SUMMARY
An epidemic of meningococcal disease caused by serogroup B meningococci expressing the P1.7-2,4 PorA protein began in New Zealand in 1991. The PorA type has remained stable. Different porB have been found in association with the P1.7-2,4 PorA, although type 4 has been most common. The clonal origins of B:P1.7-2,4 meningococci isolated from cases during 1990 to the end of 2003 were analysed. In 1990, the year immediately preceding the recognized increase in disease rates, all three subclones (ST-41, ST-42, and ST-154) of the ST-41/44 clonal complex occurred among the five isolates of B:P1.7-2,4. The two sequence types, ST-42 and ST-154, continued to cause most disease throughout New Zealand. Isolates belonging to subclone ST-41 were mostly identified early in the epidemic and in the South Island. 16S rRNA typing indicated that isolates belonging to the subclones ST-41 and ST-154 share a common ancestor, with those typing as ST-42 more distantly related with some genetically ambiguous. It is possible that ST-41 and ST-154 may have evolved one from the other but evolution to ST-42 is more difficult to explain. It is possible that one or more of the ST types could have been introduced into New Zealand prior to the first detection of clinical cases in 1990. Genetic diversity may have occurred during carriage in the community.
INTRODUCTION
New Zealand has experienced an ongoing epidemic of meningococcal disease since mid-1991 [1]. The highest case numbers occurred in 2001 with 650 cases reported, giving a population rate of 17·4/100 000 [2]. Elevated rates of disease continued to the end of 2004. Globally, most epidemics of group B disease have been caused by isolates belonging to a limited number of hypervirulent clonal complexes [3, 4], previously referred to as clonal lineages [5]. Hypervirulent clones of the ET-5 complex, first identified in the mid-1970s [3], caused epidemics or localized outbreaks in a number of countries [3, 6–9]. Epidemics in Spain, Cuba, and Brazil were caused by meningococci with strain type B:4:P1.19,15 (serogroup B, serotype 4, serosubtype P1.19,15) [3, 6, 8]; those in Norway and Oregon by strain type B:15:P1.7,16 [3, 9] and in Iquique, Chile by strain type B:15:P1.7,3 [7]. The epidemic of disease in New Zealand is attributable to strain type B:4:P1.7-2,4 [1, 2] belonging to Lineage III, now identified by multilocus sequence typing (MLST) as the ST-41/44 clonal complex. This strain type was first recognized in The Netherlands in the early 1980s [10] and subsequently reported in Belgium [11]. Monoclonal antibodies used in serosubtyping recognize only the VR2 epitope (P1.4) of this strain due to a 3-bp deletion in the VR1 loop preventing recognition of the P1.7 epitope. The VR1 with this deletion is variously described as P1.7b [12] or P1.7h [13], and more recently P1.7-2 where P1.7h, P1.7b, and P1.7-2 describe the same deletion. Sequencing enables definition of the VR2, P1.7-2. From 1990 to the end of 2003, a total of 2358 serogroup B case isolates were obtained and 85·7% (2021/2358) of these were serosubtype P1.4 [14]. Most (1758, 87·0%) of the 2021 B:P1.4 meningococci were PorB type 4, although 4·6% (93/2021) were type 14 and 4·5% (90/2021) had the porB VR1-19, VR2-D, VR3-7, and VR4-14a sequences [15].
This study analysed the clonal derivation of the B:P1.7-2,4 meningococci causing New Zealand’s epidemic. We hypothesized that the meningococcal disease epidemic started following the introduction into New Zealand of a subclone, or subclones, of the hypervirulent ST-41/44 with phenotype B:4:P1.7-2,4. To investigate this, case isolates were examined using both multilocus restriction typing (MLRT) [16] validated against MLST [4], and 16S rRNA typing [17]. Clonal analysis was undertaken on isolates obtained in the year immediately preceding the epidemic (1990) and on isolates obtained from 1991 to 2003.
