Abstract
Evenly spaced nucleosomes directly correlate with condensed chromatin and gene silencing. The ATP-dependent chromatin assembly factor (ACF) forms such structures in vitro and is required for silencing in vivo. ACF generates and maintains nucleosome spacing by constantly moving a nucleosome towards the longer flanking DNA faster than the shorter flanking DNA. But how the enzyme rapidly moves back and forth between both sides of a nucleosome to accomplish bidirectional movement is unknown. We show that nucleosome movement depends cooperatively on two ACF molecules, suggesting that ACF functions as a dimer of ATPases. Further, the nucleotide state determines whether the dimer closely engages one vs. both sides of the nucleosome. Three-dimensional reconstruction by single particle electron microscopy of the ATPase-nucleosome complex in an activated ATP state reveals a dimer architecture in which the two ATPases face each other. Our results suggest a model in which the two ATPases work in a coordinated manner, taking turns to engage either side of a nucleosome, thereby allowing processive bidirectional movement. This novel dimeric motor mechanism differs from that of dimeric motors such as kinesin and dimeric helicases that processively translocate unidirectionally and reflects the unique challenges faced by motors that move nucleosomes.
Chromatin-remodeling motors play essential roles in organizing the chromatin state for regulating eukaryotic genomes, yet how they carry out their myriad activities is poorly understood. Their substrate, the nucleosome, contains 147 bp of DNA wrapped in ~1.5 turns around an octamer of histone proteins. Even the smallest movement of the histone octamer relative to the DNA presumably requires a coordinated process of breaking and reforming the many histone-DNA contacts. The ACF chromatin-remodeling complex exemplifies the task, as it is able to move nucleosomes to create evenly spaced nucleosomal arrays that contain equal DNA on either side of each nucleosome1-10. These evenly spaced arrays are important for packaging the underlying DNA into silent chromatin structures in vivo1-10.
ACF is part of the ISWI family of remodeling complexes. The ATPase subunits of ISWI complexes can move nucleosomes by themselves while the accessory subunits modulate this basic activity11-15. The human ACF complex consists of one ATPase subunit, SNF2h and one accessory subunit, Acf16,7. SNF2h is part of the SF2 family of DExx box proteins that includes helicases and nucleic acid translocases16. The ATPase domain of SNF2h has two RecA-like domains, which are thought to form a cleft within which ATP binds. SNF2h also has an alpha-helical extension comprised of three additional domains, HAND, SANT and SLIDE which are thought to play a role in binding flanking DNA17,18. We showed previously that ACF generates a dynamic equilibrium in which nucleosomes with equal flanking DNA on either side accumulate8. Our data implied that ACF achieves the dynamic equilibrium by constantly sampling either side of the nucleosome. This sampling mechanism raised the question of how ACF efficiently switches back and forth between both sides of a nucleosome. We hypothesized that understanding how the ATP state affects interactions of the enzyme with the nucleosome would provide insight into the sampling process.
Previous work has shown that ISWI enzymes require the H4 N-terminal tail for maximal activity19-23. The role of the H4 tail is not known, but it has been hypothesized that an acidic patch on the ATPase domain of ISWI enzymes may interact with the basic patch, K16R17H18R19, on the H4 tail17,19,20,21,24,25. These previous observations imply that the ATPase subunit contacts the H4 tail and that the contacts may change during the ATPase cycle. We therefore used changes in the mobility of the H4 tail as a handle to follow how changes in the nucleotide state alter interactions between SNF2h and the nucleosome. We used electron paramagnetic resonance (EPR) spectroscopy for these studies26. We covalently attached a maleimide spin label to a cysteine introduced in place of an alanine at position 15 on the H4 tail (A15C-MSL, Supplementary Fig.1b), which is directly adjacent to the basic patch. Thermal fluctuations cause a spin label attached to a protein to undergo motion in a spatial region defined by the adjacent protein surface. The resulting EPR spectrum is a highly sensitive measure of the region accessible to the probe. Conformational changes can thus be detected via changes in probe mobility, and these are monitored as changes in the EPR spectrum. EPR can also resolve and quantify multiple states and is particularly powerful in monitoring transitions between unstructured and structured regions of proteins. The A15C-MSL nucleosomes were assembled using an asymmetric DNA template comprising the 601 positioning sequence with 60 bp of flanking DNA on one side (0-601-60, Fig. 1a and Supplementary Fig. 1a)27. The presence of the probe did not alter the maximal rate of nucleosome remodeling by SNF2h (data not shown).
