SUMMARY
To clarify whether prevalence or special pathogenicity is more important in determining urinary tract infection (UTI) causation, we compared the biotype, phylogenetic group, and virulence genes of Escherichia coli urine strains from 11 women with acute lower UTI with those of the host's dominant intestinal E. coli strain(s). Twenty-one unique E. coli clones were identified. For three women, the single faecal clone identified was also the host's urine clone, whereas for eight women faecal samples yielded 1 or 2 distinct non-urine clones (total, n=10), either with (n=3) or without (n=5) the concurrent urine clone. The eight urine clones from the latter eight women exhibited significantly greater inferred virulence, according to virulence gene content and phylogenetic background, than did the hosts' 10 corresponding ‘faecal only’ clones. In contrast, the three urine clones that were detected as the host's sole faecal clone exhibited significantly lower inferred virulence than the other eight urine clones, and were statistically indistinguishable from the 10 ‘faecal only’ clones. In conclusion, special pathogenicity is an important determinant of UTI pathogenesis in women, although prevalence may occasionally allow less virulent strains to cause UTI.
INTRODUCTION
Urinary tract infection (UTI) is a common and costly health problem among women that is usually due to Escherichia coli. [1] The causative E. coli strain often can be found in the woman's faecal flora at the time of a UTI episode [2, 3]. This observation has suggested the ‘faecal–perineal–urethral’ hypothesis for UTI pathogenesis in women, according to which the host's own faecal flora is the immediate external reservoir from which E. coli strains emerge to cause UTI [3, 4].
This phenomenon, and the finding that the most common O antigens among E. coli UTI isolates are also the most prevalent O antigens among faecal E. coli from healthy individuals, has suggested the ‘prevalence’ hypothesis, which posits that UTI occurs when ordinary faecal E. coli are in the right place at the right time in sufficient numbers to enter the urinary tract and cause infection [5]. In contrast, the statistically greater prevalence among UTI-source E. coli, compared with faecal E. coli from healthy hosts, of phylogenetic group B2, certain O antigens, and suspected or proven virulence factors such as adhesins, siderophores, toxins, and polysaccharide coatings, has suggested the ‘special pathogenicity’ hypothesis [6]. This hypothesis asserts that UTI pathogenesis is driven by the enhanced virulence capability of the causative strain, rather than by simple mass action. The term ‘uropathogenic E. coli’ is often used to describe strains with a presumed heightened ability to cause UTI [7]. Because such strains can cause infections also at non-urinary extraintestinal sites, the inclusive designation ‘extraintestinal pathogenic E. coli’ (ExPEC) also has been applied to them [8].
However, most of the epidemiological data underlying the ‘special pathogenicity’ hypothesis derive from comparisons of UTI isolates and intestinal isolates from unrelated hosts [9, 10], which leaves open the possibility that unmeasured host or environmental differences, rather than differences in virulence per se, may explain the observed bacterial differences. In addition, many experimental studies evaluating the importance of putative virulence factors (e.g. P fimbriae, cytotoxic necrotizing factor, capsule, or lipopolysaccharide) in relation to a strain's ability to cause UTI [11, 12], or other types of extraintestinal infection, have found that a single bacterial trait does not necessarily play a decisive, or even discernible role in virulence [13–16].
Therefore, we sought to compare, among women with UTI due to E. coli, the host's urine strain with the host's dominant intestinal E. coli strain(s) at the time of the infection, according to clonal identity, phylogenetic background, and (putative) virulence genes, to clarify whether prevalence or special pathogenicity is more important in determining UTI causation, and whether this relationship varies with host characteristics [6, 17, 18]. We also assessed in what proportion of women with E. coli UTI the urine clone can be readily detected as a dominant intestinal clone, consistent with the faecal–perineal–urethral hypothesis of UTI pathogenesis [3].