METHODS
Meningococcal isolates
Meningococci used in this study originated from clinical cases of meningococcal disease in New Zealand and were referred to the Meningococcal Reference Laboratory, Institute of Environmental Science and Research (ESR), under New Zealand’s national surveillance programme. All meningococci were assigned a unique isolate number.
Frozen (−70 °C) copies of meningococci obtained between mid-1989 and the end of 1991 were non-viable due to a freezer failure. Prior to freezing, these isolates had been serogrouped only. Non-viable suspensions were serotyped and serosubtyped using whole-cell ELISA [18]. From 1992 meningococci were routinely serogrouped, serotyped, and serosubtyped [18] and were maintained at −70 °C in glycerol broth suspensions (trypticase soy broth, 15% v/v glycerol). Cultures were grown on 5% sheep blood agar plates (Fort Richard Laboratories, Auckland, New Zealand) at 36 °C, in an atmosphere of 5% CO2, for 18 h. Non-serotypable meningococci were typed by use of porB PCR–amplicon restriction endonuclease analysis [19] and non-serosubtypable meningococci by porA PCR followed by sequencing [20].
DNA extraction
Genomic DNA was purified from live meningococci using cetyltrimethyl ammonium bromide (CTAB; Sigma, St. Louis, MO, USA) followed by phenol and chloroform extractions [21]. DNA was quantitated using PicoGreen fluorescent dye (Molecular Probes, Eugene, OR, USA) in a fluorometer (BMG LabTechnologies, Offenburg, Germany). Stock DNA was stored at −70 °C and 5 ng/μl dilutions, made in TE buffer (10 mm Tris, 1 mm EDTA; pH 8·0), were stored at 4 °C. DNA was extracted from non-viable frozen cell suspensions as described for viable cells, except the volume was increased to ∼400 μl by addition of TE buffer. Cell suspensions were heated to 100 °C for 10 min to lyse any whole cells and genomic DNA purified. PCR products were amplified using Qiagen Master Mix (Qiagen, Hilden, Germany). Internal fragments of MLST genes were amplified using primers described previously [4] with the addition of primers to amplify fumC (fumC-A1: 5′-CAC CGA ACA CGA CAC GAT GG-3′ and fumC-A2: 5′-ACG ACC AGT TCG TCA AAC TC-3′). 16S rRNA PCR products were amplified using primers 8F and 1492R [18]. Each 25 μl reaction contained 1·5 mm MgCl2, 200 mm of each dNTP, two primers (at 1·0 μm for MLST genes and 0·4 μm for 16S rRNA), and 5 ng DNA. Reactions were cycled in a GeneAmp¯ 9700 thermocycler (Applied Biosystems, Foster City, CA, USA). Amplification products (2 μl) were run on a 2% agarose gel (Agarose LE, Roche, Mannheim, Germany) using 0·5× Tris-borate-EDTA (TBE) running buffer before staining in an ethidium bromide solution (1 μg/ml) and visualizing under UV light. The size of the amplification product was compared to a 100-bp molecular size marker (Invitrogen, Carlsbad, CA, USA).
MLRT
The genetic relatedness of a 15% sample (342 isolates) of serogroup B meningococci expressing the P1.4 PorA isolated from cases of meningococcal disease, 1990–2003, was examined by use of MLRT [16]. The MLRT profiles for abcZ, fumC, gdh, and pdhC alleles were assessed for all meningococci in the sample. These alleles were chosen for discriminating between ST-42, ST-154, ST-155, and ST-1460, which were the sequence types of four New Zealand strains deposited in the MLST database before this work commenced. The strains NZ91/40, NZ98/58, NZ98/53, and NZ98/172 had MLST database identification numbers 653, 1003, 1004, and 598 respectively. When abcZ, fumC, gdh, or pdhC multilocus restriction profiles differed from profiles found in ST-42, ST-154, or ST-155 meningococci, the restriction profiles for adk, aroE, and pgm were also assessed.
