Abstract
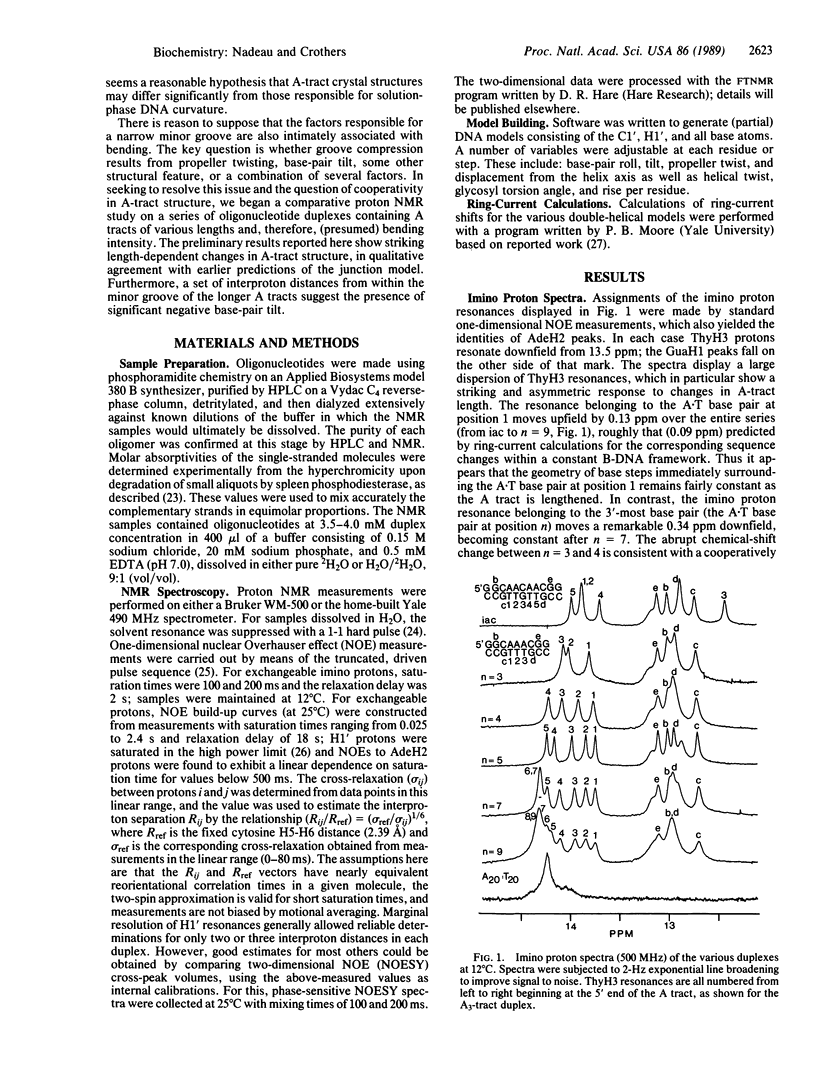
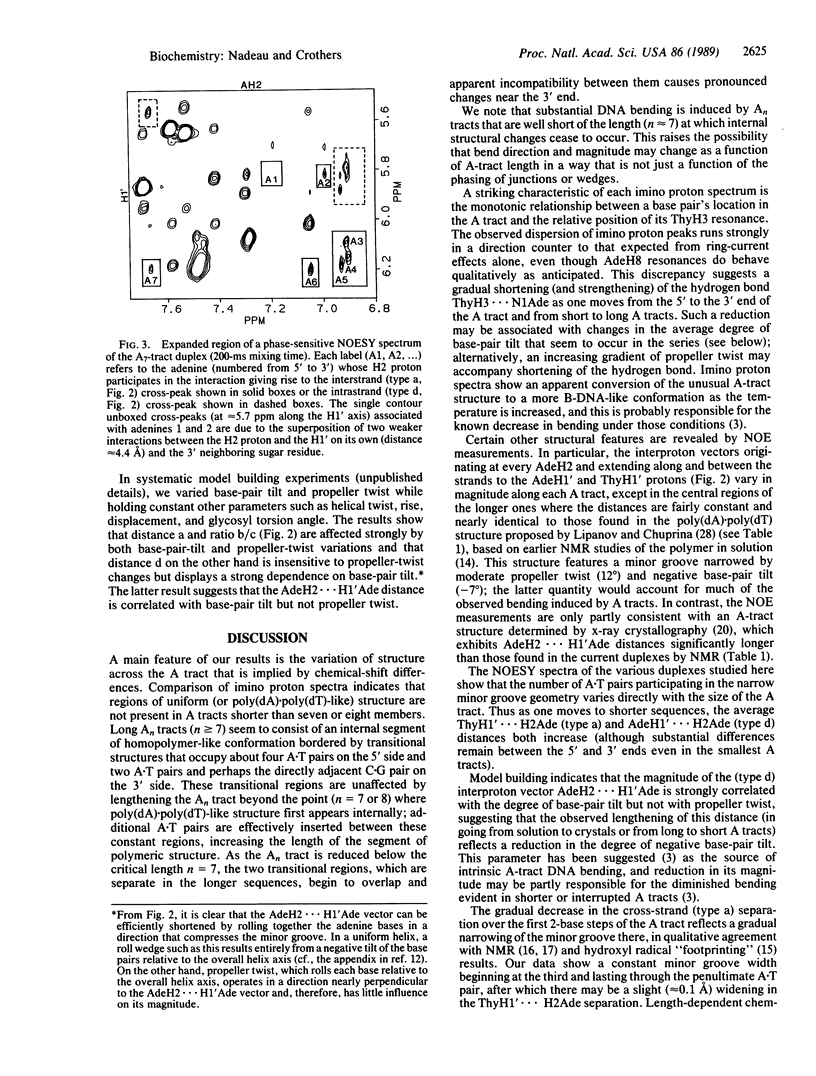
We report proton NMR studies on DNA oligonucleotides that contain A tracts of lengths known to produce various degrees of bending. Spectra of duplexes in the series 5'-(GGCAnCGG).(CCGTnGCC) (n = 3, 4, 5, 7, 9) reveal substantial structural changes within the An.Tn tract as its length is increased. Chemical-shift comparisons show that A tracts with fewer than about seven members do not contain regions of uniform [or poly(dA).poly(dT)-like] structure. Long An tracts (n greater than or equal to 7) appear to consist of an internal segment of homopolymeric conformation flanked by regions of transitional structure that occupy about four A.T pairs on the 5' side and two A.T pairs perhaps the directly adjacent G.C pair on the 3' side. In shorter duplexes (n less than 7), these two transitional regions overlap and an apparent mutual incompatibility causes length-dependent changes that are most pronounced near the 3' end. Throughout the series, there is a striking monotonic relationship between the location of an A.T pair in the A tract and the relative position of its ThyH3 resonance. The direction of the chemical-shift dispersion is opposite to that expected from consideration of ring-current effects alone; this discrepancy suggests a gradual decrease in ThyH3...N1Ade hydrogen-bond length as one moves from the 5' to the 3' end of the A tract and from short to long A tracts. Nuclear Overhauser effect measurements reveal that the interproton distances AdeH2...H1'Ade and AdeH2...H1'Thy vary along each A tract, except in the central regions of the longer ones where they are fairly constant and in good agreement with the poly(dA).poly(dT) structure proposed by Lipanov, A.A. & Chuprina, V. P. [(1987) Nucleic Acids Res. 15, 5833-5844]. This model features a substantial negative base-pair tilt, which has been suggested previously as the source of A-tract bending. In contrast, the nuclear Overhauser effect distances are inconsistent with at least one known crystallographic A-tract structure [DiGabriele, A. D., Sanderson, M. R. & Steitz, T. A. (1989) Proc. Natl. Acad. Sci. USA 86, 1816-1820], which lacks appreciable base-pair tilt.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alexeev D. G., Lipanov A. A., Skuratovskii IYa The structure of poly(dA).poly(dT) as revealed by an X-ray fibre diffraction. J Biomol Struct Dyn. 1987 Jun;4(6):989–1012. doi: 10.1080/07391102.1987.10507693. [DOI] [PubMed] [Google Scholar]
- Arnott S., Chandrasekaran R., Hall I. H., Puigjaner L. C. Heteronomous DNA. Nucleic Acids Res. 1983 Jun 25;11(12):4141–4155. doi: 10.1093/nar/11.12.4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behling R. W., Kearns D. R. 1H two-dimensional nuclear Overhauser effect and relaxation studies of poly(dA).poly(dT) Biochemistry. 1986 Jun 3;25(11):3335–3346. doi: 10.1021/bi00359a037. [DOI] [PubMed] [Google Scholar]