PATIENTS AND METHODS
Patients
Eleven ambulatory women were studied. Subject recruitment was through the clinical laboratory at Hospital Vall d'Hebron of Barcelona. During the study period (February 2002–February 2003), women who provided urine samples to the laboratory for urinalysis and culture for suspected acute cystitis were invited to participate in the study, which entailed providing informed consent, clinical data, and a self-collected rectal swab. Consecutively consenting women who met the following criteria were included in this analysis: bacteriologically documented UTI with E. coli as a sole microorganism, clinical and laboratory criteria for cystitis, and E. coli isolated from the rectal swab. Clinical criteria for cystitis included the presence of dysuria, urgency, and frequency, with or without suprapubic pain or gross haematuria, and absence of flank pain or fever >38°C. Laboratory criteria for cystitis included growth of a single E. coli colony type in numbers ⩾103 c.f.u./ml plus microscopic pyuria.
Pyuria and cultures
Pyuria was defined as >5 white blood cells/field (400×) in a centrifuged urine sediment (original volume, 10 ml). Quantitative urine culture was done by plating known volumes of urine on chromogenic UTI medium (Oxoid GmbH, Wesel, Germany). The isolated strain was identified by conventional methods [19]. Concurrent with urine collection, and prior to antibiotic administration, a rectal swab with Amies transport medium (Venturi Transystem, Brescia, Italy) was collected by the subject. The presence of visible faecal staining was confirmed. The rectal swab was inoculated onto a MacConkey agar plate and incubated at 35–37°C for 48 h. From the terminal streak area of each plate a minimum of two isolated colonies suspected to be E. coli were chosen. If multiple morphologies were noted, all unique morphotypes were sampled. Each selected colony was identified using conventional methods [19]. Since, on a statistical basis, a clone would need to be comparatively prevalent within the faecal flora to be recovered from the terminal streak area [20], the colonies selected were considered to represent dominant faecal clone(s).
Biotyping
Each urine and stool E. coli isolate was biotyped using nine biochemical tests: β-glucoronidase, indole production, ornithine decarboxylase, dulcitol, saccharose, raffinose, salicin and melibiose fermentation, and esculin hydrolysis. A unique three-digit code (biotype) was assigned for each resulting distinct combination of positive and negative results.
Molecular methods
For each E. coli isolate, the major phylogenetic group of origin (A, B1, B2, and D) was determined by a triplex PCR method [21]. Detection of virulence genes was done using an established multiplex PCR assay [22, 23]. Fifteen virulence genes were characterized, including: adhesins papA (P fimbriae structural subunit), papG alleles I, II and III (P adhesin variants), fimH (type 1 fimbriae), afa/draBC (Dr-binding adhesins), and sfa/focDE (S and F1C fimbriae); toxins hlyA (haemolysin) and cnf1 (cytotoxic necrotizing factor 1); siderophores iutA (aerobactin) and fyuA (yersiniabactin); capsule synthesis specific for group II (K1, K5, K12, etc.) kpsMII; serum resistance-associated traT; invasion of brain endothelium ibeA; and malX, a coding region near the terminus of a pathogenicity-associated island (PAI) from E. coli strain CFT073. Appropriate positive and negative controls were included. The virulence score was the number of virulence genes detected, with pap elements counting collectively as a single trait. The results of such in vitro testing predict experimental virulence in vivo [24, 25].
Clonal analysis
E. coli isolates from the same host that exhibited the same biotype and phylogenetic group, and isolates from different hosts that in addition exhibited the same virulence profile, were considered to putatively represent the same clone. This was selectively confirmed by comparing genomic profiles as generated by repetitive element PCR [26].
Statistical methods
Comparisons of proportions were tested by using Fisher's exact test. Virulence score were compared by using the Mann–Whitney U test. P values <0·05 were considered statistically significant.