16S rRNA–RFLP
Restriction of 16S rRNA PCR products with MspI (New England BioLabs, Beverly, MA, USA) was carried out in 8 μl reactions containing 4·0 μl 16S rRNA PCR product, 1× NEBuffer 2, 4 U MspI, and DNase/RNase free water. Digests were incubated in a 37 °C water bath for 4 h. The 16S rRNA restriction profiles were determined by electrophoresing 8 μl of digestion product on a 2% agarose gel. The size of the digestion products was compared to a 100-bp molecular size marker (Invitrogen) and to products from meningococci with known 16S rRNA sequence (strains NZ91/49 and NZ93/8) [17].
Fluorescent-based sequencing and analysis
MLST was performed as described previously [4] with the addition of primers to sequence fumC (fumC-S1: 5′-TCG GCA CGG GTT TGA ACA GC-3′; fumC-S2: 5′-CAA CGG CGG TTT CGC GCA AC-3′). The 16S rRNA PCR product was sequenced using primers 8F and 1492R [17]. Sequencing was carried out using a 3100 Genetic Sequencer (Applied Biosystems). Sequence data analysis was carried out using the DNASTAR sequence analysis programs (DNASTAR Inc., Madison, WI, USA).
Statistical tests
The binomial confidence interval computation by the score method [22] was used to compare the proportions of samples when the characteristics assessed were not independent.
RESULTS
Retrospective molecular description of isolates prior to 1991
Retrospective serosubtyping of stored suspensions of isolates from 1988 and 1989 failed to identify any meningococci with the P1.7-2,4 PorA. However, five group B meningococci with PorA P1.7-2,4 were identified among isolates from 1990. Four were from cases in Auckland and the fifth was from the South Island. Three were PorB type 4 and restriction type (RT)-42, one was PorB type 14 and RT-41 and the South Island isolate was PorB type 4, but RT-154 Thus, all three subclones of the ST-41/44 complex (ST-41, ST-42, and ST-154) were present in 1990.
Case isolates identified in 1991
In 1991, meningococci from 16 cases of disease were defined as B:4:P1.7-2,4 and two as B:nt:P1.7-2,4. Of the 15 non-viable cell suspensions available for MLRT, one was RT-41, eight were RT-42, four were RT-154, and two had restriction profiles similar to RT-42 but differed at one locus.
MLRT analysis of sample of B:P1.4 meningococci isolated 1990–2003
Additional to isolates obtained during 1990–1991, a 15% sample (321 isolates) of case isolates obtained from 1992 to 2003 was analysed. The sample included a random 10% sample of meningococci with phenotype B:4:P1.4 (191 isolates) and a 50% sample of B:P1.4 meningococci with a PorB type other than type 4 (130 isolates). A sample of those with alternative PorB types was included to investigate their genetic relatedness to isolates typing as B:4:P1.4.
MLRT data showed that regardless of the PorB type, isolates expressing the P1.7-2,4 PorA showed little diversity in multilocus restriction type. RT-42 and RT-154 accounting for 51·5% (176/342) and 36·3% (124/342) of the sample respectively. Only 16 (4·7%) isolates sampled were RT-41. Of these, three were from Timaru, a town on the east coast of the South Island. Subsequent analysis of 11 other isolates from Timaru cases showed a further nine were RT-41. The 26 remaining isolates had restriction profiles that differed from RT-41, RT-42, and RT-154. These meningococci were mostly isolated after 1995. All except three (strains NZ99/24, NZ98/66, and NZ02/281) had MLRT profiles differing at only one of the seven MLST loci. Strains NZ99/24 and NZ02/281 differed at two of the seven loci when compared to RT-42. Strain NZ98/66 proved very unusual, differing at all seven MLST loci when compared to ST-42 and ST-154. The combination of alleles found in this meningococcus was assigned ST-2672 (see Neisseria Multi Locus Sequence Typing website: http://pubmlst.org/neisseria). Strain NZ98/66 was also unusual in that it contained a porB type distinct from types 4, 14, and 19,D,7,14a (VR1,VR2,VR3,VR4).
