Abstract
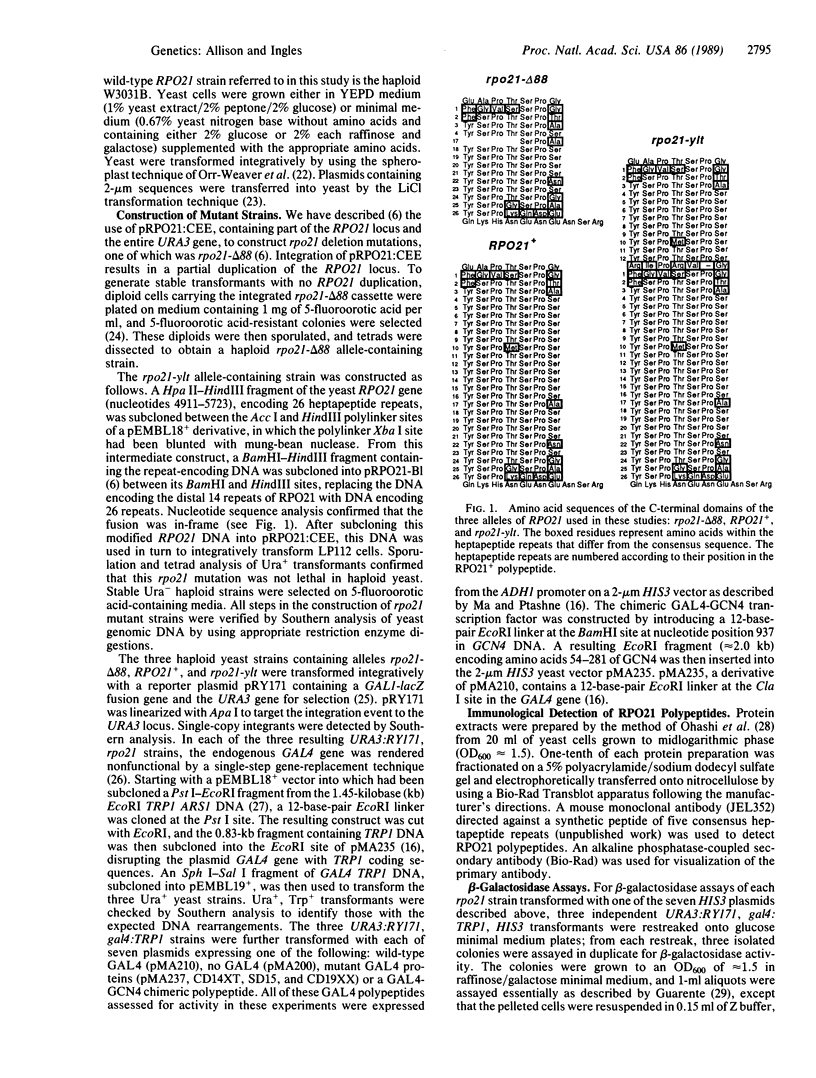
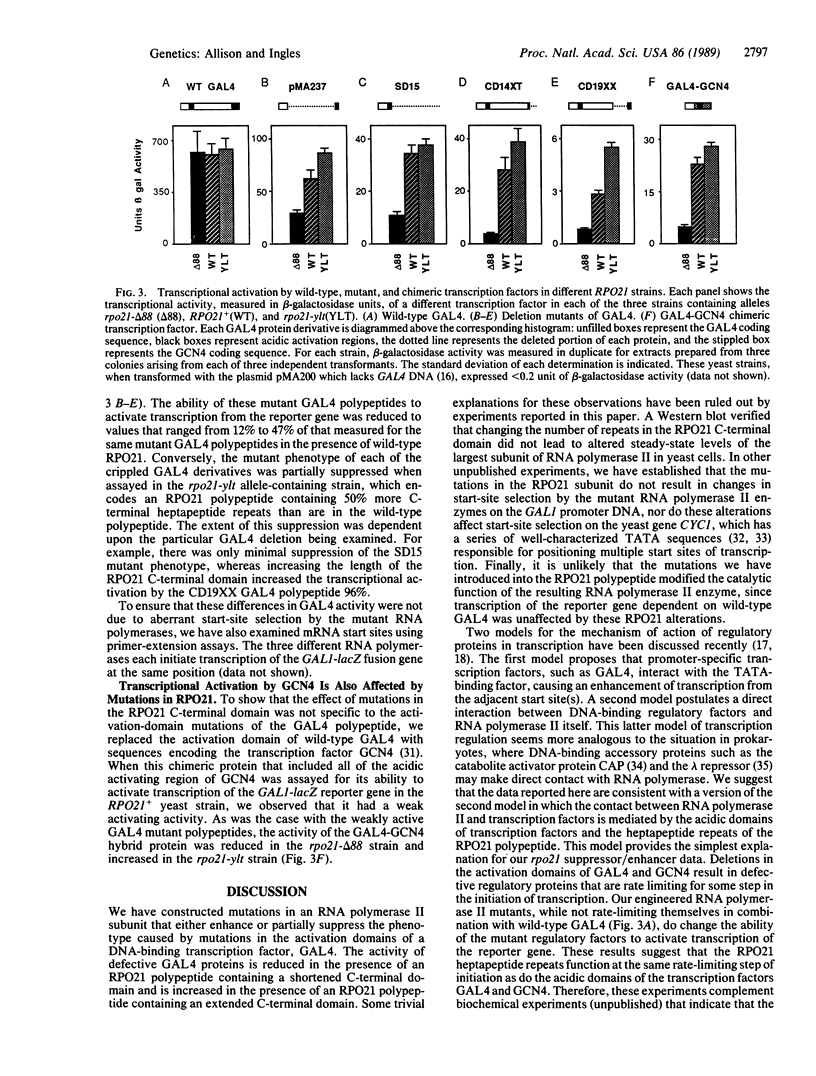
The activation domains of eukaryotic DNA-binding transcription factors, such as GAL4, may regulate transcription by contacting RNA polymerase II. One potential site on RNA polymerase II for such interactions is the C-terminal tandemly repeated heptapeptide domain in the largest subunit (RPO21). We have changed the number of heptapeptide repeats in this yeast RPO21 C-terminal domain and have expressed these mutant RNA polymerase II polypeptides in yeast cells containing either wild-type or defective GAL4 proteins. Although the number of RPO21 heptapeptide repeats had no effect on the activity of wild-type GAL4, changing the length of the C-terminal domain modified the ability of mutant GAL4 proteins to activate transcription. Shorter or longer RPO21 C-terminal domains enhanced or partially suppressed, respectively, the effects of deletions in the transcriptional-activation domains of GAL4. The same RPO21 mutations also affected transcriptional activation by a GAL4-GCN4 chimera. These data suggest that the activation domains of DNA-binding transcription factors could interact, either directly or indirectly, with the heptapeptide repeats of RNA polymerase II.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allison L. A., Moyle M., Shales M., Ingles C. J. Extensive homology among the largest subunits of eukaryotic and prokaryotic RNA polymerases. Cell. 1985 Sep;42(2):599–610. doi: 10.1016/0092-8674(85)90117-5. [DOI] [PubMed] [Google Scholar]
- Allison L. A., Wong J. K., Fitzpatrick V. D., Moyle M., Ingles C. J. The C-terminal domain of the largest subunit of RNA polymerase II of Saccharomyces cerevisiae, Drosophila melanogaster, and mammals: a conserved structure with an essential function. Mol Cell Biol. 1988 Jan;8(1):321–329. doi: 10.1128/mcb.8.1.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomei M. S., Halden N. F., Cullen C. R., Corden J. L. Genetic analysis of the repetitive carboxyl-terminal domain of the largest subunit of mouse RNA polymerase II. Mol Cell Biol. 1988 Jan;8(1):330–339. doi: 10.1128/mcb.8.1.330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D., LaCroute F., Fink G. R. A positive selection for mutants lacking orotidine-5'-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197(2):345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Brent R., Ptashne M. A eukaryotic transcriptional activator bearing the DNA specificity of a prokaryotic repressor. Cell. 1985 Dec;43(3 Pt 2):729–736. doi: 10.1016/0092-8674(85)90246-6. [DOI] [PubMed] [Google Scholar]
- Buratowski S., Hahn S., Sharp P. A., Guarente L. Function of a yeast TATA element-binding protein in a mammalian transcription system. Nature. 1988 Jul 7;334(6177):37–42. doi: 10.1038/334037a0. [DOI] [PubMed] [Google Scholar]
