SUMMARY
This study investigates the distribution of pulsed-field gel electrophoresis (PFGE) profiles within Salmonella enterica serotype Enteritidis phage type (PT) 4 and S. Typhimurium definitive phage type (DT) 104, from cases of human infection in nine European countries from 2000 to 2004. Isolates were subtyped using standardized methods and gel images submitted by each participating country to the coordinating centre (Health Protection Agency Centre for Infections, London, UK), where they were entered into a central database, developed within BioNumerics software, and designated using an agreed nomenclature. S. Enteritidis PT4 (n=3637) was differentiated into 38 different profiles. Simpson’s index of diversity (D) of profiles ranged from 0·2 to 0·4. Profile SENTXB.0001 represented at least 80% of all profiles in each country. S. Typhimurium DT104 (n=1202) was differentiated into 28 different profile types. Simpson’s D was at least 0·6 in all countries except in Austria and Italy. In both these countries over 74% of S. Typhimurium DT104 profiles were STYMXB.0013. Profile STYMXB.0061, was predominant in Denmark, Spain, Finland and England & Wales where it represented between 36% and 45% of profiles. Profile STYMXB.0001 represented nearly half of all profiles in Scotland and 23% in England & Wales. PFGE is proving useful for further discrimination within S. Enteritidis PT4 and S. Typhimurium DT104. Ascertainment of international outbreaks involving common serotypes and phage types may be increased by the timely pooling of PFGE profiles within a central database readily accessible to all participating countries.
INTRODUCTION
The prompt ascertainment and investigation of outbreaks is important in the national and international prevention and control of salmonellosis. The International Surveillance Network for Enteric Infections (Enter-net), which encompasses 17 European and five non-European countries, was established to monitor human gastrointestinal infections caused by Salmonella and Vero cytotoxin-producing Escherichia coli (VTEC) O157, including their antimicrobial resistance [1]. For salmonellosis, the primary methods for differentiation of strains within Europe are serotyping, followed by phage typing for epidemiologically important serotypes [2], and antibiogram determination. These methods have been harmonized in all participating countries and the results pooled in a timely manner with other relevant information. These processes have been key to the success of Enter-net, which has detected and aided in the investigation of numerous international outbreaks [3].
Salmonella enterica serotypes Enteritidis and Typhimurium are the most commonly reported serotypes from cases of human infections in Europe. Between 2000 and 2002, these two serotypes represented 53% and 20% respectively of the 232 442 strains reported to Enter-net. Increasingly, subdivision within phage types is becoming necessary to elucidate outbreaks during investigations, whether these are national or international. Since the 1990s some phage types of epidemiological importance have predominated. Of all strains of S. Enteritidis phage typed during this period (n=87 962), 46% were phage type (PT) 4 and of the S. Typhimurium strains (n=18 758), 33% were definitive phage type (DT) 104 (Enter-net, unpublished data). For these phage types, any geographically or temporally scattered epidemiologically linked cases may go unnoticed or may be detected late. It is, therefore, necessary to consider further discrimination within common phage types for meaningful surveillance. Pulsed-field gel electrophoresis (PFGE) is currently the gold standard method for molecular subtyping of Salmonella [4, 5], and possibly also for the rapid identification of unusual Salmonella serotypes. The method involves cutting genomic DNA at specific sites to generate fragments of different sizes followed by determination of their molecular weights by running them through an electrically charged gel matrix.
Salm-gene, a EU research project, was established to evaluate the added value of using molecular subtyping tools (PFGE essentially) for outbreak recognition of S. Enteritidis and S. Typhimurium, within the existing international surveillance of Salmonella. The project involved the participation of nine national reference laboratories in Europe: Austria, Denmark, Finland, Germany, Italy, The Netherlands, Scotland, Spain, England & Wales with France acting as software compatibility advisor.
We describe the distribution of pulsed-field profiles (PFPs) within isolates of S. Enteritidis PT4 and S. Typhimurium DT104 reported to Salm-gene from the nine participating European countries between January 2000 and September 2004.