Correlation between the multilocus restriction type and porB type
Each of the PorB types 4, 14 and porB type19,D,7,14a, was associated with all three restriction types. However, from 1990 to 2003 meningococci with type 19,D,7,14a porB were more likely to be RT-42 [89% (95% CI 77–95)] than RT-154 [11% (95% CI 5–23)]. From 1990 to 1999 PorB type 14 was more likely to be RT-154 [75% (95% CI 62–92)] than RT-42 [25% (95% CI 9–38)]. From 2000 through 2003 PorB type 14 was common in RT-42 meningococci. However, when the total period 1990–2003 was considered there was no significant difference between the association of PorB type 14 with either RT-42 (95% CI 30–59) or RT-154 (95% CI 40–69).
Analysis using 16S rRNA typing
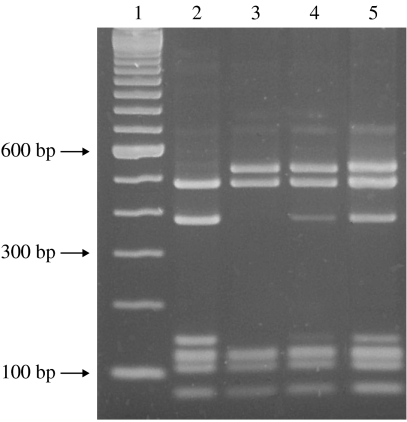
To indicate which subclone might have been the catalyst for New Zealand’s epidemic the 16S rRNA genes from meningococci with known MLRT were analysed. In previous work carried out by Sacchi and co-workers [17] two New Zealand case isolates (strains NZ91/49 and NZ93/8) given the designations NZ3966 and NZ3968 respectively were analysed by 16S rRNA sequencing. Both strain NZ91/49 and strain NZ93/8 were phenotype B:4:P1.7-2,4 but had different sequence types. Strain NZ91/49 was ST-2136, which differs from ST-154 at fumC, whereas strain NZ93/8 was ST-154. Their 16S rRNA sequences differed from each other by four substitutions at positions 717, 814, 984, and 1009, as defined by Sacchi et al. [17]. Digestion of the16S rRNA PCR product from strains NZ91/49 and NZ93/8 by MspI differentiated their 16S rRNA sequences (Fig.). The 16S rRNA amplicons from 148 meningococci with known MLRT were digested with MspI to assess differences in their 16S rRNA genes (Table). The sample included all meningococci with known MLRT isolated from 1990 to 1994 inclusive and every third meningococcus with known MLRT isolated from 1995 to 2003. Of the total, 91·2% (135/148) had the same 16S rRNA restriction profile as either strain NZ91/49 or NZ93/8 (Table). Most (58/59, 98·3%) meningococci with RT-154 or RT-154-like profiles had the same 16S rRNA restriction profile as strain NZ93/8. All nine meningococci with RT-41 or restriction profiles like RT-41 had the same 16S rRNA restriction profile as strain NZ93/8. Most meningococci (61/80, 76·3%) with RT-42 and restriction profiles like RT-42 had the same 16S rRNA restriction profile as strain NZ91/49 (Table). Seven RT-42 meningococci, with the same 16S rRNA restriction profile as strain NZ93/8 (ST-154), were confirmed by 16S rRNA sequencing as identical to strain NZ93/8 (Table). These seven meningococci were isolated in 1996, 1999, 2001 (two isolates), 2002, and 2003 (two isolates).
Fig.
16S rRNA RFLP patterns observed following gel electrophoresis of MspI digests of the 16S rRNA PCR product. Wells loaded with digested 16S rRNA PCR product was amplified from strains NZ91/49 (lane 2), NZ93/8 (lane 3), NZ91/24 (lane 4), and NZ90/21 (lane 5). Digestion products (lanes 2–5) were compared with molecular weight markers (Invitrogen) (lane 1).
Table.