- Cadena D. L., Dahmus M. E. Messenger RNA synthesis in mammalian cells is catalyzed by the phosphorylated form of RNA polymerase II. J Biol Chem. 1987 Sep 15;262(26):12468–12474. [PubMed] [Google Scholar]
- Cavallini B., Huet J., Plassat J. L., Sentenac A., Egly J. M., Chambon P. A yeast activity can substitute for the HeLa cell TATA box factor. Nature. 1988 Jul 7;334(6177):77–80. doi: 10.1038/334077a0. [DOI] [PubMed] [Google Scholar]
- Corden J. L., Cadena D. L., Ahearn J. M., Jr, Dahmus M. E. A unique structure at the carboxyl terminus of the largest subunit of eukaryotic RNA polymerase II. Proc Natl Acad Sci U S A. 1985 Dec;82(23):7934–7938. doi: 10.1073/pnas.82.23.7934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahmus M. E., Kedinger C. Transcription of adenovirus-2 major late promoter inhibited by monoclonal antibody directed against RNA polymerases IIO and IIA. J Biol Chem. 1983 Feb 25;258(4):2303–2307. [PubMed] [Google Scholar]
- Fischer J. A., Giniger E., Maniatis T., Ptashne M. GAL4 activates transcription in Drosophila. Nature. 1988 Apr 28;332(6167):853–856. doi: 10.1038/332853a0. [DOI] [PubMed] [Google Scholar]
- Fitzpatrick V. D., Ingles C. J. The Drosophila fushi tarazu polypeptide is a DNA-binding transcriptional activator in yeast cells. Nature. 1989 Feb 16;337(6208):666–668. doi: 10.1038/337666a0. [DOI] [PubMed] [Google Scholar]
- Giniger E., Ptashne M. Transcription in yeast activated by a putative amphipathic alpha helix linked to a DNA binding unit. Nature. 1987 Dec 17;330(6149):670–672. doi: 10.1038/330670a0. [DOI] [PubMed] [Google Scholar]
- Guarente L. UASs and enhancers: common mechanism of transcriptional activation in yeast and mammals. Cell. 1988 Feb 12;52(3):303–305. doi: 10.1016/s0092-8674(88)80020-5. [DOI] [PubMed] [Google Scholar]
- Guarente L. Yeast promoters and lacZ fusions designed to study expression of cloned genes in yeast. Methods Enzymol. 1983;101:181–191. doi: 10.1016/0076-6879(83)01013-7. [DOI] [PubMed] [Google Scholar]
- Hahn S., Hoar E. T., Guarente L. Each of three "TATA elements" specifies a subset of the transcription initiation sites at the CYC-1 promoter of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8562–8566. doi: 10.1073/pnas.82.24.8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochschild A., Irwin N., Ptashne M. Repressor structure and the mechanism of positive control. Cell. 1983 Feb;32(2):319–325. doi: 10.1016/0092-8674(83)90451-8. [DOI] [PubMed] [Google Scholar]
- Hope I. A., Struhl K. Functional dissection of a eukaryotic transcriptional activator protein, GCN4 of yeast. Cell. 1986 Sep 12;46(6):885–894. doi: 10.1016/0092-8674(86)90070-x. [DOI] [PubMed] [Google Scholar]
- Horikoshi M., Carey M. F., Kakidani H., Roeder R. G. Mechanism of action of a yeast activator: direct effect of GAL4 derivatives on mammalian TFIID-promoter interactions. Cell. 1988 Aug 26;54(5):665–669. doi: 10.1016/s0092-8674(88)80011-4. [DOI] [PubMed] [Google Scholar]