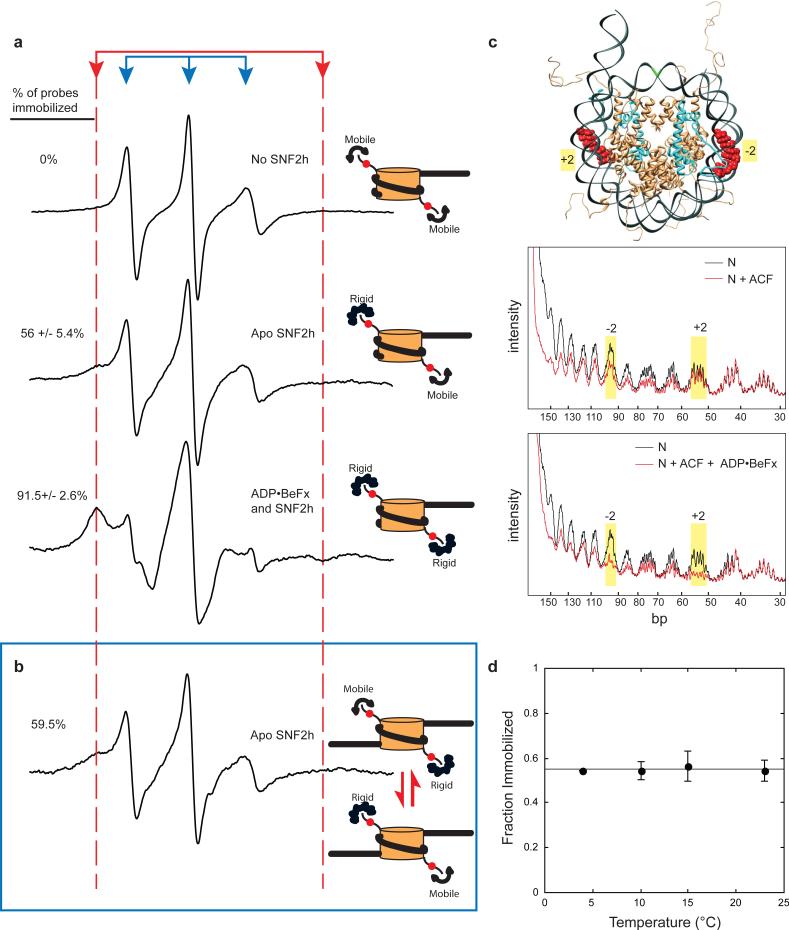
Figure 1.
ATP state regulates immobilization of the histone H4 tail and proximal interactions. (a) Left panels: EPR spectra of MSL labeled 0-601-60 nucleosomes. Right panels: Schematic interpretation of EPR spectra, based on data from (a), (c), & (d). Binding of Apo SNF2h to the nucleosome decreases the mobility of half the H4 tails. SNF2h binding in the presence of ADP•BeFx decreases the mobility of both H4 tails. (b) EPR spectrum of Apo SNF2h bound to spin-labeled 60-601-60 nucleosomes reveals that only one of the two H4 tails is immobilized. (c) Hydroxyl radical foot-printing of ACF on 0-601-60 nucleosomes. Top panel: schematic of mononucleosome structure with 12 bp of flanking DNA on one side, with dyad in green, histone H4 in blue, and the region surrounding SHL (-2) and (+2) in red. Middle panel: Protection patterns for nucleosomes alone (black line, N) compared to nucleosomes bound by Apo-ACF (red line, N+ACF). Bottom panel: nucleosomes alone (black line, N) compared to nucleosomes bound by ACF in the presence of ADP·BeFx (red line, N+ACF+ ADP·BeFx). Yellow bars highlight protection in the SHL (-2) and (+2) regions. (d) Temperature dependence of probe immobilization in the Apo SNF2h-nucleosome complex. Slope of the straight line = 2.1×10-4 ± 8×10-4 immobilized fraction/°C. Error represents s.e.m.
In the absence of SNF2h, the EPR spectrum of the A15C-MSL probe indicated a highly mobile probe (Fig. 1a, top spectrum). The high mobility of the probe in unbound nucleosomes suggested that the H4 N-terminal tails are largely unstructured. Next, we determined how binding of SNF2h altered the mobility of the H4 tail. When the nucleosomes were saturated with SNF2h in the absence of nucleotide (apo state), the EPR spectrum shows two sets of spectral components as indicated by the arrows (Fig 1a, middle spectrum). The inner spectral components (blue arrows) are indicative of a highly mobile probe whereas the wider set of spectral components (highlighted by the red dashed lines) and the broadening of the central peak are indicative of a second state with more restricted mobility. A given peak height in the left-most immobilized spectral component represents 4.1 times more spins than the same peak height for the mobile component. Deconvolution of the spectra indicated that about half of the H4 tails were in each of the two states (56±5.4% in the immobilized state, see Supplementary Fig. 2 for fitting and quantification method)28. In the presence of ADP, the immobilized subpopulation also constituted half of the probes (spectra not shown). Our attempts to trap the SNF2h-nucleosome complex in the ATP state using ATP analogs were unsuccessful as these analogs either supported low levels of remodeling (AMP-PNP, ATPγS) or did not bind with detectable affinity (AMP-PCP). We were however able to mimic an activated ATP state using the analog, ADP•BeFx. In contrast to the data in the apo state and with ADP, almost all of the probe on the H4 tail became immobilized in the presence of SNF2h and ADP•BeFx (91.5±2.6% of probe in the immobilized peak). This change is shown by the increase in spectral intensity of the immobilized component (left-most peak, Fig. 1a, bottom spectrum). This dramatic increase in the amount of probes immobilized indicated that both H4 tails were bound by SNF2h in the presence of ADP•BeFx. Together, these data indicate that SNF2h induces nucleotide-dependent changes in the H4 tail conformation such that in the apo and ADP states, half the H4 tails are immobilized and in an activated ATP state mimicked by ADP•BeFx, all H4 tails are bound.