Comparison of the 16S rRNA–RFLP and multilocus restriction type (MLRT) results in a sample of New Zealand case isolates obtained from 1990 to 2003
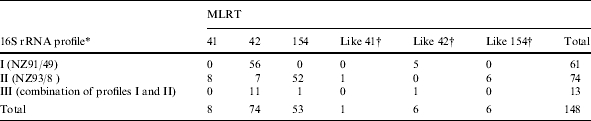
Profile I, same as 16S rRNA profile of NZ91/49 (ST-2136); Profile II, same as 16S rRNA profile of NZ93/8 (ST-154); Profile III, combination of profiles I and II (see Fig.).
Restriction profiles identical to those found in meningococci with RT-41, RT-42, or RT-154 except for one MLST locus.
Thirteen meningococci including strain NZ90/21, which was the first B:P1.7-2,4 case isolate identified in 1990, had different 16S rRNA restriction profiles to those found either in strain NZ91/49 or strain NZ93/8. Their restriction profiles appeared to be a combination of the 16S rRNA restriction profiles found in strains NZ91/49 and NZ93/8 (Fig.). All 13 had 16S rRNA restriction profiles similar to strain NZ93/8 (ST-154), although there was an additional band at 400 bp (Fig., lanes 4 and 5). For five of these strains this band was weak. For the remaining eight the 400-bp band had the same intensity as the two higher-molecular-weight bands. Sequence data from strain NZ90/21 showed that the data in positions 718, 815, 985, and 1010 were ambiguous (single amino-acid codes of Y, Y, R, and Y respectively). The nucleotides found in these positions in strains NZ91/49 and NZ93/8 were C, C, G, T and T, T, A, C respectively. The other seven meningococci had single amino-acid codes of C, C, G, T (strains NZ91/40 and NZ92/18) T, T, G, T (strain NZ00/189), Y, Y, R, Y (strain NZ96/4), T, T, R, T (strain NZ94/234), and T, T, R, Y (strain NZ00/134). The 16S rRNA data from strain NZ00/215 differed at a number of loci when compared to other New Zealand case isolates. These 16S rRNA sequences were submitted to GenBank database under accession numbers AY648028–AY648034.
DISCUSSION
The absence of B:P1.7-2,4 meningococci prior to 1990 and their identification in concert with increased levels of meningococcal disease, signalled the emergence of a new strain in New Zealand. Meningococci expressing the P1.7-2,4 PorA, accounted for 85·7% (2021/2358) of all group B meningococci isolated from cases of disease during 1990–2003 [14]. A similar rate continued to the end of 2004 (data not shown). Expression of the P1.7-2,4 PorA served as a marker of the ST-41/44 clonal complex with 99·7% (341/342) of B:P1.7-2,4 meningococci tested belonging to this complex. The exception was strain NZ98/66 which may have acquired DNA encoding the P1.7-2,4 PorA by horizontal transfer and recombination. Most isolates belonged to one of three subclones (ST-41, ST-42, ST-154) of the ST-41/44 clonal complex. Of these ST-42 was dominant.
Most B:P1.7b,4 meningococci expressed type 4 PorB although case isolates with porB other than type 4 have been identified throughout the epidemic. In a separate study it was shown that meningococci with alternative porB genes had either diverged from the original porB type 4 or horizontal transfer and recombination in the porB had occurred [15].
