Abstract
Context: Generalized glucocorticoid resistance syndrome is a rare familial or sporadic condition characterized by partial insensitivity to glucocorticoids, caused by mutations in the glucocorticoid receptor (GR) gene. Most of the reported cases are adults, demonstrating symptoms associated with mineralocorticoid and/or adrenal androgen excess caused by compensatively increased secretion of the adrenocorticotropic hormone.
Patient: We identified a new 2-yr-old female case of generalized glucocorticoid resistance syndrome. The patient (TJ) presented with a generalized seizure associated with hypoglycemia and hypokalemia. She also had hypertension and premature pubarche, whereas dexamethasone effectively suppressed these clinical manifestations.
Results: The patient’s GR gene had a heterozygotic mutation (G→A) at nucleotide position 2141 (exon 8), which resulted in substitution of arginine by glutamine at amino acid position 714 in the ligand-binding domain (LBD) of the GRα. Molecular analysis revealed that the mutant receptor had significantly impaired transactivation activity with a 2-fold reduction in affinity to ligand. It showed attenuated transactivation of the activation function (AF)-2 and reduced binding to a p160 nuclear receptor coactivator. Computer-based structural analysis revealed that replacement of arginine by glutamine at position 714 transmitted a conformational change to the LBD and the AF-2 transactivation surface, resulting in a decreased binding affinity to ligand and to the LXXLL coactivator motif.
Conclusions: Dexamethasone treatment is effective in controlling the premature pubarche, hypoglycemia, hypertension, and hypokalemia in this child case, wherein arginine 714 plays a key role in the proper formation of the ligand-binding pocket and the AF-2 surface of the GRα LBD.
A child developed generalized glucocorticoid resistance and presented with hypoglycemia-associated seizures due to a novel glucocorticoid receptor mutation with glutamine replacing arginine at position 714.
Generalized glucocorticoid resistance syndrome is a rare familial or sporadic condition characterized by partial, end-organ insensitivity to glucocorticoids (1,2,3). The patients have compensatory elevations in circulating levels of cortisol and ACTH, which maintain circadian rhythmicity but show resistance to exogenously introduced glucocorticoids (1). The affected subjects reported are mostly adults and do not demonstrate overt clinical evidence of hypo- or hypercortisolism, although their excess ACTH secretion often results in increased production of adrenal steroids with androgenic and/or mineralocorticoid activity that sometimes develop hirsutism, infertility, hypertension, and hypokalemia in the affected subjects (1,2).
The molecular basis of generalized glucocorticoid resistance syndrome has been ascribed to mutations in the human glucocorticoid receptor (GR) gene. This gene consists of nine exons and encodes human (h) GRα (1,2). hGRα consists of three distinct structural units, the N-terminal, middle DNA-binding, and C-terminal ligand-binding (LBD) domains, and functions as a hormone-activated transcription factor of glucocorticoid target genes (3,4,5). Upon binding to glucocorticoids, hGRα shuttles from the cytoplasm into the nucleus, binds to glucocorticoid response elements (GREs) in the regulatory regions of glucocorticoid-responsive genes (3,4), and initiates transcription of regulated coding sequences by attracting numerous cofactor complexes and molecules (3,6). Among them, p160-type nuclear receptor coactivators physically interact with the transactivation function (AF)-1 and -2 of hGRα, respectively, through their C-terminal portion and the middle, multiple LXXLL coactivator motif-containing nuclear receptor-binding domain (NRB).
In patients with generalized glucocorticoid resistance syndrome, inactivating mutations within the all domains of the hGRα and a 4-bp deletion at the 3′-boundary of exon 6 of the hGR gene have been previously described (1). In this study, we demonstrate a child case of pure generalized glucocorticoid resistance syndrome, which is caused by a novel heterozygous point mutation in the hGR gene. Importantly, the patient’s clinical symptoms have been well maintained with use of dexamethasone in combination with antihypertensive medicines for more than 6 yr. We performed a series of examinations to identify molecular defects of her mutant receptor.