- Ingles C. J., Himmelfarb H. J., Shales M., Greenleaf A. L., Friesen J. D. Identification, molecular cloning, and mutagenesis of Saccharomyces cerevisiae RNA polymerase genes. Proc Natl Acad Sci U S A. 1984 Apr;81(7):2157–2161. doi: 10.1073/pnas.81.7.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin N., Ptashne M. Mutants of the catabolite activator protein of Escherichia coli that are specifically deficient in the gene-activation function. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8315–8319. doi: 10.1073/pnas.84.23.8315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M. A model fungal gene regulatory mechanism: the GAL genes of Saccharomyces cerevisiae. Microbiol Rev. 1987 Dec;51(4):458–476. doi: 10.1128/mr.51.4.458-476.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kakidani H., Ptashne M. GAL4 activates gene expression in mammalian cells. Cell. 1988 Jan 29;52(2):161–167. doi: 10.1016/0092-8674(88)90504-1. [DOI] [PubMed] [Google Scholar]
- Keegan L., Gill G., Ptashne M. Separation of DNA binding from the transcription-activating function of a eukaryotic regulatory protein. Science. 1986 Feb 14;231(4739):699–704. doi: 10.1126/science.3080805. [DOI] [PubMed] [Google Scholar]
- Lech K., Anderson K., Brent R. DNA-bound Fos proteins activate transcription in yeast. Cell. 1988 Jan 29;52(2):179–184. doi: 10.1016/0092-8674(88)90506-5. [DOI] [PubMed] [Google Scholar]
- Lue N. F., Kornberg R. D. Accurate initiation at RNA polymerase II promoters in extracts from Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8839–8843. doi: 10.1073/pnas.84.24.8839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Przibilla E., Hu J., Bogorad L., Ptashne M. Yeast activators stimulate plant gene expression. Nature. 1988 Aug 18;334(6183):631–633. doi: 10.1038/334631a0. [DOI] [PubMed] [Google Scholar]
- Ma J., Ptashne M. Deletion analysis of GAL4 defines two transcriptional activating segments. Cell. 1987 Mar 13;48(5):847–853. doi: 10.1016/0092-8674(87)90081-x. [DOI] [PubMed] [Google Scholar]
- McNeil J. B., Smith M. Transcription initiation of the Saccharomyces cerevisiae iso-1-cytochrome c gene. Multiple, independent T-A-T-A sequences. J Mol Biol. 1986 Feb 5;187(3):363–378. doi: 10.1016/0022-2836(86)90439-0. [DOI] [PubMed] [Google Scholar]
- Moyle M., Hofmann T., Ingles C. J. The RPO31 gene of Saccharomyces cerevisiae encodes the largest subunit of RNA polymerase III. Biochem Cell Biol. 1986 Aug;64(8):717–721. doi: 10.1139/o86-098. [DOI] [PubMed] [Google Scholar]
- Mémet S., Gouy M., Marck C., Sentenac A., Buhler J. M. RPA190, the gene coding for the largest subunit of yeast RNA polymerase A. J Biol Chem. 1988 Feb 25;263(6):2830–2839. [PubMed] [Google Scholar]
- Nonet M., Sweetser D., Young R. A. Functional redundancy and structural polymorphism in the large subunit of RNA polymerase II. Cell. 1987 Sep 11;50(6):909–915. doi: 10.1016/0092-8674(87)90517-4. [DOI] [PubMed] [Google Scholar]
