SUMMARY
The subtypes of Campylobacter isolates from human infections in two Danish counties were compared to isolates from retail food samples and faecal samples from chickens, pigs and cattle. During a 1-year period, 1285 Campylobacter isolates from these sources were typed by two methods: ‘Penner’ heat-stable serotyping and automated ribotyping (RiboPrinting). C. jejuni was the dominating species, but C. coli was more prevalent among food and chicken isolates (16%) compared to human isolates (4%). In total, 356 different combined sero-ribotypes (subtypes) were found. A large subtype overlap was seen between human isolates and isolates from food (66%), chickens (59%) and cattle (83%). This was verified by PFGE typing of 212 isolates representing selected subtypes. All frequent (n>3) subtypes found in food were also present in humans. Sixty-one per cent of the isolates from domestically acquired infections had subtypes that were also found in food as opposed to 31% of travel-associated infections. The results showed differences in the various Campylobacter populations, e.g. the Danish population as reflected in the domestically acquired infections and the Danish-produced food was more uniform than the isolates originating from outside the country. The study shows that most C. jejuni subtypes found in poultry food samples, broiler chickens, and cattle were represented in the domestically acquired cases, indicating that C. jejuni from these reservoirs are likely sources of human infections in Denmark.
INTRODUCTION
Campylobacter is a common bacterial enteropathogen in developed countries. In recent years, this pathogen has been the most frequently isolated bacterial pathogen from cases of gastroenteritis in Denmark with an incidence of ∼82 cases/100 000 [1]. Campylobacter spp. are widespread in the environment and are present in the intestinal tract of many mammalian species and birds, including domestic farm animals such as cattle, sheep, pigs, and poultry, and also pets like cats and dogs. Few general outbreaks of campylobacteriosis are reported, and most infections are, therefore, assumed to be sporadic. However, on the basis of subtyping results, we found that 25% of culture-confirmed cases in two counties in Denmark were part of clusters, suggesting common sources of infection (V. Fussing et al., unpublished observations).
Despite the high number of human cases of campylobacteriosis the knowledge of the pathogenesis and epidemiology of Campylobacter infections are still incomplete. The small number of identified outbreaks has typically been traced to contaminated milk or water [2]. The source of infection remains unidentified for most sporadic cases, although most Campylobacter case-control studies have pointed at handling or consumption of raw or undercooked poultry as risk factors. Other risk factors have inconsistently been pointed out, e.g. untreated water, raw milk, and contact with pets or farm animals [2]. A Danish case-control study on sporadic campylobacteriosis identified the following risk factors: foreign travel, daily contact with domestic animals, and consumption of undercooked poultry, red meat at barbecues, and unpasteurized milk [3]. Together these risk factors could account for ∼42% of the cases, leaving the majority of cases unexplained. Microbiological studies strengthen the results of the epidemiological studies. Phenotypic and molecular typing methods are useful tools in studying the epidemiology of bacterial infections as identical subtypes of clinical isolates indicate a common source of infection. Furthermore, identical subtypes of isolates from food and clinical isolates within a limited time-frame suggest a possible source of infection. Specifically for Campylobacter, the usefulness of subtyping has been hampered by the lack of clearly host-specific subtypes and the apparent lack of relation between subtype and clinical manifestations in humans [4–6]. Recent studies using multi-locus sequence typing (MLST) of house-keeping genes, however, indicated that there might be some host-specific sequence type complexes [7].
Previously, mostly small-scale investigations have been carried out for comparing Campylobacter subtypes of clinical isolates with isolates from one or more possible sources, such as food, chickens, veterinary cases, and water samples [8–11]. Recently a large-scale study was reported from England showing the distribution of serotypes among human clinical isolates over a 2-year period and the relationship between risk exposures and serotypes [12]. However, in this study no food or animal isolates were included.
Here, we present the results of a 1-year study of Campylobacter subtypes in food samples and animal faecal samples compared to subtypes of human clinical isolates in two geographically distinct areas of Denmark. The patients’ home county, time of disease onset, and recent travel activity as well as the time for the food product to be found in retail stores were taken into consideration.