Subjects and Methods
Case reports
The patient (TJ) was born at 35 wk of gestation with intrauterine growth retardation (body weight at birth 1899 g). She developed generalized tonic-clonic seizures followed by unconsciousness occurring several days after emesis and diarrhea at the age of 2 yr 10 months. After being admitted to the hospital, laboratory tests revealed that she had severe hypoglycemia (plasma glucose 18 mg/dl) and hypokalemia (serum potassium 2.6 mmol/liter). Her blood pressure was also elevated (138/90 mm Hg). Endocrinological examination identified elevated levels of serum cortisol (81.2 ng/dl, normal range (nr) 3–21) and plasma ACTH (431 pg/ml, nr 1–21), and strikingly high excretion of urinary free cortisol (646 μg/d, nr 1.4–20). Plasma aldosterone level and plasma renin activity were 4.2 ng/dl (nr 1–21) and less than 0.6 ng/ml·h (nr < 6.0), respectively. She had elevated levels of serum dehydroepiandrosterone (DHEA) (730 ng/dl, nr 50–540) and androstenedione (7.6 ng/ml, nr 0.1–0.3), whereas her urinary 17-ketosteroid secretion was 5.1 μg/d (nr 0.1–3). Mild clitoromegaly was also observed. Her bone age was 8 yr at her chronological age of 3 yr 11 months. Under the clinical diagnosis of generalized glucocorticoid resistance, the patient was treated with dexamethasone, which normalized her blood pressure and serum potassium levels in combination with antihypertensive medicines. Her serum DHEA and DHEA sulfate levels and 24 h urinary free cortisol excretion were successfully suppressed to 540 ng/dl, 54 ng/dl (nr < 92), and 51.8 μg/d under the treatment with 1 and 2.5 mg/d dexamethasone, respectively. Her growth curve (body weight and height) and picture taken at 8 yr of age are shown in Supplemental Fig. 1, A and B, respectively, published as supplemental data on The Endocrine Society’s Journals Online web site at http://jcem.endojournals.org. The patient’s bone age was successfully slowed to 12 yr at her chronological age of 9 yr 11 month under dexamethasone treatment.
Materials and methods
Materials and methods used in this study are described in Supplemental Data.
Results
Identification of a novel heterozygotic point mutation in the patient’s hGR gene
We found a novel heterozygotic point mutation replacing guanine (G) by adenine (A) at nucleotide position 2141 (exon 8) from the translation start site by sequencing the patient’s hGR gene using her genomic DNA (supplemental Fig. 2). The identified mutation resulted in replacement of arginine (R) by glutamine (Q) at amino acid position 714 of the hGRα protein. We confirmed presence of this mutation in the patient’s mRNA.
hGRαR714Q is defective in stimulating a glucocorticoid-responsive promoter
We tested the transcriptional activity of hGRα harboring R to Q replacement at amino acid 714 (R714Q) on the glucocorticoid-responsive mouse mammary tumor virus (MMTV) promoter in hGRα-defective HCT116 cells. hGRαR714Q demonstrated a reduced transcriptional activity on this promoter, shifting mainly the dexamethasone titration curve of the transcriptional activity rightward, changing EC50 from −8.44 ± 0.23 to −6.88 ± 0.31 log M (P < 0.01) (Fig. 1A, top panel). Wild-type hGRα and the hGRαR714Q mutant receptor were expressed similarly from their expression plasmids in HCT116 cells, suggesting that the mutation did not affect stability of both mutated GRα mRNA and protein (Fig. 1A, bottom panel). We also found that hGRαR714Q acted as a dominant-negative receptor for the wild-type GRα on the latter’s transactivation of the glucocorticoid-responsive genes (Supplemental Fig. 3). Hydrocortisone induced significantly reduced hGRαR714Q transcriptional activity of the MMTV promoter, which was equivalent to that by dexamethasone, whereas progesterone and deoxycorticosterone did not activate hGRαR714Q at all (data not shown). These results indicate that the heterozygotic point mutation (R714Q) found in the patient’s hGR gene underlies the glucocorticoid resistance observed in this patient, whereas ligand specificity of the mutant receptor is preserved.
Figure 1.