MATERIALS AND METHODS
Strain source
All strains from cases of human salmonellosis in the nine countries that were referred to the national reference laboratory were eligible for subtyping using PFGE. Strains included any sporadic isolates and outbreak strains, with the proviso that only a single isolate from any outbreak within a country was represented.
Selection of strains
At the start of the project all nine laboratories were asked to subtype retrospectively, using PFGE, a selection of strains of the most common S. Enteritidis and S. Typhimurium phage types from isolates that had been reported to them up to December 2001. This created a library of unique strain profiles and provided the background of profiles in strains in circulation prior to the study. Strains were selected in the order they had been reported, starting from the most recent, and working back temporally until a commonly agreed target number was reached.
‘Real-time’ subtyping was implemented from January 2003 and required each laboratory to submit to the Salm-gene coordinating centre (Health Protection Agency, England & Wales), up to three PFGE gels a week, representing 20–45 isolates in all. A protocol for selecting strains to be subtyped was sent to all laboratories. This involved all isolates of S. Enteritidis and S. Typhimurium if the total number was less than the PFGE typing capacity. If the number of strains reported nationally exceeded local subtyping capacity, a simple random sampling method was applied. The sample number varied by serotype and was in proportion to the number of strains within each serotype. For example, assuming the typing capacity was 48 isolates, and 73 (70%) strains of S. Enteritidis and 31 (30%) strains of S. Typhimurium were reported to the national laboratory, 34 S. Enteritidis and 14 S. Typhimurium strains would be selected arbitrarily from these strains.
PFGE subtyping
As described previously laboratory procedures for PFGE were standardized in all nine laboratories using the Bio-Rad CHEF¯ system [6]. The following harmonized protocol was used for the studies described:
After overnight growth on TSA or nutrient broth, cells were harvested by centrifugation and resuspended in 500 μl cell suspension buffer [100 mm Tris, 100 mm EDTA (pH 8·0)]. The final cell density for plug preparation was 0·38–0·44 at 450 nm (McFarland No. 5). Proteinase K (0·5 mg/ml final concentration) was added to the cell suspension followed by mixing lysis of cell suspension 1:1 with agarose (Bio-Rad chromosomal grade). The resultant plugs were washed at least twice in distilled water and 2–3 times in TE buffer. Electrophoresis conditions were as follows: ramp initial 2 s; final 64 s; 6 V/cm; 14°C, 22 h (Bio-Rad CHEF DRII¯), 20 h (Bio-Rad CHEF DRIII¯), 18 h (Bio-Rad CHEF Mapper¯). DNA macrorestriction fragments were resolved on 1% agarose gels (Bio-Rad Pulsed-Field Certified¯ or Seakem Gold¯) with a S. Braenderup strain H9812 (kindly supplied by PulseNet USA, CDC [4]) as a molecular reference marker. To ensure compatibility the electrophoresis conditions were identical to those used by PulseNet USA.
Electronic submission of PFPs
Gel images were transmitted in tag image file format (TIFF) by email together with additional microbiological and epidemiological information on each strain, including phage type when available, to the Salm-gene coordinating centre. All gels that were considered to be of acceptable quality (in terms of image clarity, band definition and use of Braenderup reference profile strains) were entered and analysed into the Salm-gene database, created within the BioNumerics software (v. 3.1, beta, Applied Maths, Sint-Martens-Latem, Belgium). This involved naming each pattern based on a comparison with the profiles of the library (see below).
Profile designation
The designation involved assigning a six-letter code to the new pattern together with a four- digit numerical identifier [6]. The first pattern for S. Enteritidis digested with the enzyme XbaI was designated SENTXB.0001 and similarly for S. Typhimurium digested with the same enzyme, STYMXB.0001. A profile differing by at least one band to a pattern in the library was assigned a new name and added to the library.
Data analysis
All strains of S. Enteritidis and S. Typhimurium entered into the database up to 30 September 2004 were included in the analysis. This represents a subset of isolates reported to the national laboratories between January 2000 and September 2004.