METHODS
Sampling and isolation
As part of a national surveillance programme, 2084 food samples from retail outlet stores were analysed for Campylobacter by the regional veterinary and food control authorities. Imported as well as Danish food products were sampled. The geographic regions included in this study were covered by two regional laboratories, i.e. one covering Funen county and the other covering three counties of the greater Copenhagen area including Copenhagen county. Samples were enriched under microaerobic conditions for 24 h at 42°C in Mueller–Hinton broth supplemented with 0·25 mg/l sodium pyrovate, 0·25 mg/l sodium metabisulphite, 0·25 mg/l ferrosulphate, 30 mg/l cefaperazone, and 50 mg/l trimethoprim lactate. A total of 10 μl of the enriched culture was streaked on modified charcoal cefoperazone deoxycholate agar (mCCDA) and incubated for 24–48 h at 42°C. One isolate from each positive food sample was included in the study.
In the study period, broiler chickens from farms located in Funen county were sampled at slaughter and analysed for Campylobacter. One isolate per flock was included (49 isolates). No commercial broiler farms were located in Copenhagen county. After the study period, C. jejuni isolates from cattle and pigs sampled at slaughterhouses located throughout Denmark were included in the study; these were sampled as part of the national Campylobacter surveillance programme as described previously [13]. All available C. jejuni isolates from cattle sampled from spring 2001 to summer 2002 were included (58 isolates). As only a few C. jejuni isolates from pigs are obtained each year in this programme, isolates from 1999 to 2003 were included (26 isolates) to obtain a reasonable number of isolates from this source. As cattle and pig isolates were included in the study on different conditions, these data are not included in all data analysis (e.g. species distribution).
All human isolates from patients with a culture-confirmed Campylobacter infection in Copenhagen county (1 May 2001 to 10 June 2002) and Funen county (1 May 2001 to 30 April 2002) were included in the study.
Identification and subtyping
Isolates were identified to the species level by the use of simple biochemical tests [13]. Isolates considered to be other species than C. coli or C. jejuni were tested in a species-specific real-time PCR assay differentiating C. jejuni, C. coli, C. upsaliensis, and C. lari [14]. Hippurate-negative isolates with typical C. jejuni serotypes were also tested by PCR, and when positive in the C. jejuni PCR assay, they were assigned to C. jejuni.
Campylobacter isolates were typed by ‘Penner’ heat-stable serotyping (full set of 47 C. jejuni sera and 19 C. coli sera) and automated ribotyping (RiboPrinting, Qualicon, Wilmington, DE, USA) as previously described [11]. Selected isolates were PFGE typed [15] using the restriction enzyme SmaI and in some cases a second enzyme, KpnI; a reference strain was run on each gel for validation. Ribotypes were assigned by the use of the software integrated in the RiboPrinter. PFGE profiles were analysed by the use of BioNumerics software (version 3.0, Applied Maths, Sint Martens, Belgium). For designation of PFGE subtypes within sero-ribotypes, any band difference in the range of the fragments of the Salmonella Braenderup strain used as DNA marker (20·5–1135 kb) was considered significant.
RESULTS
Retail food
The 2084 analysed food samples comprised 777 raw beef samples, 643 pork samples, 460 chicken samples, and 204 turkey samples. The poultry products were sold as whole carcasses or different cuts of these, and both fresh and frozen products were included. A total of 180 thermophilic Campylobacter were obtained, of which, 67 isolates were from Funen and 113 were from Copenhagen area. The positive food samples consisted of 139 chicken samples (prevalence 38·7%), 39 turkey samples (prevalence 27·5%), and two pork samples (prevalence 0·3%). The majority of food isolates (113 isolates), were from Danish-produced food, whereas 64 isolates were from imported food from France, Italy and the United Kingdom (three food samples were of unknown origin). One hundred and forty-nine isolates were from fresh food products, 23 from frozen food, and eight from food without registered processing.
Animals
When the 1-year study period was completed, it was clear that very few non-poultry food isolates were obtained. Therefore, faecal isolates from cattle and pigs were included to represent the Campylobacter reservoirs of these food animals and thereby a potential exposure to humans. Only C. jejuni isolates were included as 95% of the human cases were caused by this species.
Humans
One isolate from each of 975 human clinical cases were included in the study. Of these, 646 were from Copenhagen and 329 from Funen.