RESULTS
Patient characteristics
For 11 of the participating women, E. coli was isolated in significant concentrations from urine and as one or more colonies from the concurrent rectal swab. These 11 women, who ranged in age from 21 to 64 years (median 30 years), constituted the study population (Table 1). At the time of enrolment, all 11 subjects had symptoms of acute cystitis. Two had an underlying condition possibly predisposing to UTI, i.e. pregnancy and prior surgery for uterine prolapse respectively. Three had a history of previous UTI (Table 1).
Table 1.
Characteristics of 11 women with symptomatic urinary tract infection (UTI) due to Escherichia coli
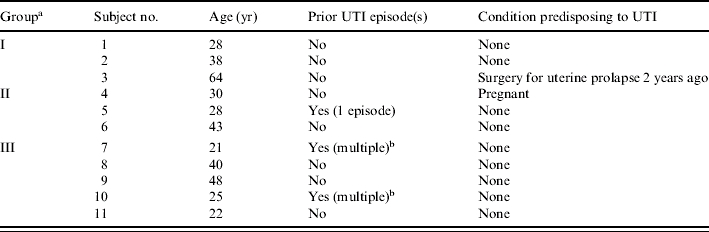
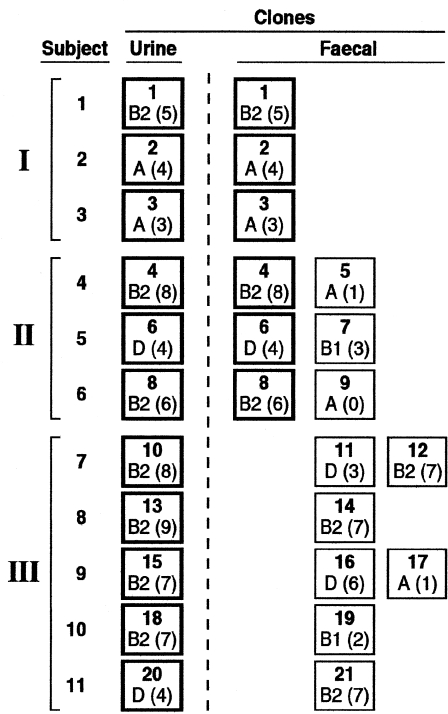
Colonization group (I, II, or III) was defined based on the correspondence of urine and faecal clones in individual subjects. In group I, a single faecal clone was detected, corresponding with the urine clone (U=F). In group II, multiple faecal clones were detected, including the subject's urine clone plus ⩾1 distinct ‘faecal only’ clone(s). In group III, the clones detected in faeces were all ‘faecal only’, i.e. did not include the urine clone (Fig.).
Multiple, ⩾2 prior UTI episodes (by history).
Clonal distribution of E. coli isolates
Eleven E. coli isolates (one per subject) were selected from the subjects' urine cultures, whereas 27 putative E. coli isolates (2–4 per subject) were selected from the corresponding stool cultures. Of the 27 faecal isolates, 24 were confirmed as E. coli. Among the multiple E. coli isolates from each subject there was precise correspondence of biotype, phylogenetic group, and virulence profile, such that if multiple colonies exhibited the same biotype, they also exhibited the same phylogenetic group and combination of virulence genes, suggesting clonality (Table 2). Genomic profiling confirmed these inferred clonal relationships (not shown). In contrast, in only one instance did isolates from different subjects exhibit the same biotype, phylogenetic group, and virulence profile, and in this instance genomic profiling showed the isolates to represent distinct, albeit closely related, host-specific genotypes. Thus, according to combined molecular and phenotypic analysis of the 35 total E. coli isolates, the 11 urine isolates represented 11 clones, whereas the 24 faecal isolates represented 16 clones (Table 2).
Table 2.