The diversity of PFGE profiles was calculated using Simpson’s index of diversity (D) as described by Grundmann et al. [7]. The value of this index ranges between 0 and 1, with 0 indicating no diversity. The 95% confidence intervals were calculated based on the variance as suggested by Grundmann et al. Access 2000 (Microsoft, Washington, USA) was used for the extraction of data and Excel 2000 (Microsoft) was used for the calculation of Simpson’s D, variance and confidence intervals.
RESULTS
S. Enteritidis
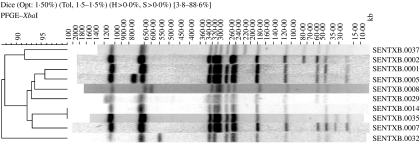
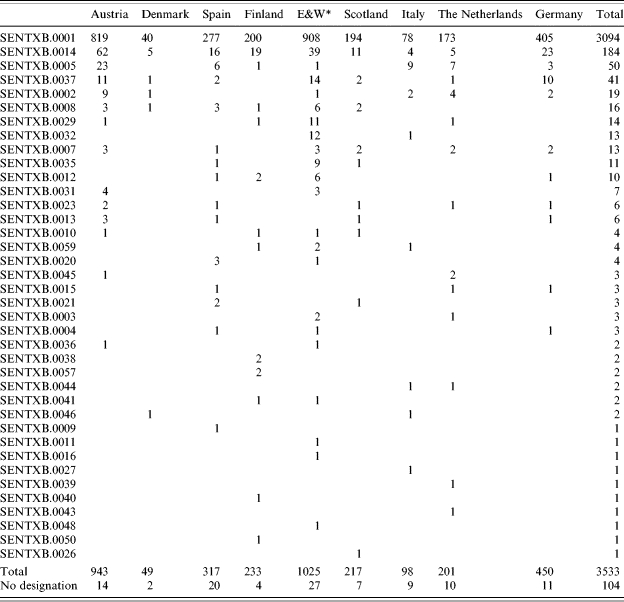
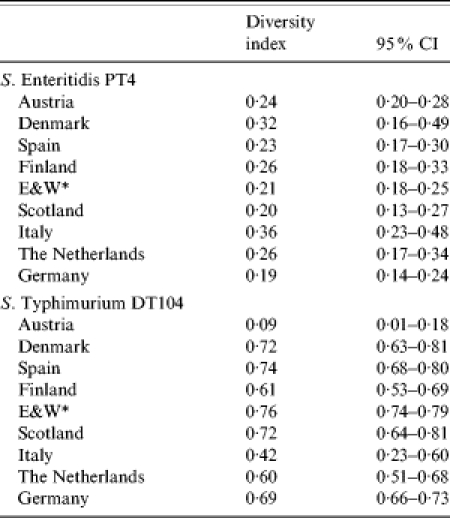
Of all strains of S. Enteritidis included in the Salm-gene database (n=11 659), 10 927 had a designated phage type, of which 3637 (33%) were PT4. Of the 3637 profiles, 3509 had been assigned a profile name by the database curator. Within the designated profiles S. Enteritidis PT4 isolates were differentiated into 38 different profile types, the 10 most common of which are illustrated in Figure 1. Profile SENTXB.0001 predominated in all countries representing at least 80% of all profiles (Table 1). Simpson’s D of profile types within PT4 ranged from 0·2 to 0·4 and was lowest in Austria, Spain, England & Wales, Scotland and Germany. Some countries had country-specific profile types. Finland had the greatest number of unique profiles (n=6), followed by England & Wales (n=3), The Netherlands (n=2), Denmark (n=1), Spain (n=1), Scotland (n=1) and Italy (n=1) (Table 1).
Fig. 1.
Ten most common profile types within S. Enteritidis PT4.
Table 1.
Number of S. Enteritidis PT4 profiles within each country
England & Wales.