hGRαR714Q has a reduced transactivation activity, about 2-fold reduction of affinity to dexamethasone, and reduced binding activity to GRIP1 in vitro. A, hGRαR714Q has a reduced transcriptional activity on the MMTV promoter in HCT116 cells. Top panel, Dexamethasone titration curves of the luciferase activity expressed from the MMTV promoter-driven reporter gene are shown. HCT116 cells were transfected with the plasmids expressing hGRα wild-type (WT) (open circles), hGRαR714Q mutant (closed circles), or a control gene (open squares) together with pMMTV-luc and pGL4.73[hRluc/SV40], and were treated with indicated concentrations of dexamethasone. Circles represent mean ± se values of the firefly luciferase activity normalized for the renilla luciferase activity in the indicated concentrations of dexamethasone. The experiment was repeated three times and representative data are shown. Bottom panel, Wild-type hGRα and hGRαR714Q are similarly expressed in HCT116 cells. Samples from HCT116 cells transfected with indicated GR-expressing plasmids were run on gels, and Western blots using anti-hGRα (top gel) or anti-β-actin (bottom gel) antibodies are shown. RLU, Relative light unit. B, hGRαR714Q has about 2-fold lower affinity to dexamethasone compared with the wild-type hGRα in whole-cell dexamethasone binding assay. Left panel shows saturation curves of radioactive dexamethasone to the wild-type hGRα (open circles) and hGRαR714Q (closed circles), and right panel demonstrates results of the Scatchard analysis examining affinity of these receptors to dexamethasone. HCT116 cells were transfected with the wild-type hGRα- or hGRαR714Q-expressing plasmid and were incubated with increasing concentrations of radioactive dexamethasone in the presence or absence of 500-fold excess amounts of cold dexamethasone. The experiment was performed three times with triplicate and mean ± se values of disassociation constant (Kd) are shown in right panel. Circles represent mean ± se values of the specific binding of radioactive dexamethasone to the receptors in A, and mean ratios of bound vs. free radioactive dexamethasone in B. C, hGRαR714Q has reduced binding activity to GRIP1 in vitro. Left panel, hGRαR714Q has reduced binding activity to various portions of GRIP1 in a GST pull-down assay. In vitro-translated and 35S-labelled wild-type hGRα (top panel) or hGRαR714Q (bottom panel) was incubated with indicated fragments of GST-fused GRIP1 in the absence or presence of 10−8 m of dexamethasone (Dex). The experiment was performed three times and representative images are shown. Results obtained in the quantitative analysis of band intensity were shown in Supplemental Fig. 6A. Right panel, hGRαR714Q has reduced binding activity to the NRB domain of GRIP1 in a GST pull-down assay. In vitro-translated and 35S-labelled wild-type hGRα or hGRαR714Q was incubated with GST-fused GRIP1-NRB in the presence of indicated concentrations of dexamethasone. The experiment was performed three times and representative images are shown. Results obtained in the quantitative analysis of band intensity were shown in Supplemental Fig. 6B.
hGRαR714Q has reduced affinity to dexamethasone, delayed nuclear translocation, and defective AF-2, whereas GRE binding is preserved
In a whole-cell dexamethasone-binding assay, dissociation constant (Kd) of the wild-type hGRα to radioactive dexamethasone was 10.95 ± 0.89 (mean ± se) nm, whereas that of the mutant receptor was 20.63 ± 2.63 nm (P < 0.01) (Fig. 1B), suggesting that hGRαR714Q has about 2-fold lower affinity to dexamethasone than the wild-type hGRα.
In the nuclear translocation study for GRαR714Q, enhanced green fluorescent protein-fused hGRα demonstrated rapid nuclear translocation similar to that of the wild-type receptor at 10−6 m of this steroid, suggesting that the molecular machinery for nuclear translocation is preserved in hGRαR714Q (Supplemental Fig. 4). Enhanced green fluorescent protein-hGRαR714Q, however, showed delayed nuclear translocation at 10−8 m of dexamethasone that would have been caused by reduced affinity to glucocorticoids. In chromatin immunoprecipitation assays, hGRαR714Q bound to GREs of endogenous glucocorticoid-induced leucine zipper protein in response to 10−6 m of dexamethasone (Supplemental Fig. 5), indicating that the GRE-binding activity of the mutant receptor is also preserved.
In glutathione-S-transferase (GST) pull-down assays (Fig. 1C and supplemental Fig. 6), hGRαR714Q demonstrated significantly reduced binding activity to the all glucocorticoid receptor-interacting protein (GRIP)-1 fragments tested, whereas its dexamethasone-dependent association with GRIP1 NRB was significantly blunted at 10−8 m of dexamethasone. hGRαR714Q also demonstrated significantly reduced binding activity to GRIP1 NRB at higher concentration (10−6 m) of dexamethasone. LBD of hGRαR714Q, hence its AF2, demonstrated defective transactivational activity (Supplemental Fig. 7). Thus, hGRαR714Q has an inherently defective AF-2 for stimulating the transcription, whereas reduced affinity of the mutant receptor to glucocorticoids may further reduce the transactivation activity of its damaged AF-2.