Clonal identity, biotype, phylogenetic group, and virulence profile of Escherichia coli isolates from urine and stool from 11 women with symptomatic urinary tract infection
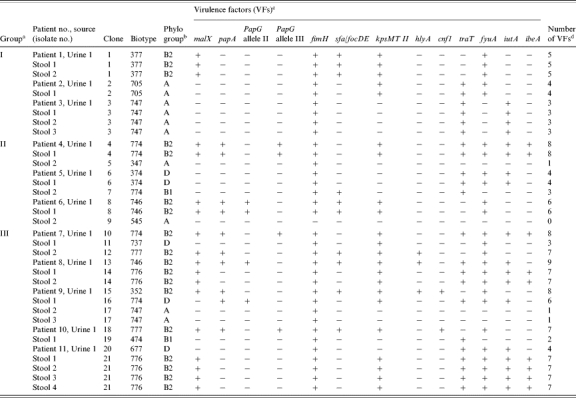
Colonization group (I, II, or III) was defined based on the correspondence of urine and faecal clones in individual subjects. In group I, a single faecal clone was detected, corresponding with the urine clone (U=F). In group II, multiple faecal clones were detected, including the subject's urine clone plus ⩾1 distinct ‘faecal only’ clone(s). In group III, the clones detected in faeces were all ‘faecal only’, i.e. did not include the urine clone. malX, marker for pathogenicity-associated island from strain CFT073; papA, P fimbriae structural subunit; papG, P fimbrial adhesin molecule; fimH, type-1 fimbriae; sfa/focDE, S and F1C fimbriae; kpsMII, group II capsule synthesis; hlyA, haemolysin; cnf1, cytotoxic necrotizing factor 1; traT, serum-resistance associated outer membrane protein; iutA, aerobactin; fyuA, yersiniabactin receptor; ibeA, invasion of brain endothelium. None of the strains showed papG allele I or afa/draBC (Dr-binding adhesins).
The number of virulence factors (VFs) was calculated for each isolate as the sum of pap (with the presence of papA and/or papG allele II or allele III counting as one unit) plus all others VFs detected.
Comparison of each subject's faecal clone(s) with the concurrent urine clone demonstrated three distinct colonization groups (Fig.). In group I (n=3 subjects), the stool culture yielded only the urine clone, i.e. without any non-urine clone. In group II (n=3 subjects), the stool culture yielded both the urine clone and an additional non-urine (‘faecal only’) clone. Thus, for the six women in these two groups (who accounted for 55% of the population) the urine clone was a dominant faecal clone at the time of the UTI episode. In contrast, in group III (n=5 subjects), the stool culture yielded one or two non-urine (‘faecal only’) clones, without the urine clone.
Fig.
Distribution of 21 Escherichia coli clones by source (urine vs. faecal) among 11 women with urinary tract infection due to E. coli. Each subject (nos. 1–11) accounts for one row of boxes (clones). Clones are shown separately for urine and faecal samples (left and right of dashed line respectively). Clones are identified as to clone number (top centre of box), phylogenetic group (bottom left of box), and virulence score (bottom right of box). Subjects are sorted by colonization group (I, II, III), depending on the correspondence of the subject's urine and faecal clones. Urine clones (boxes with bold borders) were: (i) the only clone recovered from faeces (group I; top three subjects); (ii) recovered from faeces together with ⩾1 unique non-urine clones (boxes with lighter borders) (group II; middle three subjects; middle); or (iii) absent from faeces (group III; bottom five subjects).
Host characteristics vs. colonization group
Although there were no significant differences among the three colonization groups according to patient age, group I included the oldest subject, whereas group III included the three youngest subjects. The three colonization groups did not differ appreciably according to the prevalence of predisposing conditions or prior UTI history (not shown).
Bacterial characteristics
The 21 unique E. coli clones exhibited 14 different biotypes. Their phylogenetic groups, in descending order of prevalence, were B2 (47·6%), A (23·8%), D (19%), and B1 (9·5%) (Table 2). They possessed diverse combinations of the 13 virulence genes detected within the population, with a median of five (range 0–9) such genes per clone. The most prevalent virulence genes were fimH (95%), fyuA (71%), kpsMII (62%), traT (57%), malX (48%), and iutA (43%) (Table 2).