The EPR data raised two possibilities for how SNF2h binds the nucleosome in the apo and ADP states: (a) SNF2h symmetrically binds both H4 tails and each H4 tail exists in a two-state equilibrium between mobile and immobile states (with an equilibrium constant of 1) or, (b) SNF2h asymmetrically binds only one of the two H4 tails. For model (a), we expect immobilization to increase upon lowering temperature as the highly mobile state is entropically favored whereas the structured immobile state is enthalpically favored, as seen for docking of the kinesin neck linker29. The fraction of H4 tails immobilized was unchanged, within error, from 23°C to 4°C (Fig. 1d, 54.6% immobilized at 4°C and 23°C). A van't Hoff plot of the equilibrium constant for H4 tail mobility as a function of temperature yields a ΔH of 0.76 kJ/mol, and ΔS of 4.4×10-3 kJ/mol-K (Supplementary Fig. 3), values that are substantially smaller than the favorable ΔH of 50 kJ/mol and unfavorable ΔS of 0.17 kJ/mol-K for docking of the kinesin neck linker29. These data rule out model (a) and provide strong support for the asymmetric binding of model (b). To further test model (b) we used hydroxyl radical footprinting of the same nucleosome construct to follow changes in ACF contacts as a function of nucleotide state (Fig. 1c). In the apo state, ACF binding induces asymmetric protection of nucleosomal DNA: protection is observed in the SHL(−2) region, but not the SHL(+2) region, consistent with other ISWI complexes and with model (b)9,30. In contrast, in the ADP•BeFx state, ACF binding results in significant protection in both SHL(−2) and SHL(+2) regions, consistent with the EPR data.
The asymmetry with respect to H4 tail binding observed in the apo state could arise either (a) due to the presence of asymmetric flanking DNA or (b) due to structural constraints placed by the apo state. To distinguish between these possibilities we repeated the EPR experiment using nucleosomes with 60bp of flanking DNA on both sides (60-601-60 template). Apo-SNF2h still bound only one of the two H4 tails in the context of this symmetric nucleosome (Fig. 1b). These data strongly support a model in which apo-SNF2h can only bind one H4 tail at a time, and the availability of flanking DNA biases which side of the nucleosome the enzyme binds preferentially. Together the above data suggest that the enzyme switches between an asymmetric conformation where it interacts with one H4 tail at a time in the apo state and a more symmetric conformation where it binds both H4 tails in the ADP•BeFx state.
The observation that SNF2h binds both H4 tails in the presence of ADP•BeFx suggests that either (a) one SNF2h molecule bridges both H4 tails or, (b) SNF2h binds as a dimer such that each ATPase contacts an H4 tail. To distinguish between these models we first investigated the oligomeric state of SNF2h alone. Using equilibrium analytical ultra-centrifugation we found that unbound SNF2h is a monomer (data not shown). Because several well-studied dimeric helicases dimerize upon binding their DNA substrates, we next determined if SNF2h dimerizes on nucleosomes31. If dimerization of SNF2h is tightly coupled to nucleosome binding we expected to see cooperative SNF2h binding. We measured the binding to nucleosomes by taking advantage of our observation that the fluorescence of a Cy3 dye attached near the entry site of the DNA increases upon SNF2h binding (Fig. 2a). We find that in the apo state, SNF2h binds to the nucleosome cooperatively, consistent with previous observations of cooperative binding by the Drosophila ISWI protein32. The Hill Coefficient of 1.8 suggests that at least two molecules of SNF2h bind in a manner such that binding of one molecule is strongly coupled to binding of the second (Fig. 2b). The EPR data from Figure 1 and the Hill Coefficient of 1.8, together suggest that in the apo state, SNF2h binds as a dimer but only one of the two SNF2h molecules engages an H4 tail.
Figure 2.