Species distribution and typability
C. jejuni was by far the most prevalent Campylobacter species isolated from humans (95%), retail food (85%), and chickens (80%) (Table 1). The prevalence of C. coli was fourfold higher in imported food and travel-associated cases compared to Danish-produced food and domestic cases respectively. The prevalence of C. coli in Danish pigs was 95% and in Danish cattle 5% in the years 2001–2002 as determined in the national surveillance programme [1].
Table 1.
Description of isolates included in the study. Species distribution and number of sero-ribotypes among isolates from human cases, retail food samples (mainly chicken and turkey products) and broiler chickens at slaughter. No species distribution is given for pigs and cattle as isolates from these sources were selected differently (only C. jejuni isolates were included, and these were from the whole country)
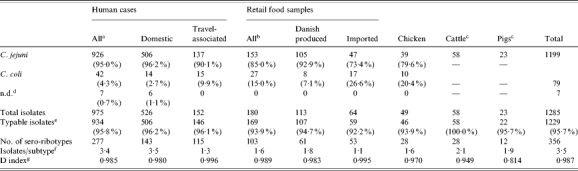
No information regarding travel was available for 297 of the human isolates.
For thee food isolates the country of origin was not registered.
Only C. jejuni were selected from cattle and pigs.
Not speciated or C. lari (one domestic human isolate).
Typable with both typing methods: serotyping and RiboPrinting.
Average number of isolates per subtype; subtype is defined as combined serotype and ribotype.
The discriminatory index, calculated as described by Hunter & Gaston [16], shows the probability of the typing system to assign different types to two unrelated strains.
In total 95·7% of all 1285 isolates were typable by the use of both serotyping and RiboPrinting. Cattle isolates (100%) and domestically acquired human isolates (96·2%) had the highest typability, and imported food products the lowest (92·2%) (Table 1). Serotyping had a typability of 97·6% and RiboPrinting 98·7%. Only isolates that were typable with both methods were assigned a combined sero-ribotype and included in the analyses using typing data.
Subtype diversity and overlap between sources
The most common C. jejuni serotypes were serotypes 2, the 4-complex, and 1,44 accounting for 58% of all isolates in the study (Table 2). A large number of different combined sero-ribotypes (defined ‘subtypes’ in the following) were found in all sources (Table 1). For all typable isolates of the study, the diversity index was 0·987 (D index as described by Hunter and Gaston [16]). The highest diversity was observed for travel-related human cases and imported food, and the lowest diversity was seen for isolates from pigs, cattle, and chickens (Table 1).
Table 2.
Serotype distribution for C. jejuni (12 most common serotypes) and number of ribotypes within each serotype (per cent of isolates in each category)
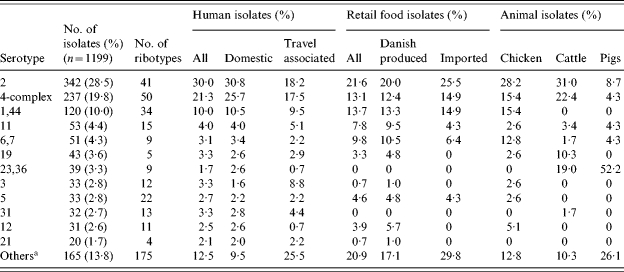
Includes isolates with non-typable serotype.
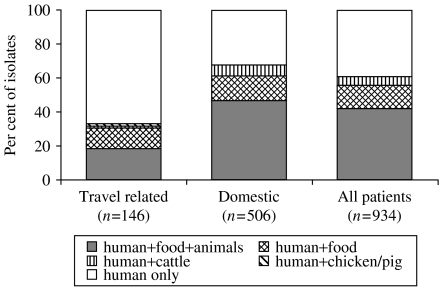
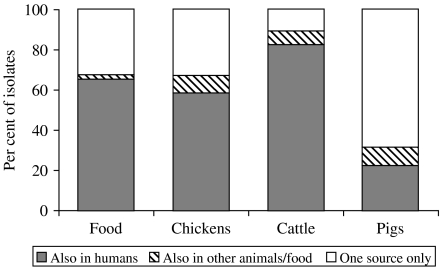
Fifty subtypes were present in both human and food isolates, representing 49% and 18% of all subtypes found in food isolates and human isolates respectively. These overlapping subtypes included 66% (111/169) of the food isolates and 56% (522/934) of the human isolates (Fig. 1). Subtyping of cattle isolates revealed a high degree of overlap between cattle and human isolates. Three-quarters (21/28) of the subtypes in cattle were also found among human isolates, representing 83% of the cattle isolates (Fig. 2). Notably, 6% of the domestic human isolates had subtypes that were isolated from cattle but not from food or other animals (Fig. 1). Most of the porcine C. jejuni isolates were serotypes 23,36 (52%) and 35 (22%). Both of these are rare serotypes in humans (<2%). The majority (65%) of chicken isolates had subtypes also present in the poultry food samples. The chicken isolates with non-overlapping subtypes showed an overrepresentation of C. coli (8/16), but a variety of different subtypes was found among these isolates.