Colonization patterns vs. bacterial and host characteristics
Virulence scores and phylogenetic group were assessed among clones representing the three colonization groups (I, II, and III), stratified within groups II and III according to urine vs. ‘faecal only’ source (Table 3). Since groups II and III urine clones did not differ significantly according to absolute virulence score, prevalence of virulence scores ⩾4, or proportion from phylogenetic group B2 or group D (not shown), groups II and III urine clones were combined for statistical analysis. The same applied to groups II and III ‘faecal only’ clones.
Table 3.
Characteristics of groups of urine and faecal Escherichia coli clones from 11 women with symptomatic urinary tract infection according to colonization behaviour
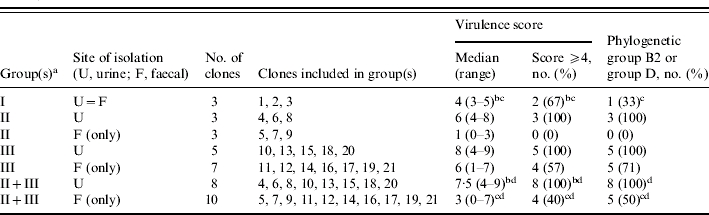
Colonization group (I, II, or III) was defined based on the correspondence of urine and faecal clones in individual subjects. In group I, a single faecal clone was detected, corresponding with the urine clone (U=F). In group II, multiple faecal clones were detected, including the subject's urine clone plus ⩾1 distinct ‘faecal only’ clone(s). In group III, the clones detected in faeces were all ‘faecal only’, i.e. did not include the urine clone.
Group I (U=F) vs. groups II+III (urine): for absolute virulence score, P=0·06 (Mann–Whitney U test); for % with virulence score ⩾4, P>0·10 (Fisher's exact test); for % from phylogenetic group B2 or group D, P=0·05 (Fisher's exact test).
Group I (urine=faecal) vs. groups II+III (faecal only), P>0·10 (Fisher's exact test).
Groups II+III (urine) vs. groups II+III (faecal only); for absolute virulence score, P=0·016 (Mann–Whitney U test); for % with virulence score ⩾4, P=0·01 (Fisher's exact test); for % from phylogenetic group B2 or group D, P=0·04 (Fisher's exact test).
Collectively, the eight combined (groups II+III) urine clones exhibited higher absolute virulence scores (P=0·016), and a significantly greater prevalence of virulence scores ⩾4 (P=0·01) and of phylogenetic group B2 or group D (P=0·04), than did the 10 combined (groups II+III) ‘faecal only’ clones from the same hosts (Table 3). They likewise exhibited a numerically (typically twofold) greater prevalence of each of the 13 individual virulence genes than did the ‘faecal only’ clones (not shown), with the differences reaching statistical significance for fyuA (100% vs. 50%, P=0·036), and borderline significance for papA (75% vs. 20%, P=0·054) and papG allele III (38% vs. 0%, P=0·07). Notably, both clones with a virulence score of ⩾8 were urine clones from groups II or III, whereas all four clones with a virulence score of ⩽2 were ‘faecal only’ clones from groups II or III. Similarly, at the individual subject level, for seven of the eight women within groups II and III the urine clone had a higher virulence score than did any of that host's concurrent ‘faecal only’ clones (Table 2).
In contrast, the three group I (urine=faecal) clones exhibited borderline significantly lower values for absolute virulence score (P=0·08) and prevalence of phylogenetic group B2 or group D (P=0·05) than did the eight combined (groups II+III) urine clones (Table 3). Indeed, they closely resembled the 10 combined (groups II+III) ‘faecal only’ clones, exhibiting no significant differences from this group according to virulence scores or phylogenetic background (Table 3).