New Zealand’s meningococcal disease epidemic progressed in a similar way to that which occurred in The Netherlands in the early 1980s [10, 23]. Multilocus enzyme electrophoresis (MLEE) was used for clonal analysis of Dutch case isolates expressing the P1.7-2,4 PorA [24]. Initially isolates were defined as electrophoretic type, ET-24 of Lineage III but over time different ET types emerged [23]. A representative strain (BZ198, MLST database identification number 409) of ET-24 meningococci from The Netherlands is deposited in the MLST database and is defined as ST-41. The identification of all three subclones, ST-41, ST-42, and ST-154 in New Zealand in 1990 does not indicate which subclone may have initiated the epidemic. The identical 16S rRNA allele found in RT-41 and RT-154 meningococci suggests that these two subclones are closely related. The distinct 16S allele found in RT-42 meningococci suggests that this subclone is more distantly related. It is of note that strain NZ90/21, which was the first B:P1.7-2,4 identified in 1990 prior to the increase in case numbers, had a 16S rRNA restriction profile that appeared to be a combination of the profiles for isolates typing as either ST-154/ST41 and those typing as ST-42 (Fig.). Evidence for mutation and/or recombination in the meningococcal 16S rRNA gene has been described previously [17]. Heterogeneity within the four copies of the 16S rRNA gene in the meningococcal genome is described [25] and was attested to in this study by the presence of ambiguous nucleotides in 16S rRNA sequence data of some ST42 isolates (Table). A possible reason for the strong band at 400 bp in these isolates (Fig.) was that two copies of the nucleotide found in ST-42 and two copies of the nucleotide found in ST-154/ST-41 were present. Those meningococci with weak 400-bp bands may have had one copy of the allele found in ST-42 and three copies of the allele found in ST-154/ST-41 allowing the dominant allele to be recognized during sequencing. Sequence heterogeneity in multiple copies of the 16S rRNA gene in bacteria and phytoplasma have been described [26–28] including one bacterium with seven different 16S rRNA genes within its 15 copies [29].
The origin of the meningococcal clone or clones that initiated the epidemic in New Zealand is unknown. In the 1980s The Netherlands was the only country to have reported large numbers of cases of disease caused by meningococci belonging to the ST-41/ST-44 clonal complex [23]. The first isolate (NZ 90/21) identified in New Zealand in 1990 was typed as ST-42 but the isolate had an ambiguous 16S rRNA profile suggesting a relationship with all three MLST types (ST-41, ST-42 and ST-154). Both the second and fourth isolates obtained from cases in 1990 were typical of ST-42 and had the 16S rRNA pattern associated with ST-42. Isolate NZ90/39 had PorB type 14, was ST-41 and had a 16S rRNA pattern typical of other ST-41 strains (Table). The remaining 1990 isolate (NZ90/45) was from a case in the South Island. It was ST-154 with the associated 16S rRNA pattern. Evolution of ST-42 to ST-41 would require a single nucleotide substitution in abcZ, whereas evolution of ST-42 to ST-154 is less likely as it would require allelic variation in the gdh gene as well as the single nucleotide substitution in abcZ. Both would require horizontal transfer of DNA encoding the 16S rRNA gene. The only difference in the MLST alleles of ST-41 and ST-154 meningococci occurs at gdh, with nine base-pair (bp) changes found at single positions within the 440-bp-differentiating gdh-9 (ST-41) and gdh-11 (ST-154). This alteration could occur following genetic horizontal transfer and recombination. The fact that the ST-154 strain identified in 1990 expressed PorB type 14 also confounds with respect to the origin of the typical epidemic strain B:4:P1.7-2,4. It is possible that the isolates of subclones ST-41 and ST42 were both introduced into New Zealand in parallel in 1990 and that ST41 evolved to ST-154. This explanation does not account for the different PorB type. The possibility that genetic exchange could have occurred during community carriage prior to detection of clinical cases cannot be ruled out, particularly as four of the first five cases occurred in the same city during the second half of 1991.
ACKNOWLEDGEMENTS
We acknowledge the laboratories throughout New Zealand that submitted meningococci as part of the national surveillance of meningococcal disease undertaken on behalf of the Ministry of Health. The serological typing of referred isolates undertaken by staff of the Invasive Pathogens Laboratory at ESR is also acknowledged. This work was supported in part by a grant from Lotteries Health Research. Kristin Dyet was an ESR Ph.D. scholar. We thank Robin Simmonds, University of Otago, for his assistance in the early stages of this work. This research made use of the Neisseria Multi Locus Sequence Typing website (http://pubmlst.org/neisseria/) located at the University of Oxford and developed by M.-S. Chan and K. Jolley.
