SUMMARY
Twenty-one Candida albicans isolates from three HIV-infected patients were collected over a period of 3 years and characterized for fluconazole susceptibility, infectivity and genetic relatedness. Fluconazole resistance was found in five isolates, four exhibited dose-dependent susceptibility and the remainder were fully susceptible to this agent. Pulsed-field gel electrophoresis of SfiI restriction digests of the genomic DNA from the isolates revealed that isolates from the same swab specimen were identical despite differences in susceptibility to fluconazole and isolates recovered over time from the three patients retained clonally related DNA fingerprints within each patient. This small-scale study confirms the persistence of oral colonization of C. albicans strains in HIV-infected patients. Clinical data also suggests that the primary infecting strain may become a persistent colonist in the oral cavity once the immune function of the patient has been restored.
Candida albicans and other opportunistic fungal pathogens are frequent colonizers of human mucosal surfaces. They are often harmless commensals in immunocompetent individuals but may be associated with minor infections such as thrush in babies and vaginal infections in women. However, in immunocompromised patients C. albicans can cause systemic infections with high mortality rates [1].
The advent of the AIDS epidemic together with improvements in therapeutics and other medical procedures have increased the size of the immunocompromised population and this has resulted in dramatic increases in the prevalence of fungal infections [2, 3]. For instance, in the United States yeast infections as a group was reported to rank as the fourth most common cause of nosocomial bloodstream infections [4, 5]. Similarly in a Taiwan hospital the prevalence of nosocomial candidemia increased 27-fold from 1981 to 1993 [6].
Oral candidiasis has been recognized as an early expression of the immunodeficiency that occurs in HIV-infected patients [7]. Indeed, unexplained oral candidiasis is considered as a clinical predictor of AIDS in previously healthy adults or worsening immunodeficiency in HIV-infected patients [8]. Oropharyngeal candidiasis due to drug-resistant fungi is a major problem for HIV-infected patients [9]. In addition, horizontal transmission of drug-resistant C. albicans has been demonstrated within a family [10, 11]. Hence, alteration of susceptibility of isolates to antifungal compounds in an HIV-infected patient may be due to horizontal transmission of drug-resistant strains or mutations in the endogenous strains. To distinguish between those two possibilities, we have conducted a molecular epidemiology study to determine the genetic relatedness of fluconazole-resistant and -susceptible isolates from the same patient and examined the clonal stability of isolates over a period of 3 years in three HIV-infected patients.
The patients were selected for this study at the outpatient infectious diseases clinics of National Taiwan University Hospital, Taipei, Taiwan. Demographic and clinical information was gathered from the patients’ medical records, which included age, most recent CD4+ lymphocyte counts, and antiretroviral and antifungal therapy as described previously [12]. Informal verbal consent was obtained from the patients and oral swabs for culture were routinely collected using dry sponge swabs (EZ Culturette, Becton Dickinson, Sparks, MD, USA). Swabs were maintained at room temperature and transported to the laboratory within 24 h where they were plated on agar media within 12 h of arrival. Swabs collected in 1999 were plated on Sabouraud dextrose with chloramphenicol and gentamicin (BBL), and those collected in 2001 and 2002 were plated on Chromagar Candida (BBL). All plates were incubated at 30°C and three independent colonies from each positive culture were selected. Additional colonies were selected if more than one morphotype was present. The minimum inhibitory concentration (MIC) of fluconazole for every isolate was determined and the genomic DNA was isolated for the purpose of genotyping.
The MIC of fluconazole was determined by the microdilution broth method, according to the guidelines of the National Committee for Clinical Laboratory Standards (NCCLS), document M27-A [13]. RPMI medium 1640 (31800-022, Invitrogen Corp., Carlsbad, CA, USA) was used for dilution and growth of yeast cultures and growth was measured spectrophotometrically at 600 nm. The MIC was defined as the lowest concentration of agent that reduced the culture broth turbidity by 50% after 48 h at 35°C. Isolates with an MIC ⩾64 μg/ml were considered resistant to fluconazole; an MIC of ⩽8 μg/ml was defined as susceptible and isolates with MICs of 16 or 32 μg/ml were termed susceptible-dose dependent.