SNF2h and ACF function as dimers of ATPases. (a) Schematic of nucleosome structure with dye attachment sites for (b) and (d). The DNA is end-labeled with Cy3 (blue) on the shorter flanking DNA. The octamer is labeled with Cy5 at H2A-120C (yellow). (b) Cy3 fluorescence intensity of the construct shown in (a) as a function of SNF2h concentration, using nucleosomes with 78bp of flanking DNA on one side. A representative replicate curve is shown. Data are fit to the general equation for cooperative binding (see Methods). Hill Coefficient (n) = 1.8 ±0.17; K1/2 = 353±30nM. (c) Schematic of FRET-based nucleosome remodeling assay. Rate constant of remodeling is measured by following the decrease in FRET between Cy3 and Cy5 in the presence of ATP. (d) Left panel: Nucleosome remodeling rate constant as a function of SNF2h concentration for nucleosomes with 78bp of flanking DNA. Right panel: Nucleosome remodeling rate constant as a function of ACF concentration for nucleosomes with 20 bp of flanking DNA. Hill Coefficient (n) = 1.8±0.1; K’1/2 = 281±32 nM for SNF2h and Hill Coefficient (n) = 1.9±.3; K’1/2 = 26±3 nM for ACF. Each panel represents global fits to data obtained from three independent experiments. (e) SNF2h binds as a cooperative dimer to the nucleosome in the absence of nucleotide (black circles), and in the presence of ADP (blue squares). In the presence of ADP•BeFx, SNF2h binds non-cooperatively (red triangles). These binding measurements were carried out with nucleosomes containing 40bp of flanking DNA on one side and a Cy3 label on the short DNA end. Binding of SNF2h to these nucleosomes is ~2-fold weaker relative to the nucleosomes used in (b)9,46. A representative replicate curve with each nucleotide analogue is shown (left panel), and the average K1/2 and Hill Coefficient from three replicates is shown (table). Errors represent s.e.m.
To determine whether two SNF2h molecules were necessary to mediate maximal nucleosome remodeling, we measured the dependence of chromatin remodeling activity on SNF2h concentration using a FRET-based method (Fig. 2c). The rate constant of remodeling also depends cooperatively on SNF2h concentration with a Hill Coefficient of 1.8 (Fig. 2d, left panel). We next determined if the entire ACF complex also functions most effectively as a dimer. We analogously saw a cooperative dependence of the remodeling rate constant on ACF concentration with a Hill Coefficient of 1.9 (Fig. 2d, right panel). Together, these data strongly suggest that the predominant functional form of ACF is a dimer of ATPases.
A hallmark of dimeric motors such as kinesin and the E. coli Rep helicase is that they cycle between states in which one motor subunit is engaged with the substrate and states in which both motor subunits are transiently engaged31. By working in coordinated pairs, one motor subunit can serve as an anchor to the other moving motor to prevent dissociation from the substrate. The ATP state helps regulate the affinity of the motor for the substrate. Our EPR results suggest that a SNF2h dimer analogously cycles between at least two states, one in which only one ATPase engages an H4 tail and another in which both ATPases engage the two H4 tails. To determine if these different conformational states reflect states with different affinities, we measured the affinity of SNF2h for the nucleosome in different ATP states (Fig. 2e). In the presence of ADP, SNF2h bound cooperatively but with slightly weaker affinity than in the apo state. In both the apo and ADP states, the high cooperativity of binding indicates that binding of one SNF2h molecule by itself is very weak and requires the presence of another SNF2h molecule to increase its overall affinity. The cooperativity could arise either from direct SNF2h-SNF2h contacts or could be mediated through a conformational change in the nucleosome without direct SNF2h-SNF2h contacts. In the presence of ADP•BeFx, the K1/2 for SNF2h binding is ~3-fold lower than that in the apo state indicating a stronger binding affinity. Further, SNF2h binding in the presence of ADP•BeFx is not cooperative (Hill Coefficient =1). These data suggest that in presence of ADP•BeFx the affinity of each SNF2h molecule is sufficiently high such that binding of one SNF2h molecule is no longer highly dependent on the presence of the other.