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Martin DR, Walker SJ, Baker MG, Lennon DR. New Zealand epidemic of meningococcal disease identified by a strain with phenotype B:4:P1.4. J Infect Dis. 1998;177:497–500. doi: 10.1086/517385. [DOI] [PubMed] [Google Scholar]
- 2.Dyet K, Devoy A, McDowell R, Martin D. New Zealand’s epidemic of meningococcal disease described using molecular analysis: implications for vaccine delivery. Vaccine. 2005;23:2236–2238. doi: 10.1016/j.vaccine.2005.01.050. [DOI] [PubMed] [Google Scholar]
- 3.Caugant DA. Population genetics and molecular epidemiology of Neisseria meningitidis. Acta Pathol Microbiol Immunol Scand. 1998;106:505–525. [PubMed] [Google Scholar]
- 4.Maiden MC, Bygraves JA, Feil E et al. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci. 1998;95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Caugant D, Bol P, Houby EA, Zanen HC, Froholm LO. Clones of serogroup B Neisseria meningitidis causing systemic disease in the Netherlands, 1985–1986. J Infect Dis. 1998;162:867–874. doi: 10.1093/infdis/162.4.867. [DOI] [PubMed] [Google Scholar]
- 6.Caugant D, Froholm L, Bovre K, Holten E. Intercontinental spread of a genetically distinctive complex of clones of Neisseria meningitidis causing epidemic disease. Proc Natl Acad Sci USA. 1998;83:4927–4931. doi: 10.1073/pnas.83.13.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cruz C, Pavez G, Aguilar E et al. Serotype-specific outbreak of group B meningococcal disease in Iquique, Chile. Epidemiol Infect. 1990;105:119–126. doi: 10.1017/s0950268800047713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sacchi CT, Pessoa LL, Ramos SR et al. Ongoing group B Neisseria meningitidis epidemic in Sao Paulo, Brazil, due to increased prevalence of a single clone of the ET-5 complex. J Clin Microbiol. 1992;30:1734–1738. doi: 10.1128/jcm.30.7.1734-1738.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diermayer M, Hedberg K, Fischer M, Perkins B, Reeves M, Fleming D. Epidemic serogroup B meningococcal disease in Oregon, the evolving epidemiology of the ET-5 strain. J Amer Med Assoc. 1999;281:1493–1497. doi: 10.1001/jama.281.16.1493. [DOI] [PubMed] [Google Scholar]
- 10.Scholten RJ, Bijlmer HA, Poolman JT et al. Meningococcal disease in The Netherlands, 1958–1990: a steady increase in the incidence since 1982 partially caused by new serotypes and subtypes of Neisseria meningitidis. Clin Infect Dis. 1993;16:237–246. doi: 10.1093/clind/16.2.237. [DOI] [PubMed] [Google Scholar]
- 11.Van Looveren M, Vandamme P, Hauchecorne M et al. Molecular epidemiology of recent Belgian isolates of Neisseria meningitidis serogroup B. J Clin Microbiol. 1998;36:2828–2834. doi: 10.1128/jcm.36.10.2828-2834.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wedege E, Dalseg R, Caugant DA, Poolman JT, Frøholm LO. Expression of an inaccessible P1.7 subtype epitope on meningococcal class 1 proteins. J Med Microbiol. 1993;38:23–28. doi: 10.1099/00222615-38-1-23. [DOI] [PubMed] [Google Scholar]
- 13.van der Ley P, van der Biezen J, Poolman JT. Construction of Neisseria meningitidis strains carrying multiple chromosomal copies of the porA gene for use in the production of a multivalent outer membrane vesicle vaccine. Vaccine. 1995;13:401–407. doi: 10.1016/0264-410x(95)98264-b. [DOI] [PubMed] [Google Scholar]