Yeast isolates were each grown on Sabouraud dextrose agar plates for 48 h at 37°C and colonies were suspended in 100 mm Tris–HCl, 100 mm EDTA (pH 8·0). The DNA was prepared as described previously [14]. Pulsed-field gel electrophoresis (PFGE) was performed with a Biometra Rotaphor¯ (Whatman Biometra, Göttingen, Germany) [15]. Dendrogram analysis was performed by using Bionumerics software version 3.0 (Applied Maths, Kortrijk, Belgium). The Jaccard coefficient was used to analyse the similarities of the band patterns and the unweighted pair group method with average linkages (UPGMA) was used for cluster analysis. Isolates exhibiting >80% relatedness (<6 bands difference) were considered to be clonally related.
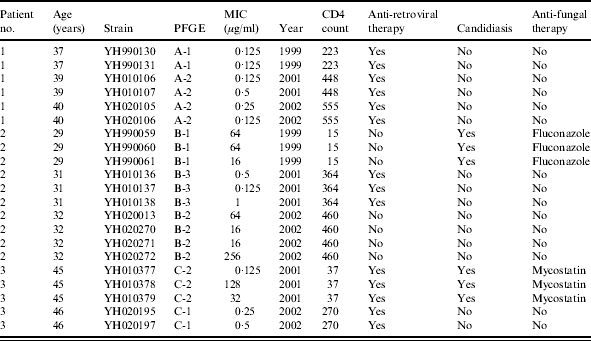
The Table summarizes the characteristics of the 21 isolates of C. albicans recovered from the three patients studied. Six isolates were available for patient no. 1, 10 from patient no. 2 and five from patient no. 3. The ages of the subjects at the first survey were 37, 29 and 45 years respectively. Each of the patients showed an improvement in CD4 count over the sampling period but patient nos. 2 and 3 had the lowest counts on first isolation of C. albicans. They all had received antiretroviral therapy but patient no. 2 did not commence antivirals until 2 years after first presentation and then only received treatment for 1 year. Patient nos. 2 and 3 both had clinical candidiasis and received fluconazole and mycostatin treatment until the infection was cleared. Nevertheless they continued to harbour C. albicans in the oral cavity.
Table.
Characteristics of 21 Candida albicans isolates recovered from three HIV-infected patients
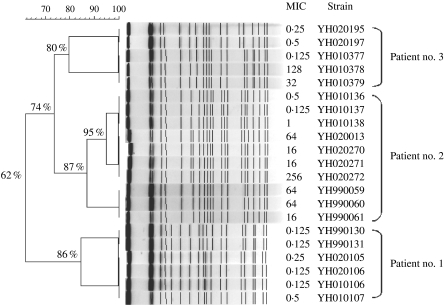
All isolates from patient no. 1 were fully susceptible to fluconazole and two distinct DNA types were identified within this patient. Patient no. 2 on the other hand yielded isolates over the time period ranging from frank resistance through dose-dependent susceptibility to fully susceptible. Three different DNA profiles were evident but isolates of the same clonal group had widely different MIC values for fluconazole. Indeed, three isolates from patient no. 2 (YH020270, YH020272 and YH020272) were all recovered from the same swab specimen but exhibited a 16-fold difference in MIC despite being of the same DNA profile. A similar situation was seen with three isolates from patient no. 3 in 2001 which displayed a 100-fold difference in susceptibility to fluconazole. Moreover, two isolates were recovered from patient no. 3 in 2002 although he had no obvious symptoms of candidiasis. These isolates had almost identical DNA fingerprints and although some band differences were evident they still exhibited nearly 80% relatedness to earlier isolates from this patient leading to the conclusion that the later isolates were indistinguishable from those first isolated from this patient with clinical candidiasis (Fig.). This observation was also true for patient no. 2 who retained the same strain of C. albicans despite resolution of clinical symptoms.
Fig.
Cluster analysis of the 21 C. albicans isolates based on the patterns of SfiI restriction endonuclease analysis of genomic DNA. The dendrogram was constructed by Bionumerics software, with 2% optimization and 0·8% position tolerance, using UPGMA based on the Jaccard similarity coefficients.