Our finding that two ATPases are required for maximal nucleosome remodeling raises the question of how the nucleosome structure accommodates two SNF2h molecules. Other dimeric motors, such as kinesin and helicases, are oriented such that each ATPase subunit can take turns translocating on the polymeric substrate in the same direction31. We used negative stain electron microscopy to visualize the complex of SNF2h with nucleosomes in the presence of ADP•BeFx (Fig. 3 and Supplementary Fig. 4 and Supplemental Methods). Nucleosomes with 60bp of flanking DNA (0-601-60) were incubated with SNF2h concentrations comparable to the K1/2 for nucleosomes in the presence of ADP•BeFx, adsorbed to a glow discharged carbon film, and negatively stained with uranyl formate. The specimen was imaged at tilt angles of 60° and 0° (Supplementary Fig. 5). A total of 10,059 pairs of particles were interactively selected from 100 image pairs. Classification of particles from images of untilted specimen shows three distinct classes (Fig. 3a, b) that can be clearly recognized as a nucleosome by itself and a nucleosome with either one or two SNF2h molecules bound. On average ~70% of the complexes contained two SNF2h molecules bound. The singly bound SNF2h molecules could reflect the use of non-saturating SNF2h, a technical necessity to prevent particle crowding on the grid. In the complexes with two SNF2h molecules bound, the flanking DNA was not clearly visible in the two-dimensional (2D) class averages, possibly because the flanking DNA is flexible and gets averaged out. An alternative possibility is that in most of the complexes, the flanking DNA is rearranged due to interaction with a domain of SNF2h. Consistent with this possibility we do not clearly observe the extended HAND-SANT-SLIDE domain that has been shown to interact with flanking DNA in the apo state17. We hypothesize that there may be a conformational rearrangement of the HAND-SANT-SLIDE domain in the presence of ADP•BeFx. Further, no large region of direct contact between the two SNF2h molecules is apparent, consistent with the Hill Coefficient of 1.0 in this state (Fig. 2e).
Figure 3.
Visualization of SNF2h bound to the nucleosome in the presence of ADP•BeFx using EM. (a) three different views of the 3-D reconstruction of dimeric SNF2h bound to the nucleosome (left panels) and corresponding representative 2-D class averages (right panels). The crystal structure of the core mononucleosome was placed manually into the 3D reconstruction. Histone H4 is highlighted in red. The isosurface of the 3-D reconstruction at high threshold is shown in blue, and low threshold in grey. (b) Left panel: representative 2-D class average of negative stain EM images of unbound nucleosomes. Right panel: representative 2-D class average of one SNF2h bound to a nucleosomes. (c) Representative 2-D class averages of SNF2h alone. Numbers used to calculate a particular class average shown in lower left corner.
In the 2D class averages, the region of each SNF2h monomer that interacts with the nucleosome appears to contain two globular lobes (Fig. 3a). These lobes may represent the two RecA-related ATP binding folds observed in SF2 family motors33,34. The two lobes are also apparent in the 2D class averages of SNF2h alone (Fig. 3c and Supplementary Fig. 6). Three-dimensional (3D) reconstructions of the nucleosome with two (or one) SNF2h bound were calculated using the well-established random conical tilt approach to a resolution of ~ 27 Å without the explicit application of any two-fold symmetry (Fig. 3a and Supplementary Figs 5 and 7)35. The two SNF2h molecules face each other on the nucleosome, and seem to obey the 2-fold symmetry of the nucleosome with one putative ATPase domain at SHL(+2) and one at SHL(−2) (Fig. 3a, 2-fold or opposing symmetry most apparent in the middle panel). Consistent with the EPR and footprinting data in Figure 1, each SNF2h monomer seems to directly contact one H4 N-terminal tail and the SHL(−2)/(+2) regions in the ADP•BeFx state. While the overall architecture appears almost symmetric, given the 27 Å resolution, any local structural asymmetries that may exist between the two SNF2h molecules cannot be resolved.
The above architecture raises the question of how the dimeric partners cooperate rather than compete in a “tug of war.” Our findings suggest an “alternating action” model schematized in Figure 4. In this model, each ATPase takes turns in engaging the flanking DNA on either side and the corresponding H4 tail at SHL(−2) or (+2)9,17. An ability of the two ATPases to take turns, as suggested by our observation that the apo state of the enzyme engages only one H4 tail at a time, would help avoid a “tug of war” situation. The ATPase that engages the longer DNA hydrolyzes ATP faster, as previously shown8,36. This ATPase becomes the leading ATPase and sets the direction of nucleosome movement by translocating on nucleosomal DNA37,38. The leading ATPase generates a DNA loop/wave that can propagate across the histone octamer as suggested previously39-41. The second, subordinate ATPase could then further act as another anchor to stabilize the intermediate while the leading ATPase is translocating (Fig. 4, mimicked by ADP•BeFx). Interaction with the second H4 tail may help in the binding of the subordinate ATPase. In the simplest version of this model, the subordinate ATPase does not bind or hydrolyze ATP once the leading ATPase fires42. This division of labor between identical subunits is analogous to hexameric helicases where occupancy of one ATPase subunit regulates the affinity of an adjacent subunit for nucleotide43. Whether the communication between the two ATPases is direct or through the nucleosome remains an important future question. A variation of this model in which the non-leading ATPase also hydrolyzes ATP is described in Supplementary Figure 8. Successive rounds of sampling and translocation would then equalize the DNA on either side of a nucleosome. A dimer-based mechanism is also indicated by single-molecule data showing that dimeric ACF complexes can switch the direction of nucleosome translocation several times without dissociation (supporting manuscript by Blosser et al.). Our results help explain the significance of previous observations that two Drosophila ACF molecules can bind in the context of DNA and provide a mechanistic explanation for the processive action of ISWI complexes3,44,45.