S. Typhimurium
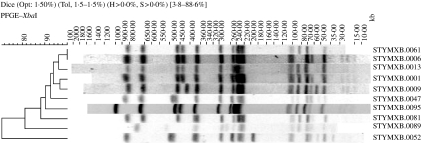
Overall, 5817 strains of S. Typhimurium were entered into the Salm-gene database. DT104 represented 26% (n=1202) of all strains with a designated phage type (n=4631). Of these DT104 strains, 1060 were assigned a PFGE profile type. Twenty-eight different profile types were identified, the 10 most common of which are shown in Figure 2. There were differences between countries in the distribution and diversity of profile types. Simpson’s D was at least 0·6 except in Austria and Italy (0·09 and 0·42 respectively) (Table 2). In both these countries the majority of profiles were STYMXB.0013. This profile was found in all other countries but more frequently in Germany and The Netherlands, where it represented 45% and 74% respectively of all profiles in these countries. Profile STYMXB.0061, was predominant in Denmark, Spain, Finland and England & Wales where it represented between 36% and 45% of profiles. Profile STYMXB.0001 represented nearly half of all profiles in Scotland and a high proportion of profiles (23%) in England & Wales (Table 3).
Fig. 2.
Ten most common profile types within S. Typhimurium DT104.
Table 2.
Diversity of profiles of S. Enteritidis PT4 and S. Typhimurium DT104 within each country
England & Wales.
Table 3.
Number of S. Typhimurium DT104 profiles within each country
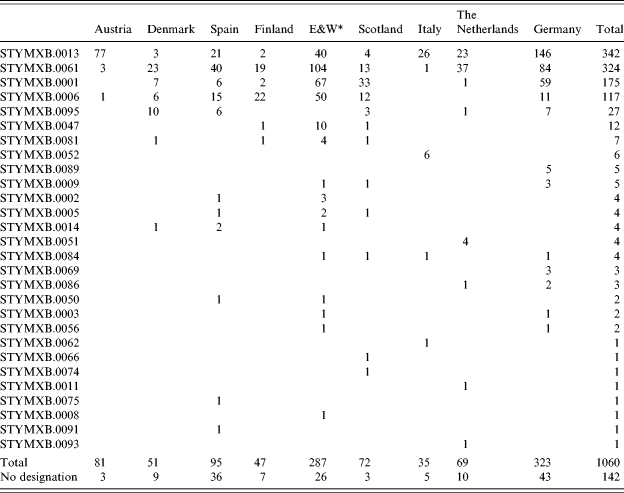
England & Wales.
A remaining 104 profiles of S. Enteritidis and 142 profiles of S. Typhimurium had no matching patterns in the library at the time of analysis. These strains could not be designated due to late submission and time constraints.
DISCUSSION
Phage typing, based on common protocols, is vital for improving outbreak ascertainment and for outbreak investigations of S. Enteritidis and S. Typhimurium. For common phage types within these serovars, such as PT4 and DT104 respectively, further discriminatory methods are necessary to differentiate these strains. Molecular typing methods are commonly used as a tool to help with outbreak investigations, PFGE being currently the gold standard for Salmonella. This method has been implemented through standardized protocols in the United States [4] and several countries in Europe, with the view of enhancing Salmonella outbreak ascertainment internationally.
Both PT4 and DT104 are recognized as highly clonal organisms [8]. As the present study has demonstrated, these phage types have been differentiated into several PFGE subtypes in all nine countries.
As can be seen from Figures 1 and 2, differences between profile types are mainly in the number and/or position of bands of high (450–1135 kb) and low (20–100 kb) molecular mass. In some cases the differences are minor (1–2 band difference). Although previous studies have concluded subtypes based on minor band differences may not be sufficiently discriminatory [9], minor band differences might be key in identifying outbreak strains and in their differentiation from sporadic cases [10, 11]. In this respect discrimination within phage type for investigating Salmonella outbreaks may be dependent on changes in a single band. This is contrary to the recommendations of Tenover et al. for other organisms [12]. Furthermore, the influence of plasmids may have some effect on PFGE profiles and whenever possible this should also be taken into account [11].
There was little variance with the genetic diversity of PT4 between the nine countries (Simpson’s D=0·2–0·4). This was in contrast to DT104. Countries with the highest diversity were Denmark, Finland, Germany, England & Wales, Scotland, The Netherlands and Spain (Simpson’s D=0·6–0·8) for DT104. The lower diversity observed in Austria (Simpson’s D=0·2) and Italy (Simpson’s D=0·4) may be due to fewer imported infections and/or fewer imports of contaminated food.