DISCUSSION
This molecular epidemiological analysis of concurrent E. coli clones from the urine and faeces of women with UTI yielded three main findings. First, consistent with previous studies [2, 3, 6], the urine clone is usually one of the host's dominant faecal clones at the time of a UTI episode. Second, among women with UTI who have one or more distinct ‘faecal only’ clones, the urine clones typically exhibit significantly greater inferred virulence that do the corresponding ‘faecal only’ clones. Third, among women with UTI in whom faecal cultures yield only the urine clone, the urine clones usually exhibit lower inferred virulence than do urine clones from women with distinct non-urine (‘faecal only’) faecal clones, instead resembling ‘faecal only’ clones. These findings support the faecal–perineal–urethral hypothesis for UTI pathogenesis and suggest that special pathogenicity is the main driving force for UTI. However, they also suggest that in some women, high prevalence within the faecal reservoir may allow comparatively low-virulence strains to cause UTI.
We found that in six (55%) of the 11 women the host's urine clone was readily detectable as a dominant faecal clone, with or without additional faecal clones. This is consistent with the widely accepted notion that the faecal microflora is the reservoir from which E. coli clones emerge to cause UTI [4]. Concordance between urine and concurrent faecal clones has been documented in previous studies of women and men with acute UTI [2, 3, 27]. In contrast, of particular interest here were the five women in whom the urine clone was not encountered among the several colonies selected for analysis from the stool culture. Conceivably, in these subjects the urine clone actually was present in the stool, but was missed because of limited sampling [28]. Alternatively, the urine clone might truly have been absent from stool at the time of the study. If so, it may have resided there previously, even as a dominant clone, but with subsequent migration to the (more immediately relevant for UTI pathogenesis) vaginal and periurethral reservoirs, which we did not sample [4, 29]. Or it might never have been an intestinal resident, instead having been inoculated directly into the vaginal or periurethral area, perhaps via sexual contact [30].
Among the eight hosts with one or more distinct ‘faecal only’ clones in addition to the urine clone, the urine clones exhibited significantly greater inferred virulence than did the corresponding ‘faecal only’ clones. This was true both at the population level and, in a by-subject analysis, for seven of the eight individual women. This strongly suggests that special pathogenicity was a crucial driving principle underlying UTI in these subjects and, conversely, that the virulence genes and phylogenetic groups we analysed are relevant surrogate markers for in vivo virulence potential. Interestingly, we found that, of the individual virulence genes studied, a relative newcomer, fyuA (yersiniabactin system), for which a direct contribution to virulence has been demonstrated experimentally in mice [31], exhibited the statistically strongest association with urine vs. faecal source.
Also of note, the subgroup with the numerically highest virulence scores, and the greatest prevalence of (virulence-associated) phylogenetic group B2 [32], were the group III urine clones. These clones were not detected in the host's faeces, consistent with the possibility of them representing minority faecal clones that had successfully out-competed one or more high-prevalence faecal clones for entry into the urinary tract. Taken together, the data suggest the possibility of a gradient of virulence among urine clones, with virulence being inversely proportional to the clone's relative prevalence within the faecal reservoir, i.e. monoclonally dominant (low virulence), co-dominant (moderate-to-high virulence), or minority/absent (high virulence).
The single host whose urine clone appeared to be less virulent than the concurrent faecal clone was of interest. This subject (no. 11) had no known compromising conditions or prior UTI history, was relatively young, and presented with acute cystitis. Yet her urine clone was from group D and had only four virulence genes, whereas her sole detected faecal clone was from group B2 and had seven virulence genes. Possible explanations for this seeming paradox include an unrecognized host defence defect, absence of the (higher virulence) faecal clone from the more immediate vaginal and/or periurethral reservoirs, and imprecision of the molecular markers for assessing true in vivo virulence potential. It also is possible, although unlikely, that the (more virulent appearing) faecal clone actually was present in urine as part of a polyclonal infection, but was missed by our one-colony sampling of urine cultures.