Figure 4.
Simple model for how a dimeric ACF moves nucleosomes. H4 tail is in red and the two ATPases in ACF are shown in blue and purple. Only one subunit binds ATP at a time. In the ATP state, each ATPase subunit takes turns in binding the flanking DNA. The ATPase that binds the longer flanking DNA (purple) hydrolyzes ATP faster and starts translocating DNA across the nucleosome. During and post hydrolysis, the second ATPase (blue) also engages the nucleosome, preventing loss of the DNA loop-containing intermediate (mimicked by ADP•BeFx). In the ADP state, the non-translocating monomer disengages and the translocating monomer remains engaged with the nucleosome. ATP state may also regulate the extent of any direct contacts between the two ATPase subunits and such contacts may be substantially fewer in ADP-BeFx state than in other ATP states.
We hypothesize that in contrast to kinesin and dimeric helicases, whose biological functions require unidirectional translocation along a largely uniform polymeric substrate, the biological functions of chromatin remodeling enzymes like ACF place very different demands on motor architecture. The opposing architecture of the two motors in ACF may enable ACF to rapidly and processively change the direction of nucleosome movement in order to achieve a defined spacing. It will be interesting to investigate whether bidirectional movement via dimerization is a general feature of enzymes that space nucleosomes, and whether remodeling enzymes with other activities use different strategies.
Methods Summary.
EPR measurements were performed with an EMX EPR spectrometer from Bruker Instruments (Billerica, MA). First derivative, X-band spectra were recorded in a high-sensitivity microwave cavity using 50-s, 100-Gauss wide magnetic field sweeps. EM samples were adsorbed to a glow-discharged copper grid coated with carbon film for 30 seconds followed by conventional negative stain with 0.75% uranyl formate. Images were collected using a Tecnai T12 microscope (FEI company, Hillsboro, OR) and recorded at a magnification of 52,000X with an UltraScan 4096 × 4096 pixel CCD camera (Gatan Inc, USA). Full methods are described in Supplementary Methods.
Supplementary Material
Acknowledgements
We thank J. Widom (Northwestern University) for the 601 plasmid. We thank H. Madhani, M.D. Simon, and members of the Narlikar Lab for helpful discussion and comments on the manuscript. We thank R. Howard for help with AUC; W. Ross, J. Lin, S. Hota and B. Bartholomew for advice on footprinting; and C. Cunningham for assistance with nucleosome depiction. This work was supported by grants from the Sandler Family Supporting Foundation (Sandler Opportunity Award and New Technology Award in Basic Science to Y.C, Program for Breakthrough Biomedical Research (PBBR) Award to G.J.N.), UCSF Academic Senate Shared Equipment Grant (to Y.C), grants from the National Institutes of Health (to R.C. and G.J.N). P.P. and J.G.Y. were supported by US National Science Foundation Graduate Research Fellowships. G.J.N is a Leukemia and Lymphoma Society Scholar.
Footnotes
This paper is dedicated to Daniel Herschlag in honor of his 51st birthday.
Supplementary Information accompanies the paper on www.nature.com/nature.
REFERENCES
- 1.Corona DF, et al. ISWI regulates higher-order chromatin structure and histone H1 assembly in vivo. PLoS Biol. 2007;5:e232. doi: 10.1371/journal.pbio.0050232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fyodorov DV, Blower MD, Karpen GH, Kadonaga JT. Acf1 confers unique activities to ACF/CHRAC and promotes the formation rather than disruption of chromatin in vivo. Genes Dev. 2004;18:170–183. doi: 10.1101/gad.1139604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ito T, Bulger M, Pazin MJ, Kobayashi R, Kadonaga JT. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- 4.Varga-Weisz PD, et al. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature. 1997;388:598–602. doi: 10.1038/41587. [DOI] [PubMed] [Google Scholar]
- 5.Deuring R, et al. The ISWI chromatin-remodeling protein is required for gene expression and the maintenance of higher order chromatin structure in vivo. Mol Cell. 2000;5:355–365. doi: 10.1016/s1097-2765(00)80430-x. [DOI] [PubMed] [Google Scholar]