Fig. 1.
Percentage of human isolates with subtypes (combined sero-ribotypes) found among human isolates only and isolates with subtypes also found in other sources: subtypes in humans, food, and at least one animal species; subtypes in humans and retail food; subtypes in humans and cattle; subtypes in humans and either chickens or pigs.
Fig. 2.
Percentage of food and animal isolates with subtypes (combined sero-ribotypes) also found in human patients, subtypes found in at least one other non-human source, and subtypes only found in one source.
Travel-associated human cases in relation to food and animal isolates
Clear differences were seen in the distribution of subtypes from cases that reported foreign travel prior to symptom onset and domestic cases. A larger overlap of subtypes to food and/or animals was observed for domestic cases (62%) than for travel-associated cases (31%) (Fig. 1). Isolates from travel-associated cases had a higher frequency of C. coli and a more even distribution of a large number of C. jejuni serotypes (Tables 1 and 2). Specifically, serotype 3 was significantly more common among travel-associated cases than domestic cases, and serotype 2 and 4-complex were less common in travel-associated cases (P<0·05, tested by χ2 test). Among all 1229 typable isolates, 233 isolates had a unique subtype, i.e. the subtype was only represented by one isolate from any of the sources. Forty-two per cent of the travel-associated cases had a unique type, whereas this was only the case for 12% of the domestic cases.
Subtypes exclusively found in either humans or food
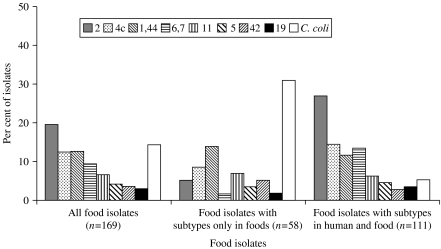
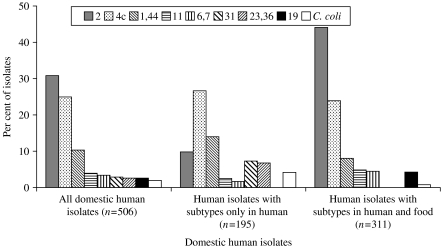
To further investigate possible differences between types found in domestically acquired human infections and food sold in Denmark, the subtypes were classified as ‘shared subtypes’ (subtypes found in both domestically acquired infections and food), ‘human subtypes’ (subtypes found in humans, but not in food), and ‘food subtypes’ (subtypes found in food, but not in humans). The total number of subtypes in this subset of data was 196: 41 of these were shared subtypes, 53 were food subtypes and 102 were human subtypes. Some noteworthy species and serotype differences were found between shared subtypes and subtypes specific for either humans or food (Figs 3 and 4). The food subtypes were dominated by C. coli (31% of the isolates). However, no combined sero-ribotype of either C. coli or C. jejuni was particularly prevalent among the food subtypes (1–3 isolates per subtype), and the isolates were almost evenly distributed among a large number of serotypes, i.e. most of the generally highly prevalent serotypes were underrepresented among the food subtypes (Fig. 3). This was in particular the case for serotype 2 as only 3% of the isolates with a food subtype were serotype 2 (compared to 20% of all typable food isolates). Likewise, serotype 2 was relatively infrequent (10%) among human subtypes compared to all human domestic isolates (31%) (Fig. 4). Serotypes 1,44, 31, and 23,36 were overrepresented among the human subtypes; the latter two serotypes were not found among the food isolates in this study.
Fig. 3.
Distribution of C. coli and the eight most common C. jejuni serotypes among all food isolates; food isolates with combined sero-ribotypes present among food (but not humans); and food isolates with sero-ribotypes present in both humans and food.
Fig. 4.