In C. albicans, chromosomal changes can occur at high frequencies and these may be related to the duration of exposure to antifungal drugs. For example, the loss of one homologue of chromosome 4 and the gain of one copy of chromosome 3 occurred after incubation in the presence of medium with fluconazole [16]. These changes were not apparently due to the overexpression of putative drug pumps or the drug target and thus indicate that some other mechanism(s) in those isolates was operating. Thus far, two main mechanisms of drug resistance in C. albicans have been identified (1) reduction of drug accumulation by preventing the import of drug into the cell and activating the efflux of drug from the cell, and (2) alteration of the drug target which includes mutating the target of drug, over expression of the target, and bypassing the drug-targeted enzyme by alterations of other enzymes in the same enzymatic pathway [2, 3]. In our study, isolates from a patient in the same year had an identical DNA pattern despite gross differences in their susceptibility to fluconazole. Hence, our results suggest that alterations in susceptibility to fluconazole in C. albicans are not associated with major changes in chromosomal DNA, which is consistent with the reported mechanisms of drug resistance. A correlation between fluconazole susceptibility and DNA pattern was not identified in the present study. Nevertheless, we demonstrated that fluconazole-susceptible and -resistant strains from the same patient retained their clonal relationship.
Although horizontal transmission of drug-resistant C. albicans strains has been demonstrated [10, 11, 17], our data indicate that the persistence of oral colonization with C. albicans in HIV-infected patients is more suggestive of endogenous acquisition of infections [18–20]. Patient nos. 2 and 3 suffered from candidiasis in 1999 and 2001 respectively. For patient no. 2, the infecting strains from the candidiasis stage in 1999 and colonizing strains collected in 2001 and 2002 had ∼87% relatedness. The isolates recovered from symptomatic infection in patient no. 3 in 2001 and the isolate recovered in the absence of symptoms were also highly related. These data suggest that infecting strains and colonizing strains are from the same clone and it is possible that colonizing strains emerge due to minor changes in the chromosome of infecting strains and vice versa, which may not be reflected by our genotyping methods. Whether clonally related infecting strains and colonizing strains from the same patient have a similar degree of virulence in animal models warrants further investigation.
In the present study, patient no. 2 in 1999 and patient no. 3 in 2001 had developed candidiasis. Coincidently, the CD4+ count of patient no. 2 in 1999 was 15 cells/mm3 and that of patient no. 3 in 2001 was 37 cells/mm3. Thus, our data are consistent with the observation that progressive cell-mediated immunodeficiency with a CD4+ lymphocyte count <200 cells/mm3 is a risk factor for development of candidiasis [12, 21]. To establish an infection, opportunistic pathogens, including C. albicans, have to evade the immune system, survive and multiply in the host, and spread to new tissues. Further investigation is needed to explain how these organisms switch from harmless commensals to human pathogens when the opportunity arises and how infecting opportunistic pathogens become colonists when the patients’ immune systems have been restored.
ACKNOWLEDGEMENTS
We thank Pfizer for supplying the fluconazole.
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Pappas PG, Rex JH, Lee J et al. A prospective observational study of candidemia: epidemiology, therapy, and influences on mortality in hospitalized adult and pediatric patients. Clin Infect Dis. 2003;37:634–643. doi: 10.1086/376906. [DOI] [PubMed] [Google Scholar]
- 2.White TC, Marr KA, Bowden RA. Clinical, cellular, and molecular factors that contribute to antifungal drug resistance. Clin Microbiol Rev. 1998;11:382–402. doi: 10.1128/cmr.11.2.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang YL, Lo H-J. Mechanisms of antifungal agent resistance. J Microbiol Immunol Infect. 2001;34:79–86. [PubMed] [Google Scholar]
- 4.Beck-Sague C, Jarvis WR. Secular trends in the epidemiology of nosocomial fungal infections in the United States, 1980–1990. National Nosocomial Infections Surveillance System. J Infect Dis. 1993;167:1247–1251. doi: 10.1093/infdis/167.5.1247. [DOI] [PubMed] [Google Scholar]