- 6.Poot RA, et al. HuCHRAC, a human ISWI chromatin remodelling complex contains hACF1 and two novel histone-fold proteins. Embo J. 2000;19:3377–3387. doi: 10.1093/emboj/19.13.3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bochar DA, et al. A family of chromatin remodeling factors related to Williams syndrome transcription factor. Proc Natl Acad Sci U S A. 2000;97:1038–1043. doi: 10.1073/pnas.97.3.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang JG, Madrid TS, Sevastopoulos E, Narlikar GJ. The chromatin-remodeling enzyme ACF is an ATP-dependent DNA length sensor that regulates nucleosome spacing. Nat Struct Mol Biol. 2006;13:1078–1083. doi: 10.1038/nsmb1170. [DOI] [PubMed] [Google Scholar]
- 9.Kagalwala MN, Glaus BJ, Dang W, Zofall M, Bartholomew B. Topography of the ISW2-nucleosome complex: insights into nucleosome spacing and chromatin remodeling. Embo J. 2004;23:2092–2104. doi: 10.1038/sj.emboj.7600220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun FL, Cuaycong MH, Elgin SC. Long-range nucleosome ordering is associated with gene silencing in Drosophila melanogaster pericentric heterochromatin. Mol Cell Biol. 2001;21:2867–2879. doi: 10.1128/MCB.21.8.2867-2879.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Corona DF, et al. ISWI is an ATP-dependent nucleosome remodeling factor. Mol Cell. 1999;3:239–245. doi: 10.1016/s1097-2765(00)80314-7. [DOI] [PubMed] [Google Scholar]
- 12.Aalfs JD, Narlikar GJ, Kingston RE. Functional differences between the human ATP-dependent nucleosome remodeling proteins BRG1 and SNF2H. J Biol Chem. 2001;276:34270–34278. doi: 10.1074/jbc.M104163200. [DOI] [PubMed] [Google Scholar]
- 13.Eberharter A, et al. Acf1, the largest subunit of CHRAC, regulates ISWI-induced nucleosome remodelling. Embo J. 2001;20:3781–3788. doi: 10.1093/emboj/20.14.3781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Langst G, Bonte EJ, Corona DF, Becker PB. Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell. 1999;97:843–852. doi: 10.1016/s0092-8674(00)80797-7. [DOI] [PubMed] [Google Scholar]
- 15.Ito T, et al. ACF consists of two subunits, Acf1 and ISWI, that function cooperatively in the ATP-dependent catalysis of chromatin assembly. Genes Dev. 1999;13:1529–1539. doi: 10.1101/gad.13.12.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Durr H, Flaus A, Owen-Hughes T, Hopfner KP. Snf2 family ATPases and DExx box helicases: differences and unifying concepts from high-resolution crystal structures. Nucleic Acids Res. 2006;34:4160–4167. doi: 10.1093/nar/gkl540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dang W, Bartholomew B. Domain architecture of the catalytic subunit in the ISW2-nucleosome complex. Mol Cell Biol. 2007;27:8306–8317. doi: 10.1128/MCB.01351-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grune T, et al. Crystal structure and functional analysis of a nucleosome recognition module of the remodeling factor ISWI. Mol Cell. 2003;12:449–460. doi: 10.1016/s1097-2765(03)00273-9. [DOI] [PubMed] [Google Scholar]
- 19.Clapier CR, Langst G, Corona DF, Becker PB, Nightingale KP. Critical role for the histone H4 N terminus in nucleosome remodeling by ISWI. Mol Cell Biol. 2001;21:875–883. doi: 10.1128/MCB.21.3.875-883.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Clapier CR, Nightingale KP, Becker PB. A critical epitope for substrate recognition by the nucleosome remodeling ATPase ISWI. Nucleic Acids Res. 2002;30:649–655. doi: 10.1093/nar/30.3.649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hamiche A, Kang JG, Dennis C, Xiao H, Wu C. Histone tails modulate nucleosome mobility and regulate ATP-dependent nucleosome sliding by NURF. Proc Natl Acad Sci U S A. 2001;98:14316–14321. doi: 10.1073/pnas.251421398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fazzio TG, Gelbart ME, Tsukiyama T. Two distinct mechanisms of chromatin interaction by the Isw2 chromatin remodeling complex in vivo. Mol Cell Biol. 2005;25:9165–9174. doi: 10.1128/MCB.25.21.9165-9174.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferreira H, Flaus A, Owen-Hughes T. Histone modifications influence the action of Snf2 family remodelling enzymes by different mechanisms. J Mol Biol. 2007;374:563–579. doi: 10.1016/j.jmb.2007.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zofall M, Persinger J, Bartholomew B. Functional role of extranucleosomal DNA and the entry site of the nucleosome in chromatin remodeling by ISW2. Mol Cell Biol. 2004;24:10047–10057. doi: 10.1128/MCB.24.22.10047-10057.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langst G, Becker PB. ISWI induces nucleosome sliding on nicked DNA. Mol Cell. 2001;8:1085–1092. doi: 10.1016/s1097-2765(01)00397-5. [DOI] [PubMed] [Google Scholar]