Distribution of C. coli and the eight most common C. jejuni serotypes among all domestic human isolates; domestic human isolates with combined sero-ribotypes present among human isolates (but not food); and domestic human isolates with sero-ribotypes present in both humans and food.
Confirmation of relationship by PFGE typing
All isolates assigned to eight sero-ribotypes represented by both human and food isolates were further genotyped by PFGE using one enzyme (SmaI), and isolates belonging to one specific subtype (serotype 6,7; ribotype C-13) were PFGE typed using an additional enzyme (KpnI) as the majority of these isolates were not digested by SmaI. In total, 212 isolates were PFGE typed, including 148 human isolates, 39 retail food isolates, 13 cattle isolates, 10 chicken isolates and 2 pig isolates. The number of isolates per sero-ribotype ranged from 13 to 56, and PFGE could further divide each of these into 3–18 distinct PFGE profiles. Seventy per cent of the human isolates had sero-ribo-PFGE types that were also found among at least one of the other sources. For the most common sero-ribotype of the study (serotype 2; ribotype C-2) only one of the 44 human isolates had a PFGE type identical to a food isolate, but 28 human isolates matched the profile of cattle isolates. A large majority of the retail food isolates (82%), broiler chicken isolates (100%), cattle isolates (62%), and pig isolates (50%) had PFGE profiles that were also found among isolates from patients. One of the two pork isolates of the study was part of a large cluster of C. jejuni isolates (serotype 6,7, ribotype C-13) and the pork isolate had the same PFGE type as 13 human isolates and nine retail poultry products and chicken isolates. The second pork isolate (C. jejuni serotype 1,44) was not PFGE typed, but three human isolates had the same sero-ribotype.
DISCUSSION
In this study the subtypes of Campylobacter isolates from food and food animals were compared to the subtypes of human clinical isolates. Two methods were used for subtyping a total of 1285 isolates. These methods, heat-stable serotyping and automated ribotyping, are methods that are well-suited for analysing a large number of isolates due to the ease of performance of both methods. Furthermore, serotyping is a definitive method, and RiboPrinting can as well be considered a definitive method in the sense that new profiles are directly compared to a database of stored profiles. The discriminatory power of these two methods in combination was high, but was further enhanced by the use of PFGE for clusters of sero-ribotypes. We previously compared the performance of six typing methods for C. jejuni and found that serotyping had a relatively low discriminatory power, RiboPrinting had a medium discriminatory power, and PFGE was among the most discriminatory of methods [11]. The methods were also found to give stable types during in vitro and in vivo passage of C. jejuni strains [17]. Because of the diverse nature of Campylobacter populations and a weak clonal structure due to frequent recombination [18–20] it is advantageous to use methods that are not too discriminatory. The applied methods in the present study were able to identify definitive subtypes that could easily be handled when comparing isolates from different sources. As the purpose of this study was to compare isolates with an unknown epidemiological linkage, only isolates that had an identical subtype were grouped together in the analysis. PFGE typing was used to study some of the sero-ribotypes in detail, and thereby further determine the likelihood of isolates being epidemiologically related.
The prevalence of Campylobacter in retail food products is generally found to be much higher for poultry products compared to pork and beef products [1, 21, 22] although cattle and pigs harbour Campylobacter in the intestinal tract at similar frequencies as poultry [13, 21]. In this study, 35% of retail poultry products, 0·3% of pork and 0% of beef samples were contaminated with Campylobacter. As so few Campylobacter isolates from non-poultry products were obtained, C. jejuni isolates from pig and cattle faecal samples at slaughter were included in the study to represent these Campylobacter reservoirs.
The distribution and overlap of Campylobacter sero-ribotypes among isolates from human patients, poultry products, chickens, cattle and pigs were analysed. Comparison of subtypes among poultry products and human isolates revealed a large degree of subtype overlap, as 66% of the food isolates and 56% of all human isolates had a subtype also found in humans and food respectively. This was further substantiated by PFGE typing as almost all PFGE-typed food and chicken isolates had profiles that were indistinguishable from profiles of human isolates. Other epidemiological data also support the assumption that poultry is an important source of human infections, e.g. case-control studies in developed countries indicating that 10–40% of the sporadic cases are caused by consumption of poultry [2], and the unintentional ‘intervention study’ performed in Belgium during the dioxin crisis (withdrawal of poultry products), which resulted in a 40% decrease in Campylobacter cases [23]. A large overlap was found between subtypes from chickens and retail poultry products, showing that the Campylobacter strains found in the intestines of poultry, as expected, also could be found in the food products of poultry origin. More C. coli was present in chickens than Danish-produced food, but the subtyping data could not indicate any chicken strains that seemed to be less able to survive the slaughter or processing environment.