- 5.Pfaller MA, Jones RN, Messer SA, Edmond MB, Wenzel RP. National surveillance of nosocomial blood stream infection due to species of Candida other than Candida albicans: frequency of occurrence and antifungal susceptibility in the SCOPE Program. SCOPE Participant Group. Surveillance and Control of Pathogens of Epidemiologic. Diagn Microbiol Infect Dis. 1998;30:121–129. doi: 10.1016/s0732-8893(97)00192-2. [DOI] [PubMed] [Google Scholar]
- 6.Chen YC, Chang SC, Sun CC, Yang LS, Hsieh WC, Luh KT. Secular trends in the epidemiology of nosocomial fungal infections at a teaching hospital in Taiwan, 1981 to 1993. Infect Control Hosp Epidemiol. 1997;18:369–375. doi: 10.1086/647628. [DOI] [PubMed] [Google Scholar]
- 7.Holmberg K, Meyer RD. Fungal infections in patients with AIDS and AIDS-related complex. Scand J Infect Dis. 1986;18:179–192. doi: 10.3109/00365548609032326. [DOI] [PubMed] [Google Scholar]
- 8.Klein RS, Harris CA, Small CB, Moll B, Lesser M, Friedland GH. Oral candidiasis in high-risk patients as the initial manifestation of the acquired immunodeficiency syndrome. N Engl J Med. 1984;311:354–358. doi: 10.1056/NEJM198408093110602. [DOI] [PubMed] [Google Scholar]
- 9.Vanden Bossche H, Marichal P, Odds FC. Molecular mechanisms of drug resistance in fungi. Trends Microbiol. 1994;2:393–400. doi: 10.1016/0966-842x(94)90618-1. [DOI] [PubMed] [Google Scholar]
- 10.Dromer F, Improvisi L, Dupont B et al. Oral transmission of Candida albicans between partners in HIV-infected couples could contribute to dissemination of fluconazole-resistant isolates. AIDS. 1997;11:1095–1101. doi: 10.1097/00002030-199709000-00003. [DOI] [PubMed] [Google Scholar]
- 11.Makarova NU, Pokrowsky VV, Kravchenko AV et al. Persistence of oropharyngeal Candida albicans strains with reduced susceptibilities to fluconazole among human immunodeficiency virus-seropositive children and adults in a long-term care facility. J Clin Microbiol. 2003;41:1833–1837. doi: 10.1128/JCM.41.5.1833-1837.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hung CC, Yang YL, Lauderdale TL et al. Colonization of human immunodeficiency virus-infected outpatients in Taiwan with Candida species. J Clin Microbiol. 2005;43:1600–1603. doi: 10.1128/JCM.43.4.1600-1603.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.National Committee of Clinical Laboratory Standards. 1997. Reference method for broth dilution antifungal susceptibility testing of yeasts; approved standard. M27A.
- 14.Hsu MC, Chen KW, Lo HJ et al. Species identification of medically important fungi by use of real-time LightCycler PCR. J Med Microbiol. 2003;52:1071–1076. doi: 10.1099/jmm.0.05302-0. [DOI] [PubMed] [Google Scholar]
- 15.Chen KW, Lo HJ, Lin YH, Li SY. Comparison of four molecular typing methods to assess genetic relatedness of Candida albicans clinical isolates in Taiwan. J Med Microbiol. 2005;54:249–258. doi: 10.1099/jmm.0.45829-0. [DOI] [PubMed] [Google Scholar]
- 16.Perepnikhatka V, Fischer FJ, Niimi M et al. Specific chromosome alterations in fluconazole-resistant mutants of Candida albicans. J Bacteriol. 1999;181:4041–4049. doi: 10.1128/jb.181.13.4041-4049.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu J, Ramos AR, Vilgalys R, Mitchell TG. Clonal and spontaneous origins of fluconazole resistance in Candida albicans. J Clin Microbiol. 2000;38:1214–1220. doi: 10.1128/jcm.38.3.1214-1220.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pittet D, Monod M, Filthuth I, Frenk E, Suter PM, Auckenthaler R. Contour-clamped homogeneous electric field gel electrophoresis as a powerful epidemiologic tool in yeast infections. Am J Med. 1991;91:256S–263S. doi: 10.1016/0002-9343(91)90378-b. [DOI] [PubMed] [Google Scholar]
- 19.Reagan DR, Pfaller MA, Hollis RJ, Wenzel RP. Characterization of the sequence of colonization and nosocomial candidemia using DNA fingerprinting and a DNA probe. J Clin Microbiol. 1990;28:2733–2738. doi: 10.1128/jcm.28.12.2733-2738.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Voss A, Hollis RJ, Pfaller MA, Wenzel RP, Doebbeling BN. Investigation of the sequence of colonization and candidemia in nonneutropenic patients. J Clin Microbiol. 1994;32:975–980. doi: 10.1128/jcm.32.4.975-980.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Feigal DW, Katz MH, Greenspan D et al. The prevalence of oral lesions in HIV-infected homosexual and bisexual men: three San Francisco epidemiological cohorts. AIDS. 1991;5:519–525. doi: 10.1097/00002030-199105000-00007. [DOI] [PubMed] [Google Scholar]