- Burkhoff A. M., Tullius T. D. The unusual conformation adopted by the adenine tracts in kinetoplast DNA. Cell. 1987 Mar 27;48(6):935–943. doi: 10.1016/0092-8674(87)90702-1. [DOI] [PubMed] [Google Scholar]
- Calladine C. R., Drew H. R., McCall M. J. The intrinsic curvature of DNA in solution. J Mol Biol. 1988 May 5;201(1):127–137. doi: 10.1016/0022-2836(88)90444-5. [DOI] [PubMed] [Google Scholar]
- Chuprina V. P. Anomalous structure and properties of poly (dA).poly(dT). Computer simulation of the polynucleotide structure with the spine of hydration in the minor groove. Nucleic Acids Res. 1987 Jan 12;15(1):293–311. doi: 10.1093/nar/15.1.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll M., Frederick C. A., Wang A. H., Rich A. A bifurcated hydrogen-bonded conformation in the d(A.T) base pairs of the DNA dodecamer d(CGCAAATTTGCG) and its complex with distamycin. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8385–8389. doi: 10.1073/pnas.84.23.8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiGabriele A. D., Sanderson M. R., Steitz T. A. Crystal lattice packing is important in determining the bend of a DNA dodecamer containing an adenine tract. Proc Natl Acad Sci U S A. 1989 Mar;86(6):1816–1820. doi: 10.1073/pnas.86.6.1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fratini A. V., Kopka M. L., Drew H. R., Dickerson R. E. Reversible bending and helix geometry in a B-DNA dodecamer: CGCGAATTBrCGCG. J Biol Chem. 1982 Dec 25;257(24):14686–14707. [PubMed] [Google Scholar]
- Giessner-Prettre C., Pullman B., Borer P. N., Kan L. S., Ts'o P. O. Ring-current effects in the Nmr of nucleic acids: a graphical approach. Biopolymers. 1976 Nov;15(11):2277–2286. doi: 10.1002/bip.1976.360151114. [DOI] [PubMed] [Google Scholar]
- Giessner-Prettre C., Pullman B., Caillet J. Theoretical study on the proton chemical shifts of hydrogen bonded nucleic acid bases. Nucleic Acids Res. 1977 Jan;4(1):99–116. doi: 10.1093/nar/4.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagerman P. J. Sequence-directed curvature of DNA. Nature. 1986 May 22;321(6068):449–450. doi: 10.1038/321449a0. [DOI] [PubMed] [Google Scholar]
- Katahira M., Sugeta H., Kyogoku Y., Fujii S., Fujisawa R., Tomita K. One- and two-dimensional NMR studies on the conformation of DNA containing the oligo(dA)oligo(dT) tract. Nucleic Acids Res. 1988 Sep 12;16(17):8619–8632. doi: 10.1093/nar/16.17.8619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kime M. J., Moore P. B. Nuclear Overhauser experiments at 500 MHz on the downfield proton spectrum of a ribonuclease-resistant fragment of 5S ribonucleic acid. Biochemistry. 1983 May 24;22(11):2615–2622. doi: 10.1021/bi00280a004. [DOI] [PubMed] [Google Scholar]