Although 63% of the domestic human isolates belonged to subtypes found in retail food, several fairly common subtypes in domestically acquired infections were not represented among the food and chicken isolates. This could be due to the limited number of isolates included in the study or it could indicate that sources other than poultry are epidemiologically relevant for transmission of Campylobacter to humans. Several studies have pointed at other possible sources than poultry, including cattle and water, as reservoirs for human infections [7, 24, 25]. In this study, 83% of the C. jejuni isolates from cattle had sero-ribotypes also found among human isolates and in many cases PFGE typing confirmed this relationship. Therefore, despite the fact that a low prevalence of Campylobacter was found in beef products, it is possible that cattle and/or beef are sources of human infections. In contrast, a small overlap in subtypes was seen between human and pig isolates, and C. jejuni from Danish pigs are, therefore, not likely to be a significant source of human infections. Likewise, recent MLST results showed that most C. jejuni isolates from pigs in United Kingdom belonged to a sequence type complex that was not found in human isolates [26]. The possible role of the much more prevalent C. coli in pigs was not investigated in this study, but a recent British study showed that C. coli from poultry clustered separately from C. coli from pigs when typed by AFLP [27]. Unfortunately, no C. coli from humans were included in their study.
The subtypes were classified into three groups on the basis of their presence in food and domestically acquired infections: food subtypes, human subtypes and shared subtypes. The most common serotype in humans was serotype 2, and this serotype was equally common in food, chickens, and cattle. In addition, the ribotypes within serotype 2 were to a large extent the same in humans as in the other sources (shared subtypes). Thus, it is reasonable to assume that most of the serotype 2 strains found in the poultry and cattle reservoirs are pathogenic to humans. Prevalent food subtypes could be explained as Campylobacter subtypes that people are exposed to, but these do not cause disease (possible less virulent strains). The food subtypes were mostly C. coli, and this could indicate that C. coli generally are less pathogenic to humans than C. jejuni. Furthermore, the few C. coli isolates from domestically acquired human infections did not seem to originate from poultry food products. Some C. jejuni subtypes present in food were not found in humans, but each of these subtypes was only found with low frequency in food. Therefore, it would not be reasonable, based on this finding, to point out any specific C. jejuni strains that might be avirulent or less virulent to humans.
Travel-associated cases had a higher diversity of types than domestic cases, indicating that travel-associated cases were exposed to a wider range of Campylobacter strains than the domestic cases. Also, the travel-associated cases seem to be exposed to strains that are less common in Denmark. Interestingly, C. coli was more common among patients traveling abroad (10%) the week before onset of symptoms than among domestic cases (3%). C. coli was also more common in travel-associated cases in England [12]. This could indicate that C. coli is more common in the sources that people are exposed to during foreign travel – this is supported by the higher frequency of C. coli in imported food than in Danish-produced food.
A large diversity of subtypes was seen among Campylobacter isolates from human cases and possible sources and reservoirs of Campylobacter. It is well-known that Campylobacter typing by the methods used in this study or other hitherto described typing methods are not suited for attribution analysis of Campylobacter cases. The reason is the lack of host specificity and the weak clonal structure of Campylobacter. Although this study also documents a large diversity of Campylobacter, the results clearly show that there are differences in the various populations of Campylobacter. Notably, the Danish population of Campylobacter as reflected in the domestically acquired cases and the Danish-produced poultry products are more uniform than the Campylobacter populations outside Denmark, as isolates from travel-associated cases and imported food clearly show a higher diversity of subtypes. The distribution of C. jejuni subtypes present in pigs is distinctly different from the distribution in other sources. Campylobacter spp. in pigs are usually dominated by C. coli – also a distinct difference from the other reservoirs where C. coli is the minority species. Therefore, it is not likely that pigs are important reservoirs for C. jejuni causing infections in Denmark. However, the significant overlap of subtypes in poultry food products, chicken, and cattle with subtypes in domestically acquired human cases show that C. jejuni from these reservoirs are possible sources of human infection in Denmark.