Certain profile types predominated within some countries. Within The Netherlands, Austria, Finland, Denmark, Germany, Italy, England & Wales, Scotland and Spain, the majority of profiles of S. Enteritidis PT4 were SENTXB.0001 although several other less common subtypes could also be observed. A similar distribution of profile patterns has been reported previously, although the restriction endonuclease used for PFGE was not always XbaI [13–15]. The predominant subtype SENTXB.0001 may be identical to those recognized in these studies. This is not unlikely, considering the rapidity by which this phage type is known to have spread internationally [8, 16]. Further comparisons using the same enzyme and similar gel running times are required before this can be verified.
Some neighbouring countries shared the same predominant profile of S. Typhimurium DT104; for example in Austria and Italy, profile STYMXB.0067 represented the majority of profiles (74 and 90% respectively). The profiles of DT104 shared by neighbouring countries, suggests they are sharing local reservoirs or sources of multi-drug resistant DT104. In contrast Denmark, Spain, Finland, England & Wales and The Netherlands STYMXB.0061 predominated representing at least 36% of all profiles within each country. This profile was less frequently found in Austria, Scotland, Italy and Germany. Multi-drug resistant DT104 is known to have disseminated globally [17] and it is highly probable that this clone is represented in these subtypes.
Similarities in the occurrence and distribution of subtypes were apparent between countries. For example Austria and Germany shared eight subtypes of S. Enteritidis PT4 out of the 14 and 11 profiles observed in these countries respectively. Twenty-six of the PT4 subtypes were shared by at least two countries. The other 38 were unique to the reporting country. Because of the very low number of these profiles (in many cases only one isolate of this type was reported), we are unable to make conclusive remarks regarding their specificity. The present study has provided a timely analysis, at molecular level, of a representative sample of human Salmonella isolates across Europe. The laboratory protocols used in Salm-gene have been developed in line with those of PulseNet USA. This is a national subtyping network for the USA for foodborne pathogens using PFGE [4] and similar networks have been extended in Canada, and recently in Asia and Latin America. The use of the S. Braenderup control strain H9812 in this study has ensured compatibility between the Salm-gene and PulseNet network. As part of the PulseNet Europe ring-trial, the PulseNet network has been extended to 28 institutes in 19 European countries within and out of Salm-gene (PulseNet Europe) in late 2004, and the Salm-gene database is providing the foundation of this network for Salmonella. Combined with case and phenotypic information, the rapid pooling of PFGE profiles into a central database is readily accessible to Enter-net, Salm-gene and PulseNet Europe participating countries. This will facilitate the timely ascertainment of international outbreaks involving common serotypes and phage types within Europe, and for countries with phage typing expertise, will also negate the expensive and time-consuming transfer of isolates through the postal system for subsequent molecular typing.
ACKNOWLEDGEMENTS
We are grateful to Mrs L. R. Ward of the HPA Laboratory of Enteric Pathogens for phage typing of strains of S. Enteritidis and S. Typhimurium described in this study (England & Wales), and supplying the necessary phage typing reagents (other countries); to Dr B. Swaminathan of the Centers for Disease Control and Prevention, Atlanta, USA, for supplying and authorizing the use of the PulseNet USA reference strain of S. Braenderup H9812, and to Professor P. Grimont of the Institut Pasteur, France for his advice on software compatibility issues. Thanks are also due to Neville Verlander of the HPA Centre for Infections, London, UK, for statistical support. The Salm-gene project was funded by the EC, DG RESEARCH within key Action No. 2 (Control of Infectious Diseases) of Framework Programme 6, Proposal No. QLRT-2000-01940.