- Kintanar A., Klevit R. E., Reid B. R. Two-dimensional NMR investigation of a bent DNA fragment: assignment of the proton resonances and preliminary structure analysis. Nucleic Acids Res. 1987 Jul 24;15(14):5845–5862. doi: 10.1093/nar/15.14.5845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H. S., Crothers D. M. Calibration of DNA curvature and a unified description of sequence-directed bending. Proc Natl Acad Sci U S A. 1988 Mar;85(6):1763–1767. doi: 10.1073/pnas.85.6.1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koo H. S., Wu H. M., Crothers D. M. DNA bending at adenine . thymine tracts. Nature. 1986 Apr 10;320(6062):501–506. doi: 10.1038/320501a0. [DOI] [PubMed] [Google Scholar]
- Kopka M. L., Fratini A. V., Drew H. R., Dickerson R. E. Ordered water structure around a B-DNA dodecamer. A quantitative study. J Mol Biol. 1983 Jan 5;163(1):129–146. doi: 10.1016/0022-2836(83)90033-5. [DOI] [PubMed] [Google Scholar]
- Lipanov A. A., Chuprina V. P. The structure of poly(dA):poly(dT) in a condensed state and in solution. Nucleic Acids Res. 1987 Jul 24;15(14):5833–5844. doi: 10.1093/nar/15.14.5833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marini J. C., Levene S. D., Crothers D. M., Englund P. T. Bent helical structure in kinetoplast DNA. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7664–7668. doi: 10.1073/pnas.79.24.7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maroun R. C., Olson W. K. Base sequence effects in double-helical DNA. III. Average properties of curved DNA. Biopolymers. 1988 Apr;27(4):585–603. doi: 10.1002/bip.360270404. [DOI] [PubMed] [Google Scholar]
- Nadeau J. G., Gilham P. T. Anomalous hairpin formation in an oligodeoxyribonucleotide. Nucleic Acids Res. 1985 Nov 25;13(22):8259–8274. doi: 10.1093/nar/13.22.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson H. C., Finch J. T., Luisi B. F., Klug A. The structure of an oligo(dA).oligo(dT) tract and its biological implications. Nature. 1987 Nov 19;330(6145):221–226. doi: 10.1038/330221a0. [DOI] [PubMed] [Google Scholar]
- Prunell A., Goulet I., Jacob Y., Goutorbe F. The smaller helical repeat of poly(dA) . poly(dT) relative to DNA may reflect the wedge property of the dA . dT base pair. Eur J Biochem. 1984 Jan 16;138(2):253–257. doi: 10.1111/j.1432-1033.1984.tb07909.x. [DOI] [PubMed] [Google Scholar]
- Ulanovsky L. E., Trifonov E. N. Estimation of wedge components in curved DNA. Nature. 1987 Apr 16;326(6114):720–722. doi: 10.1038/326720a0. [DOI] [PubMed] [Google Scholar]
- Ulanovsky L., Bodner M., Trifonov E. N., Choder M. Curved DNA: design, synthesis, and circularization. Proc Natl Acad Sci U S A. 1986 Feb;83(4):862–866. doi: 10.1073/pnas.83.4.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H. M., Crothers D. M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984 Apr 5;308(5959):509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- Zinkel S. S., Crothers D. M. DNA bend direction by phase sensitive detection. Nature. 1987 Jul 9;328(6126):178–181. doi: 10.1038/328178a0. [DOI] [PubMed] [Google Scholar]