ACKNOWLEDGEMENTS
Numerous people have helped us to carry out this study. We give special thanks to the technicians at the two laboratories who performed the subtyping of isolates: Sussi Kristoffersen and Sidsel Boesen at the Danish Veterinary Institute, and Louise Storm and Michael E. Petersen at the Statens Serum Institut. We are also grateful to the two regional veterinary and food control authorities Funen and Northeast-Sealand for providing the food isolates. Peter Gerner-Smidt has kindly critically read the manuscript.
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Annual Report on Zoonoses in Denmark 2002. Copenhagen. Denmark: Danish Zoonosis Centre, Ministry of Food, Agriculture and Fisheries; 2003. [Google Scholar]
- 2.Friedman CR, Neimann J, Wegener HC, Tauxe RV., Nachamkin I, Blaser MJ. Campylobacter. 2nd edn. Washington, DC: ASM Press; 2000. Epidemiology of Campylobacter jejuni infections in the United States and other industrialized nations; pp. 3–26. [Google Scholar]
- 3.Neimann J, Engberg J, Molbak K, Wegener HC. A case-control study of risk factors for sporadic campylobacter infections in Denmark. Epidemiol Infect. 2003;130:353–366. doi: 10.1017/s0950268803008355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Endtz HP, Ang CW, van Den BN et al. Molecular characterization of Campylobacter jejuni from patients with Guillain–Barre and Miller–Fisher syndromes. J Clin Microbiol. 2000;38:2297–2301. doi: 10.1128/jcm.38.6.2297-2301.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Engberg J, Nachamkin I, Fussing V et al. Absence of clonality of Campylobacter jejuni in serotypes other than HS:19 associated with Guillain–Barré syndrome and gastroenteritis. J Infect Dis. 2001;184:215–220. doi: 10.1086/322010. [DOI] [PubMed] [Google Scholar]
- 6.Jackson CJ, Fox AJ, Wareing DR, Sutcliffe EM, Jones DM. Genotype analysis of human blood isolates of Campylobacter jejuni in England and Wales. Epidemiol Infect. 1997;118:81–89. doi: 10.1017/s0950268896007388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schouls LM, Reulen S, Duim B et al. Comparative genotyping of Campylobacter jejuni by amplified fragment length polymorphism, multilocus sequence typing, and short repeat sequencing: strain diversity, host range, and recombination. J Clin Microbiol. 2003;41:15–26. doi: 10.1128/JCM.41.1.15-26.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanninen ML, Perko-Makela P, Pitkala A, Rautelin H. A three-year study of Campylobacter jejuni genotypes in humans with domestically acquired infections and in chicken samples from the Helsinki area. J Clin Microbiol. 2000;38:1998–2000. doi: 10.1128/jcm.38.5.1998-2000.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hudson JA, Nicol C, Wright J, Whyte R, Hasell SK. Seasonal variation of Campylobacter types from human cases, veterinary cases, raw chicken, milk and water. J Appl Microbiol. 1999;87:115–124. doi: 10.1046/j.1365-2672.1999.00806.x. [DOI] [PubMed] [Google Scholar]
- 10.Nadeau E, Messier S, Quessy S. Prevalence and comparison of genetic profiles of Campylobacter strains isolated from poultry and sporadic cases of campylobacteriosis in humans. J Food Prot. 2002;65:73–78. doi: 10.4315/0362-028x-65.1.73. [DOI] [PubMed] [Google Scholar]
- 11.Nielsen EM, Engberg J, Fussing V, Petersen L, Brogren CH, On SL. Evaluation of phenotypic and genotypic methods for subtyping Campylobacter jejuni isolates from humans, poultry, and cattle. J Clin Microbiol. 2000;38:3800–3810. doi: 10.1128/jcm.38.10.3800-3810.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sopwith W, Ashton M, Frost JA et al. Enhanced surveillance of campylobacter infection in the North West of England 1997–1999. J Infect. 2003;46:35–45. doi: 10.1053/jinf.2002.1072. [DOI] [PubMed] [Google Scholar]