hGRαR714Q has structurally altered LBD
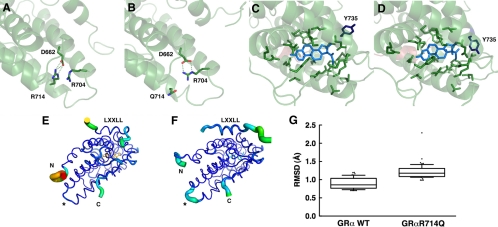
Crystallographic analyses reported previously revealed that the GRα LBD consists of 12 α-helices and four β-sheets, which fold into a three-layer structure (7). Binding of glucocorticoid to the ligand-binding pocket induces conformational changes in hGRα LBD and creates the AF-2 surface in which the helix 12 plays a critical role (4,7,8). In the three-dimensional crystallographic structural model of the wild-type hGRα LBD bound to dexamethasone, R714 is located in the C-terminal portion of the helix 10. Arginine is a basic amino acid, having a large, positively charged side chain, which protrudes into a space created by helices 7–10 by which this residue forms a salt bridge with glutamic acid (E) 662 located in helix 8 (Fig. 2A). In hGRαR714Q LBD, the arginine residue is replaced by glutamine, which has a smaller, uncharged side chain. The mutation breaks the salt bridge with E662. The simulation shows that the negative charge of E662 is instead balanced by the formation of a stable new salt bridge with R704 of helix 9 (Fig. 2B). This change in electrostatic bonding results in a 0,3 nm displacement of R704 in the mutant receptor and releases helix 10 from its former salt bridge constraint, which leads to further conformational changes in the ligand-binding pocket, most notably observed in the rotametric state of tyrosine (Y) 735 on helix 10 (Fig. 2, C and D). Mutation at residue 714 in the hGRα LBD also destabilizes the AF-2 surface and obstructs optimal binding of the LXXLL coactivator motif to this transactivation domain, as determined by a shift in root mean square deviation over the duration of the simulation (Fig. 2, E–G, and Supplemental Fig. 8).
Figure 2.
Arginine (R) to glutamine (Q) replacement at amino acid 714 of the hGRα alters three-dimensional structure of its LBD. A and B, Arginine (R) to glutamine (Q) replacement at amino acid 714 of the hGRα causes formation of a new salt bridge between arginine (R) at 704 and aspartic acid (D) at 662. In the wild-type hGRα LBD, R714 is tightly bound to D662 through an electrostatic salt bridge (A). Substitution of arginine for glutamine in the hGRαR714Q LBD results in a rearrangement of the side chains, forming a new salt bridge between R704 and D662 and displacing Q714 (B). This relaxes some constraint on helix 10 and results in structural changes throughout the LBD. C and D, hGRαR714Q LBD has altered ligand-binding pocket. Ligand-binding pockets of the wild-type hGRα (C) and hGRαR714Q (D) are shown. Arginine to glutamine replacement at amino acid 714 causes alteration in the ligand-binding pocket in hGRαR714Q LBD. The greatest change observed between the wild-type hGRα LBD and hGRαR714Q LBD is in the rotametric state of tyrosine (Y) 735. E–G, hGRαR714Q LBD has the destabilized AF-2 surface that blocks optimal binding of the LXXLL coactivator motif. The thickness and color of the Cα trace of the wild-type hGRα LBD (E) and hGRαR714Q LBD (F) indicate the areas of least (thin and blue) to most (thick and red) motion over the course of the simulation. As expected, the termini of the LBD and the bound peptide move the most. There is significant motion at the mutation site (*) in the mutant LBD (F) but not in the wild-type, suggesting the mutation caused destabilization in this structural area. The bound LXXLL peptide is thus more labile in the mutant LBD structure as seen by the thickness of its Cα trace and quantified by the box plots (bars indicate 10th and 90th percentiles, and dots show outlined results) in G, in which the median root mean square deviation (RMSD) for the helical residues containing the LXXLL motif (amino acids 743–750 of the human transcription intermediary factor 2) is 1.2 Å with the mutant LBD over the course of the simulation, whereas it is only 0.86 Å with the wild-type LBD (P < 0.0001), suggesting that the mutant structure binds the peptide with less affinity.
The arginine residue mutated in this patient is located in the same position in the GR peptide among different species, whereas it was also preserved in the other human nuclear receptors (Supplemental Fig. 9). Thus, R714 of the hGRα appears to play an important role in the formation of the three-dimensional structure and function of this receptor.