- 26.Rice S, et al. A structural change in the kinesin motor protein that drives motility. Nature. 1999;402:778–784. doi: 10.1038/45483. [DOI] [PubMed] [Google Scholar]
- 27.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 28.Naber N, Purcell TJ, Pate E, Cooke R. Dynamics of the nucleotide pocket of myosin measured by spin-labeled nucleotides. Biophys J. 2007;92:172–184. doi: 10.1529/biophysj.106.090035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rice S, et al. Thermodynamic properties of the kinesin neck-region docking to the catalytic core. Biophys J. 2003;84:1844–1854. doi: 10.1016/S0006-3495(03)74992-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schwanbeck R, Xiao H, Wu C. Spatial contacts and nucleosome step movements induced by the NURF chromatin remodeling complex. J Biol Chem. 2004;279:39933–39941. doi: 10.1074/jbc.M406060200. [DOI] [PubMed] [Google Scholar]
- 31.Lohman TM, Thorn K, Vale RD. Staying on track: common features of DNA helicases and microtubule motors. Cell. 1998;93:9–12. doi: 10.1016/s0092-8674(00)81139-3. [DOI] [PubMed] [Google Scholar]
- 32.Chin J, Langst G, Becker PB, Widom J. Fluorescence anisotropy assays for analysis of ISWI-DNA and ISWI-nucleosome interactions. Methods Enzymol. 2004;376:3–16. doi: 10.1016/S0076-6879(03)76001-7. [DOI] [PubMed] [Google Scholar]
- 33.Durr H, Korner C, Muller M, Hickmann V, Hopfner KP. X-ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA. Cell. 2005;121:363–373. doi: 10.1016/j.cell.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 34.Thoma NH, et al. Structure of the SWI2/SNF2 chromatin-remodeling domain of eukaryotic Rad54. Nat Struct Mol Biol. 2005;12:350–356. doi: 10.1038/nsmb919. [DOI] [PubMed] [Google Scholar]
- 35.Radermacher M, et al. Cryo-electron microscopy and three-dimensional reconstruction of the calcium release channel/ryanodine receptor from skeletal muscle. J Cell Biol. 1994;127:411–423. doi: 10.1083/jcb.127.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stockdale C, Flaus A, Ferreira H, Owen-Hughes T. Analysis of nucleosome repositioning by yeast ISWI and Chd1 chromatin remodeling complexes. J Biol Chem. 2006;281:16279–16288. doi: 10.1074/jbc.M600682200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zofall M, Persinger J, Kassabov SR, Bartholomew B. Chromatin remodeling by ISW2 and SWI/SNF requires DNA translocation inside the nucleosome. Nat Struct Mol Biol. 2006;13:339–346. doi: 10.1038/nsmb1071. [DOI] [PubMed] [Google Scholar]
- 38.Whitehouse I, Stockdale C, Flaus A, Szczelkun MD, Owen-Hughes T. Evidence for DNA translocation by the ISWI chromatin-remodeling enzyme. Mol Cell Biol. 2003;23:1935–1945. doi: 10.1128/MCB.23.6.1935-1945.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cairns BR. Chromatin remodeling: insights and intrigue from single-molecule studies. Nat Struct Mol Biol. 2007;14:989–996. doi: 10.1038/nsmb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Strohner R, et al. A ‘loop recapture’ mechanism for ACF-dependent nucleosome remodeling. Nat Struct Mol Biol. 2005;12:683–690. doi: 10.1038/nsmb966. [DOI] [PubMed] [Google Scholar]
- 41.Langst G, Becker PB. Nucleosome remodeling: one mechanism, many phenomena? Biochim Biophys Acta. 2004;1677:58–63. doi: 10.1016/j.bbaexp.2003.10.011. [DOI] [PubMed] [Google Scholar]
- 42.Racki L, Narlikar G. ATP-dependent chromatin remodeling enzymes: two heads are not better, just different. Curr Opin Genet Dev. 2008;18:137–144. doi: 10.1016/j.gde.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Enemark EJ, Joshua-Tor L. On helicases and other motor proteins. Curr Opin Struct Biol. 2008;18:243–257. doi: 10.1016/j.sbi.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fyodorov DV, Kadonaga JT. Dynamics of ATP-dependent chromatin assembly by ACF. Nature. 2002;418:897–900. doi: 10.1038/nature00929. [DOI] [PubMed] [Google Scholar]
- 45.Gangaraju V, Prasad P, Srour A, Kagalwala M, Bartholomew B. Conformational changes associated with template commitment in ATP-dependent chromatin remodeling by ISW2. Mol Cell. 2009;35:58–69. doi: 10.1016/j.molcel.2009.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.He X, Fan HY, Narlikar GJ, Kingston RE. Human ACF1 alters the remodeling strategy of SNF2h. J Biol Chem. 2006;281:28636–28647. doi: 10.1074/jbc.M603008200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.