160:135–145 [DOI] [PubMed] [Google Scholar]
- Scheffel A, Gruska M, Faivre D, Linaroudis A, Plitzko JM, Schüler D 2006. An acidic protein aligns magnetosomes along a filamentous structure in magnetotactic bacteria. Nature 440:110–114 [DOI] [PubMed] [Google Scholar]
- Schmid MF, Booth CR 2008. Methods for aligning and for averaging 3D volumes with missing data. J Struct Biol 161:243–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid MF, Paredes AM, Khant HA, Soyer F, Aldrich HC, Chiu W, Shively JM 2006. Structure of Halothiobacillus neapolitanus carboxysomes by cryo-electron tomography. J Mol Biol 364:526–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz CL, Sarbash VI, Ataullakhanov FI, McIntosh JR, Nicastro D 2007. Cryo-fluorescence microscopy facilitates correlations between light and cryo-electron microscopy and reduces the rate of photobleaching. J Microsc 227:98–109 [DOI] [PubMed] [Google Scholar]
- Seybert A, Herrmann R, Frangakis A 2006. Structural analysis of Mycoplasma pneumoniae by cryo-electron tomography. J Struct Biol 156:342–354 [DOI] [PubMed] [Google Scholar]
- Shimizu TS, Le Novère N, Levin MD, Beavil AJ, Sutton BJ, Bray D 2000. Molecular model of a lattice of signalling proteins involved in bacterial chemotaxis. Nat Cell Biol 2:792–796 [DOI] [PubMed] [Google Scholar]
- Stricker J, Maddox P, Salmon ED, Erickson HP 2002. Rapid assembly dynamics of the Escherichia coli FtsZ-ring demonstrated by fluorescence recovery after photobleaching. Proc Natl Acad Sci U S A 99:3171–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studdert CA, Parkinson JS 2005. Insights into the organization and dynamics of bacterial chemoreceptor clusters through in vivo crosslinking studies. Proc Natl Acad Sci 102:15623–15628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam S 2005. Bridging the imaging gap: Visualizing subcellular architecture with electron tomography. Curr Opin Struct Biol 8:316–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramaniam S, Milne JL 2004. Three-dimensional electron microscopy at molecular resolution. Annu Rev Bioph Biom 33:141–155 [DOI] [PubMed] [Google Scholar]
- Suloway C, Shi J, Cheng A, Pulokas J, Carragher B, Potter CS, Zheng SQ, Agard DA, Jensen GJ 2009. Fully automated, sequential tilt-series acquisition with Leginon. J Struct Biol [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting CS, Hsieh C, Sundararaman S, Mannella C, Marko M 2007. Cryo-electron tomography reveals the comparative three-dimensional architecture of Prochlorococcus, a globally important marine cyanobacterium. J Bacteriol 189:4485–4493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tivol WF, Briegel A, Jensen GJ 2008. An improved cryogen for plunge freezing. Microsc Microanal 14:375–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachtenberg S, Dorward LM, Speransky VV, Jaffe H, Andrews SB, Leapman RD 2008. Structure of the cytoskeleton of Spiroplasma melliferum BC3 and its interactions with the cell membrane. J Mol Biol 378:776–787 [DOI] [PubMed] [Google Scholar]
- Tsien RY 1998. The green fluorescent protein. Annu Rev Biochem 67:509–544 [DOI] [PubMed] [Google Scholar]
- Vollmer W, Holtje JV 2004. The architecture of the murein (peptidoglycan) in gram-negative bacteria: Vertical scaffold or horizontal layer(s)? J Bacteriol 186:5978–5987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis RM, Hirai T, Chalah A, Kessel M, Peters PJ, Subramaniam S 2003. Electron microscopic analysis of membrane assemblies formed by the bacterial chemotaxis receptor Tsr. J Bacteriol 185:3636–3643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner JN, Chen EY, Guberman JM, Zippilli AR, Irgon JJ, Gitai Z 2009. Quantitative genome-scale analysis of protein localization in an asymmetric bacterium. Proc Natl Acad Sci 106:7858–7863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolanin PM, Baker MD, Francis NR, Thomas DR, DeRosier DJ, Stock JB 2006. Self-assembly of receptor/signaling complexes in bacterial chemotaxis. Proc Natl Acad Sci 103:14313–14318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright ER, Iancu CV, Tivol WF, Jensen GJ 2006. Observations on the behavior of vitreous ice at ∼82 and ∼12 K. J Struct Biol 153:241–252 [DOI] [PubMed] [Google Scholar]
- Yeates TO, Kerfeld CA, Heinhorst S, Cannon GC, Shively JM 2008. Protein-based organelles in bacteria: Carboxysomes and related microcompartments. Nat Rev Micro 6:681–691 [DOI] [PubMed] [Google Scholar]
- Zhang P, Bos E, Heymann J, Gnaegi H, Kessel M, Peters PJ, Subramaniam S 2004. Direct visualization of receptor arrays in frozen-hydrated sections and plunge-frozen specimens of E. coli engineered to overproduce the chemotaxis receptor Tsr. J Microsc 216:76–83 [DOI] [PubMed] [Google Scholar]
- Zheng QS, Braunfeld MB, Sedat JW, Agard DA 2004. An improved strategy for automated electron microscopic tomography. J Struct Biol 147:91–101 [DOI] [PubMed] [Google Scholar]
- Zheng SQ, Keszthelyi B, Branlund E, Lyle JM, Braunfeld MB, Sedat JW, Agard DA 2007. UCSF tomography: An integrated software suite for real-time electron microscopic tomographic data collection, alignment, and reconstruction. J Struct Biol 157:138–147 [DOI] [PubMed] [Google Scholar]
- Zuber B, Chami M, Houssin C, Dubochet J, Griffiths G, Daffe M 2008. Direct visualization of the outer membrane of mycobacteria and corynebacteria in their native state. J Bacteriol 190:5672–5680 [DOI] [PMC free article] [PubMed] [Google Scholar]