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Fisher IST. The Enter-net international surveillance network – how it works. Eurosurveillance. 1999;4:52–55. doi: 10.2807/esm.04.05.00073-en. [DOI] [PubMed] [Google Scholar]
- 2.Threlfall EJ, Frost JA. The identification, typing and fingerprinting of Salmonella: laboratory aspects and epidemiological applications. Journal of Applied Bacteriology. 1990;68:5–16. doi: 10.1111/j.1365-2672.1990.tb02542.x. [DOI] [PubMed] [Google Scholar]
- 3.Fisher IST, Threlfall EJ. The Enter-net and Salm-gene databases of food-borne bacterial pathogens causing human infections in Europe and beyond: an international collaboration in the development of intervention strategies. Epidemiology and Infection. 2005;133:1–7. doi: 10.1017/s095026880400305x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swaminathan B et al. PulseNet: the molecular subtyping network for foodborne bacterial disease surveillance, United States. Emerging Infectious Diseases. 2001;7:382–389. doi: 10.3201/eid0703.010303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Threlfall EJ. Antimicrobial drug resistance in Salmonella: problems and perspectives in food- and water-borne infections. FEMS Microbiology Reviews. 2002;26:141–148. doi: 10.1111/j.1574-6976.2002.tb00606.x. [DOI] [PubMed] [Google Scholar]
- 6.Peters TM et al. The Salm-gene project – a European collaboration for DNA fingerprinting for food-related salmonellosis. Eurosurveillance. 2003;8:46–50. doi: 10.2807/esm.08.02.00401-en. [DOI] [PubMed] [Google Scholar]
- 7.Grundmann H, Hori S, Tanner G. Determining confidence intervals when measuring genetic diversity and the discriminatory abilities of typing methods for microorganisms. Journal of Clinical Microbiology. 2001;39:4190–4192. doi: 10.1128/JCM.39.11.4190-4192.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodrigue DC, Tauxe RV, Rowe B. International increase in Salmonella enteritidis: a new pandemic. Epidemiology and Infection. 1990;105:21–27. doi: 10.1017/s0950268800047609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Threlfall EJ et al. Application of pulsed-field gel electrophoresis to an international outbreak of Salmonella agona. Emerging Infectious Diseases. 1996;2:130–132. doi: 10.3201/eid0202.960209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Threlfall EJ, Hopkins KL, Ward LR. Diversification in Salmonella Typhimurium DT 104. Emerging Infectious Diseases. 2005;11:980–981. [Google Scholar]
- 11.Threlfall EJ et al. Emergence of new subclones of multiresistant Salmonella typhimurium DT 104 possibly associated with poultry meat. Veterinary Record. 2004;154:89–90. doi: 10.1136/vr.154.3.89. [DOI] [PubMed] [Google Scholar]
- 12.Tenover FC et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. Journal of Clinical Microbiology. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernandez J et al. Analysis of molecular epidemiology of Chilean Salmonella enterica serotype enteritidis isolates by pulsed-field gel electrophoresis and bacteriophage typing. Journal of Clinical Microbiology. 2003;41:1617–1622. doi: 10.1128/JCM.41.4.1617-1622.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lukinmaa S et al. Salmonella enteritidis phage types 1 and 4: pheno- and genotypic epidemiology of recent outbreaks in Finland. Journal of Clinical Microbiology. 1999;37:2176–2182. doi: 10.1128/jcm.37.7.2176-2182.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bakeri SA et al. Genetic diversity of human isolates of Salmonella enterica serovar Enteritidis in Malaysia. Journal of Applied Microbiology. 2003;95:773–780. doi: 10.1046/j.1365-2672.2003.02033.x. [DOI] [PubMed] [Google Scholar]
- 16.Cogan TA, Humphrey TJ. The rise and fall of Salmonella Enteritidis in the UK. Journal of Applied Microbiology. 2003;94:114S–119S. doi: 10.1046/j.1365-2672.94.s1.13.x. [DOI] [PubMed] [Google Scholar]
- 17.Davis MA, Hancock DD, Besser TE. Multiresistant clones of Salmonella enterica: the importance of dissemination. Journal of Laboratory and Clinical Medicine. 2002;140:135–141. doi: 10.1067/mlc.2002.126411. [DOI] [PubMed] [Google Scholar]