- Ohashi A., Gibson J., Gregor I., Schatz G. Import of proteins into mitochondria. The precursor of cytochrome c1 is processed in two steps, one of them heme-dependent. J Biol Chem. 1982 Nov 10;257(21):13042–13047. [PubMed] [Google Scholar]
- Orr-Weaver T. L., Szostak J. W., Rothstein R. J. Yeast transformation: a model system for the study of recombination. Proc Natl Acad Sci U S A. 1981 Oct;78(10):6354–6358. doi: 10.1073/pnas.78.10.6354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petko L., Lindquist S. Hsp26 is not required for growth at high temperatures, nor for thermotolerance, spore development, or germination. Cell. 1986 Jun 20;45(6):885–894. doi: 10.1016/0092-8674(86)90563-5. [DOI] [PubMed] [Google Scholar]
- Quiocho F. A. Carbohydrate-binding proteins: tertiary structures and protein-sugar interactions. Annu Rev Biochem. 1986;55:287–315. doi: 10.1146/annurev.bi.55.070186.001443. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Ruden D. M., Ma J., Ptashne M. No strict alignment is required between a transcriptional activator binding site and the "TATA box" of a yeast gene. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4262–4266. doi: 10.1073/pnas.85.12.4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigler P. B. Transcriptional activation. Acid blobs and negative noodles. Nature. 1988 May 19;333(6170):210–212. doi: 10.1038/333210a0. [DOI] [PubMed] [Google Scholar]
- Struhl K. Promoters, activator proteins, and the mechanism of transcriptional initiation in yeast. Cell. 1987 May 8;49(3):295–297. doi: 10.1016/0092-8674(87)90277-7. [DOI] [PubMed] [Google Scholar]
- Struhl K. The JUN oncoprotein, a vertebrate transcription factor, activates transcription in yeast. Nature. 1988 Apr 14;332(6165):649–650. doi: 10.1038/332649a0. [DOI] [PubMed] [Google Scholar]
- Tschumper G., Carbon J. Sequence of a yeast DNA fragment containing a chromosomal replicator and the TRP1 gene. Gene. 1980 Jul;10(2):157–166. doi: 10.1016/0378-1119(80)90133-x. [DOI] [PubMed] [Google Scholar]
- Webster N., Jin J. R., Green S., Hollis M., Chambon P. The yeast UASG is a transcriptional enhancer in human HeLa cells in the presence of the GAL4 trans-activator. Cell. 1988 Jan 29;52(2):169–178. doi: 10.1016/0092-8674(88)90505-3. [DOI] [PubMed] [Google Scholar]
- Yocum R. R., Hanley S., West R., Jr, Ptashne M. Use of lacZ fusions to delimit regulatory elements of the inducible divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Oct;4(10):1985–1998. doi: 10.1128/mcb.4.10.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehring W. A., Lee J. M., Weeks J. R., Jokerst R. S., Greenleaf A. L. The C-terminal repeat domain of RNA polymerase II largest subunit is essential in vivo but is not required for accurate transcription initiation in vitro. Proc Natl Acad Sci U S A. 1988 Jun;85(11):3698–3702. doi: 10.1073/pnas.85.11.3698. [DOI] [PMC free article] [PubMed] [Google Scholar]




