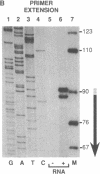
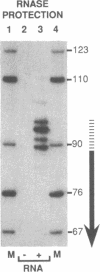
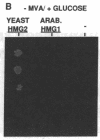
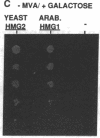
Abstract
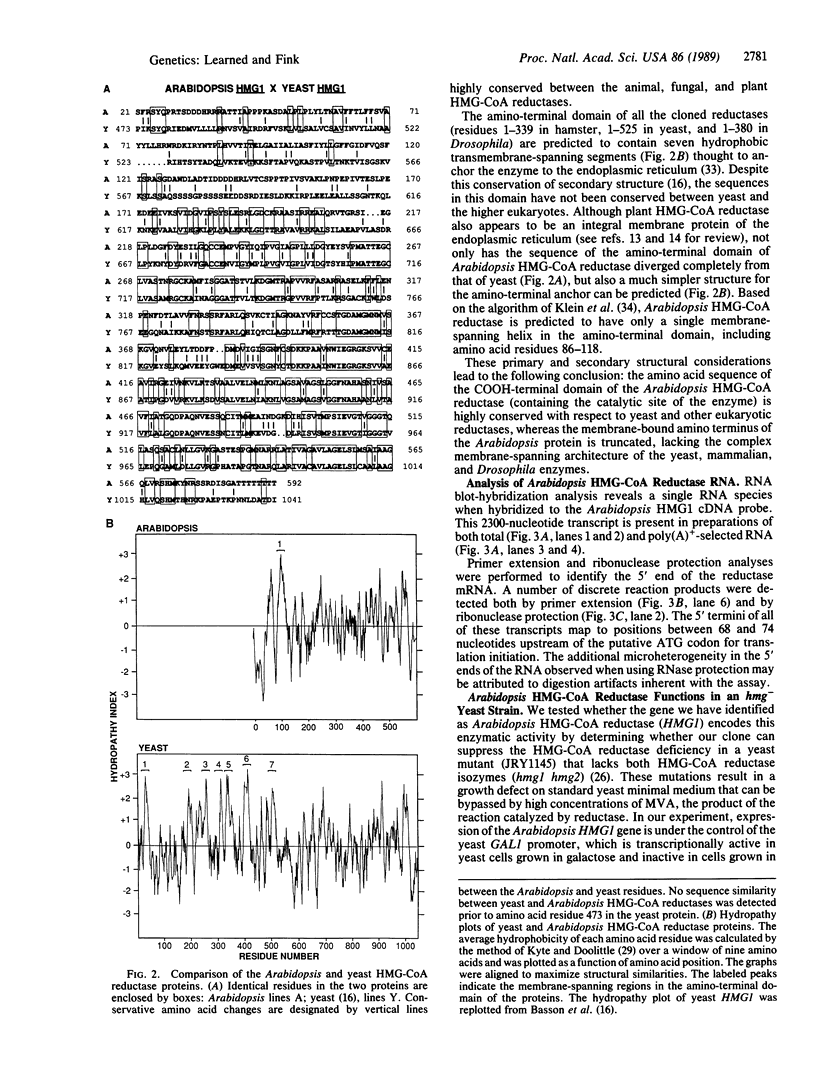
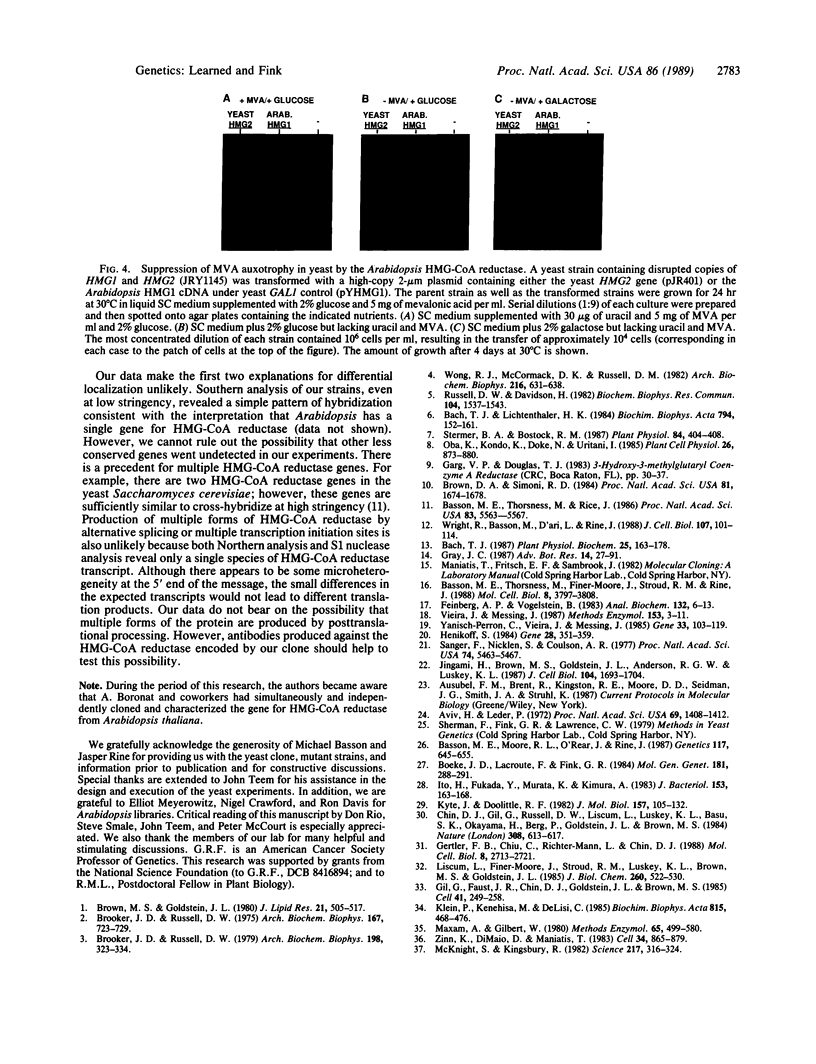
We have isolated the Arabidopsis thaliana gene (HMG1) encoding 3-hydroxy-3-methylglutaryl-CoA reductase [HMG-CoA reductase; (S)-mevalonate:NAD+ oxido-reductase (CoA-acylating), EC 1.1.1.88], the catalyst of the first committed step in isoprenoid biosynthesis. cDNA copies of the plant gene were identified by hybridization with a short, highly conserved segment of yeast HMG-CoA reductase as probe. DNA sequence analysis reveals that the COOH-terminal domain of the Arabidopsis HMG-CoA reductase (containing the catalytic site of the enzyme) is highly conserved with respect to the yeast, mammalian, and Drosophila enzymes, whereas the membrane-bound amino terminus of the Arabidopsis protein is truncated and lacks the complex membrane-spanning architecture of the yeast and animal reductases. Expression of the Arabidopsis gene from the yeast GAL1 promoter in a yeast mutant lacking HMG-CoA reductase activity suppresses the growth defect of the yeast mutant. Taken together, the sequence similarity to other cloned HMG-CoA reductase genes and the suppression of the yeast hmg- mutant provide strong evidence that the novel Arabidopsis gene we have cloned encodes a functional HMG-CoA reductase enzyme.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aviv H., Leder P. Purification of biologically active globin messenger RNA by chromatography on oligothymidylic acid-cellulose. Proc Natl Acad Sci U S A. 1972 Jun;69(6):1408–1412. doi: 10.1073/pnas.69.6.1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bach T. J., Lichtenthaler H. K. Application of modified Lineweaver-Burk plots to studies of kinetics and regulation of radish 3-hydroxy-3-methylglutaryl-CoA reductase. Biochim Biophys Acta. 1984 Jun 6;794(1):152–161. doi: 10.1016/0005-2760(84)90308-4. [DOI] [PubMed] [Google Scholar]
- Basson M. E., Moore R. L., O'Rear J., Rine J. Identifying mutations in duplicated functions in Saccharomyces cerevisiae: recessive mutations in HMG-CoA reductase genes. Genetics. 1987 Dec;117(4):645–655. doi: 10.1093/genetics/117.4.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basson M. E., Thorsness M., Finer-Moore J., Stroud R. M., Rine J. Structural and functional conservation between yeast and human 3-hydroxy-3-methylglutaryl coenzyme A reductases, the rate-limiting enzyme of sterol biosynthesis. Mol Cell Biol. 1988 Sep;8(9):3797–3808. doi: 10.1128/mcb.8.9.3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basson M. E., Thorsness M., Rine J. Saccharomyces cerevisiae contains two functional genes encoding 3-hydroxy-3-methylglutaryl-coenzyme A reductase. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5563–5567. doi: 10.1073/pnas.83.15.5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D. One and two codon insertion mutants of bacteriophage f1. Mol Gen Genet. 1981;181(3):288–291. doi: 10.1007/BF00425599. [DOI] [PubMed] [Google Scholar]
- Brooker J. D., Russell D. W. Properties of microsomal 3-hydroxy-3-methylglutaryl coenzyme A reductase from Pisum sativum seedlings. Arch Biochem Biophys. 1975 Apr;167(2):723–729. doi: 10.1016/0003-9861(75)90517-2. [DOI] [PubMed] [Google Scholar]
- Brooker J. D., Russell D. W. Regulation of microsomal 3-hydroxy-3-methylglutaryl coenzyme A reductase from pea seedlings: rapid posttranslational phytochrome-mediated decrease in activity and in vivo regulation by isoprenoid products. Arch Biochem Biophys. 1979 Nov;198(1):323–334. doi: 10.1016/0003-9861(79)90425-9. [DOI] [PubMed] [Google Scholar]
- Brown D. A., Simoni R. D. Biogenesis of 3-hydroxy-3-methylglutaryl-coenzyme A reductase, an integral glycoprotein of the endoplasmic reticulum. Proc Natl Acad Sci U S A. 1984 Mar;81(6):1674–1678. doi: 10.1073/pnas.81.6.1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown M. S., Goldstein J. L. Multivalent feedback regulation of HMG CoA reductase, a control mechanism coordinating isoprenoid synthesis and cell growth. J Lipid Res. 1980 Jul;21(5):505–517. [PubMed] [Google Scholar]
- Chin D. J., Gil G., Russell D. W., Liscum L., Luskey K. L., Basu S. K., Okayama H., Berg P., Goldstein J. L., Brown M. S. Nucleotide sequence of 3-hydroxy-3-methyl-glutaryl coenzyme A reductase, a glycoprotein of endoplasmic reticulum. Nature. 1984 Apr 12;308(5960):613–617. doi: 10.1038/308613a0. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Gertler F. B., Chiu C. Y., Richter-Mann L., Chin D. J. Developmental and metabolic regulation of the Drosophila melanogaster 3-hydroxy-3-methylglutaryl coenzyme A reductase. Mol Cell Biol. 1988 Jul;8(7):2713–2721. doi: 10.1128/mcb.8.7.2713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gil G., Faust J. R., Chin D. J., Goldstein J. L., Brown M. S. Membrane-bound domain of HMG CoA reductase is required for sterol-enhanced degradation of the enzyme. Cell. 1985 May;41(1):249–258. doi: 10.1016/0092-8674(85)90078-9. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jingami H., Brown M. S., Goldstein J. L., Anderson R. G., Luskey K. L. Partial deletion of membrane-bound domain of 3-hydroxy-3-methylglutaryl coenzyme A reductase eliminates sterol-enhanced degradation and prevents formation of crystalloid endoplasmic reticulum. J Cell Biol. 1987 Jun;104(6):1693–1704. doi: 10.1083/jcb.104.6.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein P., Kanehisa M., DeLisi C. The detection and classification of membrane-spanning proteins. Biochim Biophys Acta. 1985 May 28;815(3):468–476. doi: 10.1016/0005-2736(85)90375-x. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Liscum L., Finer-Moore J., Stroud R. M., Luskey K. L., Brown M. S., Goldstein J. L. Domain structure of 3-hydroxy-3-methylglutaryl coenzyme A reductase, a glycoprotein of the endoplasmic reticulum. J Biol Chem. 1985 Jan 10;260(1):522–530. [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- McKnight S. L., Kingsbury R. Transcriptional control signals of a eukaryotic protein-coding gene. Science. 1982 Jul 23;217(4557):316–324. doi: 10.1126/science.6283634. [DOI] [PubMed] [Google Scholar]
- Russell D. W., Davidson H. Regulation of cytosolic HMG-CoA reductase activity in pea seedlings: contrasting responses to different hormones, and hormone-product interaction, suggest hormonal modulation of activity. Biochem Biophys Res Commun. 1982 Feb 26;104(4):1537–1543. doi: 10.1016/0006-291x(82)91426-7. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stermer B. A., Bostock R. M. Involvement of 3-hydroxy-3-methylglutaryl coenzyme a reductase in the regulation of sesquiterpenoid phytoalexin synthesis in potato. Plant Physiol. 1987 Jun;84(2):404–408. doi: 10.1104/pp.84.2.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Wong R. J., McCormack D. K., Russell D. W. Plastid 3-hydroxy-3-methylglutaryl coenzyme A reductase has distinctive kinetic and regulatory features: properties of the enzyme and positive phytochrome control of activity in pea seedlings. Arch Biochem Biophys. 1982 Jul;216(2):631–638. doi: 10.1016/0003-9861(82)90253-3. [DOI] [PubMed] [Google Scholar]
- Wright R., Basson M., D'Ari L., Rine J. Increased amounts of HMG-CoA reductase induce "karmellae": a proliferation of stacked membrane pairs surrounding the yeast nucleus. J Cell Biol. 1988 Jul;107(1):101–114. doi: 10.1083/jcb.107.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Zinn K., DiMaio D., Maniatis T. Identification of two distinct regulatory regions adjacent to the human beta-interferon gene. Cell. 1983 Oct;34(3):865–879. doi: 10.1016/0092-8674(83)90544-5. [DOI] [PubMed] [Google Scholar]