- 14.Devoy AF, Dyet KH, Martin DR. Stability of PorA during a meningococcal disease epidemic. J Clin Microbiol. 2005;43:832–837. doi: 10.1128/JCM.43.2.832-837.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dyet K, Martin D. Sequence variation in the porB gene from B:P1.7-2,4 meningococci causing New Zealand’s epidemic. J Clin Microbiol. 2005;43:838–842. doi: 10.1128/JCM.43.2.838-842.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dyet KH, Simmonds RS, Martin DR. Multilocus restriction typing method to predict the sequence type of meningococci. J Clin Microbiol. 2004;42:1742–1745. doi: 10.1128/JCM.42.4.1742-1745.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sacchi CT, Whitney AM, Reeves MW, Mayer LW, Popovic T. Sequence diversity of Neisseria meningitidis 16S rRNA genes and use of 16S rRNA gene sequencing as a molecular subtyping tool. J Clin Microbiol. 2002;40:4520–4527. doi: 10.1128/JCM.40.12.4520-4527.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abdillahi H, Poolman JT. Neisseria meningitidis group B serosubtyping using monoclonal antibodies in whole cell ELISA. Microb Pathog. 1988;4:27–32. doi: 10.1016/0882-4010(88)90045-9. [DOI] [PubMed] [Google Scholar]
- 19.Dyet KH, Simmonds RS, Martin DR. Evaluation of porB PCR-amplicon restriction endonuclease analysis as a method to determine porB variable region sequences in nonserotypable meningococci. J Clin Microbiol. 2004;42:1731–1733. doi: 10.1128/JCM.42.4.1731-1733.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saunders NB, Zollinger WD, Rao VB. A rapid and sensitive PCR strategy employed for amplification and sequencing of porA from a single colony-forming unit of Neisseria meningitidis. Gene. 1993;137:153–162. doi: 10.1016/0378-1119(93)90001-j. [DOI] [PubMed] [Google Scholar]
- 21.Moore E, Arnscheidt A, Krüger A, Strömpl C, Mau M., Akkermans ADL, van Elsas JD, de Bruijn FJ. Molecular microbial ecology manual. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1999. Simplified protocols for the preparation of genomic DNA from bacterial cultures; pp. 1–15. [Google Scholar]
- 22.Vollsett SE. Confidence intervals for a binomial proportion. Stat Med. 1993;12:809–824. doi: 10.1002/sim.4780120902. [DOI] [PubMed] [Google Scholar]
- 23.Scholten RJ, Poolman JT, Valkenburg HA, Bijlmer HA, Dankert J, Caugant DA. Phenotypic and genotypic changes in a new clone complex of Neisseria meningitidis causing disease in The Netherlands, 1958–1990. J Infect Dis. 1994;169:673–676. doi: 10.1093/infdis/169.3.673. [DOI] [PubMed] [Google Scholar]
- 24.Selander R, Caugant D, Ochman H, Musser J, Gilmour M, Whittam T. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl Environ Microbiol. 1986;51:873–884. doi: 10.1128/aem.51.5.873-884.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tettelin H, Saunders NJ, Heidelberg J et al. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science. 2000;287:1809–1815. doi: 10.1126/science.287.5459.1809. [DOI] [PubMed] [Google Scholar]
- 26.Liefting LW, Andersen MT, Beever RE, Gardner RC, Forster RLS. Sequence heterogeneity in the two 16S rRNA genes of Phormium yellow leaf phytoplasma. Appl Environ Microbiol. 1996;62:3133–3139. doi: 10.1128/aem.62.9.3133-3139.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mylvaganam S, Dennis PP. Sequence heterogeneity between the two genes encoding 16S rRNA from the halophilic archaebacterium Haloarcula marismortui. Genetics. 1992;130:399–410. doi: 10.1093/genetics/130.3.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reischl U, Feldmann K, Naumann L et al. 16S rRNA sequence diversity in Mycobacterium celatum strains caused by presence of two different copies of 16S rRNA gene. J Clin Microbiol. 1998;36:1761–1764. doi: 10.1128/jcm.36.6.1761-1764.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bosshard PP, Zbinden R, Altwegg M. Paenibacillus turicensis sp. nov., a novel bacterium harbouring heterogeneities between 16S rRNA genes. Int J Syst Evol Microbiol. 2002;52:2241–2249. doi: 10.1099/00207713-52-6-2241. [DOI] [PubMed] [Google Scholar]