Discussion
Generalized seizures of our case were due to her hypoglycemia, which would have been further exacerbated by the preexisting gastroenterological symptoms. The hypokalemia was most likely caused by increased mineralocorticoid as well as cortisol secretion. Reduced glucocorticoid action in the liver, which could not stimulate sufficient glucose production most likely resulted in the hypoglycemia during the acute illness. Hypoglycemia and subsequent generalized seizures have never been reported in the adult cases of generalized glucocorticoid resistance syndrome (1); thus, these clinical manifestations should be considered especially in child cases of this syndrome.
Treatment with dexamethasone to suppress the ACTH production effectively controlled the clinical symptoms. The use of dexamethasone allowed normalization of her hypothalamic-pituitary-adrenal axis and thus appropriately slowed her growth by efficiently suppressing excess production of adrenal androgens that accelerated bone development. Our case suggests that this synthetic steroid is useful for the treatment of infants/children with generalized glucocorticoid resistance syndrome in addition to adult cases (9).
The mutation replacing arginine by glutamine altered the structure of the hGRα ligand-binding pocket and the AF-2 binding surface, resulting in a reduction of affinity of the receptor to dexamethasone and ultimately reducing the transcriptional activity of hGRαR714Q. Although R714 does not directly contribute to the formation of the ligand-binding pocket or the AF-2 surface, the R714Q mutation switches the salt bridge partner of E662 to R704, which subsequently shifts positioning of structural components of the hGRα LBD and alters the conformations of the ligand-binding pocket and the AF-2 surface.
Supplementary Material
Acknowledgments
For three-dimensional simulation of the GRαR714Q LBD, we used the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health (http://biowulf.nih.gov). We thank Drs. David Goldstein and Ted Groshong for primary clinical care of the patient, Drs. R. M. Evans, G. L. Hager, M.-J. Tsai for providing plasmids, Mr. K. Zachman for superb technical assistance, and Drs. G. P. Chrousos and E. Charmadari for critically reading and discussing this manuscript.
Footnotes
This work was supported by the Intramural Research Program of Eunice Kennedy Shriver National Institute of Child Health and Human Development and National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Disclosure Summary: All authors have nothing to disclose.
First Published Online March 24, 2010
Abbreviations: AF, Activation function; DHEA, dehydroepiandrosterone; GR, glucocorticoid receptor; GRIP, glucocorticoid receptor-interacting protein; h, human; GRE, glucocorticoid response element; GST, glutathione-S-transferase; LBD, ligand-binding domain; MMTV, mouse mammary tumor virus; nr, normal range; NRB, nuclear receptor-binding domain.
References
- Charmandari E, Kino T, Ichijo T, Chrousos GP 2008 Generalized glucocorticoid resistance: clinical aspects, molecular mechanisms, and implications of a rare genetic disorder. J Clin Endocrinol Metab 93:1563–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charmandari E, Kino T 2007 Novel causes of generalized glucocorticoid resistance. Horm Metab Res 39:445–450 [DOI] [PubMed] [Google Scholar]
- Kino T, Chrousos GP 2004 Glucocorticoid and mineralocorticoid receptors and associated diseases. Essays Biochem 40:137–155 [DOI] [PubMed] [Google Scholar]
- Chrousos GP, Kino T 2005 Intracellular glucocorticoid signaling: a formerly simple system turns stochastic. Sci STKE 2005:pe48 [DOI] [PubMed] [Google Scholar]
- Kino T, De Martino MU, Charmandari E, Mirani M, Chrousos GP 2003 Tissue glucocorticoid resistance/hypersensitivity syndromes. J Steroid Biochem Mol Biol 85:457–467 [DOI] [PubMed] [Google Scholar]
- McKenna NJ, Lanz RB, O'Malley BW 1999 Nuclear receptor coregulators: cellular and molecular biology. Endocr Rev 20:321–344 [DOI] [PubMed] [Google Scholar]
- Bledsoe RK, Montana VG, Stanley TB, Delves CJ, Apolito CJ, McKee DD, Consler TG, Parks DJ, Stewart EL, Willson TM, Lambert MH, Moore JT, Pearce KH, Xu HE 2002 Crystal structure of the glucocorticoid receptor ligand binding domain reveals a novel mode of receptor dimerization and coactivator recognition. Cell 110:93–105 [DOI] [PubMed] [Google Scholar]
- Kino T, Su YA, Chrousos GP 2009 Human glucocorticoid receptor isoform β: recent understanding of its potential implications in physiology and pathophysiology. Cell Mol Life Sci 66:3435–3448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otten BJ, Stikkelbroeck MM, Claahsen-van der Grinten HL, Hermus AR 2005 Puberty and fertility in congenital adrenal hyperplasia. Endocr Dev 8:54–66 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.