- 13.Nielsen EM, Engberg J, Madsen M. Distribution of serotypes of Campylobacter jejuni and C. coli from Danish patients, poultry, cattle and swine. FEMS Immunol Med Microbiol. 1997;19:47–56. doi: 10.1111/j.1574-695X.1997.tb01071.x. [DOI] [PubMed] [Google Scholar]
- 14.Jensen AN, Andersen MT, Dalsgaard A, Baggesen DL, Nielsen EM. Development of real-time PCR and hybridization methods for detection of thermophilic Campylobacter spp. in pig faecal samples. J Appl Microbiol. doi: 10.1111/j.1365-2672.2005.02616.x. (in press). [DOI] [PubMed] [Google Scholar]
- 15.Ribot EM, Fitzgerald C, Kubota K, Swaminathan B, Barrett TJ. Rapid pulsed-field gel electrophoresis protocol for subtyping of Campylobacter jejuni. J Clin Microbiol. 2001;39:1889–1894. doi: 10.1128/JCM.39.5.1889-1894.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hunter PR, Gaston MA. Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. J Clin Microbiol. 1988;26:2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nielsen EM, Engberg J, Fussing V. Genotypic and serotypic stability of Campylobacter jejuni strains during in vitro and in vivo passage. Int J Med Microbiol. 2001;291:379–385. doi: 10.1078/1438-4221-00136. [DOI] [PubMed] [Google Scholar]
- 18.Boer P, Wagenaar JA, Achterberg RP, Putten JP, Schouls LM, Duim B. Generation of Campylobacter jejuni genetic diversity in vivo. Mol Microbiol. 2002;44:351–359. doi: 10.1046/j.1365-2958.2002.02930.x. [DOI] [PubMed] [Google Scholar]
- 19.Dingle KE, Colles FM, Wareing DR et al. Multilocus sequence typing system for Campylobacter jejuni. J Clin Microbiol. 2001;39:14–23. doi: 10.1128/JCM.39.1.14-23.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Suerbaum S, Lohrengel M, Sonnevend A, Ruberg F, Kist M. Allelic diversity and recombination in Campylobacter jejuni. J Bacteriol. 2001;183:2553–2559. doi: 10.1128/JB.183.8.2553-2559.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pezzotti G, Serafin A, Luzzi I, Mioni R, Milan M, Perin R. Occurrence and resistance to antibiotics of Campylobacter jejuni and Campylobacter coli in animals and meat in northeastern Italy. Int J Food Microbiol. 2003;82:281–287. doi: 10.1016/s0168-1605(02)00314-8. [DOI] [PubMed] [Google Scholar]
- 22.Zhao C, Ge B, De Villena J et al. Prevalence of Campylobacter spp., Escherichia coli, and Salmonella serovars in retail chicken, turkey, pork, and beef from the Greater Washington, DC, area. Appl Environ Microbiol. 2001;67:5431–5436. doi: 10.1128/AEM.67.12.5431-5436.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vellinga A, Van Loock F. The dioxin crisis as experiment to determine poultry-related campylobacter enteritis. Emerg Infect Dis. 2002;8:19–22. doi: 10.3201/eid0801.010129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacobs-Reitsma W., Nachamkin I, Blaser MJ. Campylobacter. 2nd edn. Washington, DC: ASM Press; 2000. Campylobacter in the food supply; pp. 467–481. [Google Scholar]
- 25.Siemer BL, Harrington CS, Nielsen EM et al. Genetic relatedness among Campylobacter jejuni serotyped isolates of diverse origin as determined by numerical analysis of amplified fragment length polymorphism (AFLP) profiles. J Appl Microbiol. 2004;96:795–802. doi: 10.1111/j.1365-2672.2004.02205.x. [DOI] [PubMed] [Google Scholar]
- 26.Manning GA, Dowson CG, Bagnall MC, Ahmed IH, West M, Newell DG. Multilocus sequence typing for comparison of veterinary and human isolates of Campylobacter jejuni. J Clin Microbiol. 2003;69:6370–6379. doi: 10.1128/AEM.69.11.6370-6379.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hopkins KL, Desai M, Frost JA, Stanley J, Logan JMJ. Fluorescent amplified fragment length polymorphism genotyping of Campylobacter jejuni and Campylobacter coli strains and its relationship with host specificity, serotyping, and phage typing. J Clin Microbiol. 2004;42:229–235. doi: 10.1128/JCM.42.1.229-235.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]