Finally, we found that those urine clones with no detectable competing faecal clone(s) (group I), exhibited borderline significantly lower inferred virulence, according to virulence scores and phylogenetic background, than the urine clones that appeared to have out-competed one or more other clones in the host to enter the urinary tract (groups II and III). Indeed, the group I (urine=faecal) clones were statistically indistinguishable, according to virulence-associated characteristics, from the ‘faecal only’ clones from groups II and III. The group I urine clones appeared to be the host's sole faecal E. coli clone, so clearly predominated within the faecal flora at the time of the UTI episode. Such prevalence may have compensated for their relative lack of virulence, thereby allowing them to cause UTI even though, as a first approximation, they appeared to represent ordinary commensal strains [6].
Limitations of the study include the small sample size, the non-systematic approach to recruitment, the heterogeneity of the study population, the limited sampling of the urine and faecal E. coli populations, the absence of vaginal and periurethral sampling, and the non-longitudinal design. Strengths include the comparison of urine and faecal E. coli from individual hosts, which provides optimally matched comparison groups but has been used in few previous studies, particularly for women [6, 17, 33–35]. Other strengths include the analysis of multiple faecal colonies to detect clonal diversity and the use of sophisticated molecular typing methods, which permitted comparisons of molecularly inferred virulence [24] with observed colonization behaviour.
In summary, we found that among 11 women with symptomatic lower UTI due to E. coli that: (i) the urine clone was usually one of the host's dominant faecal clones, (ii) urine clones were usually substantially more virulent than concurrent ‘faecal only’ clones from the same host, and (iii) urine clones were less virulent if they constituted the host's sole faecal clone than if they had competing (non-urine) faecal clones. These findings support the faecal–perineal–urethral hypothesis for UTI pathogenesis in women and, while allowing for a contribution from prevalence in certain instances, indicate that special pathogenicity is the main driving force underlying UTI. They thus suggest that reduction of intestinal colonization with uropathogenic E. coli and interference with E. coli virulence mechanisms could help prevent UTI.
ACKNOWLEDGEMENTS
This work was supported by grant Fondo de Investigación Sanitaria (FIS 01/1353), Fondo de Investigación Sanitaria (FIS 02/1887) and Red Española de Investigación en Patologia Infecciosa (REIPI C3/14), Spain (G.P.); and Office of Research and Development, Medical Research Service, Department of Veterans Affairs, USA (J.R.J.).
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Russo TA, Johnson JR. Medical and economic impact of extraintestinal infections due to Escherichia coli: focus on an increasingly important endemic problem. Microbes and Infection. 2003;5:449–456. doi: 10.1016/s1286-4579(03)00049-2. [DOI] [PubMed] [Google Scholar]
- 2.Gruneberg RN. Relationship of infecting urinary organism to the faecal flora in patients with symptomatic urinary infection. Lancet. 1969;2:766–768. doi: 10.1016/s0140-6736(69)90478-4. [DOI] [PubMed] [Google Scholar]
- 3.Yamamoto S et al. Genetic evidence supporting the fecal-perineal-urethral hypothesis in cystitis caused by Escherichia coli. Journal of Urology. 1997;157:1127–1129. [PubMed] [Google Scholar]
- 4.Stamm WE et al. Urinary tract infections: from pathogenesis to treatment. Journal of Infectious Diseases. 1989;159:400–406. doi: 10.1093/infdis/159.3.400. [DOI] [PubMed] [Google Scholar]
- 5.Turck M, Petersdorf RG. The epidemiology of nonenteric Escherichia coli infections: prevalence of serological groups. Journal of Clinical Investigation. 1962;41:1760–1765. doi: 10.1172/JCI104635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plos K et al. Intestinal carriage of P fimbriated Escherichia coli and the susceptibility to urinary tract infection in young children. Journal of Infectious Diseases. 1995;171:625–631. doi: 10.1093/infdis/171.3.625. [DOI] [PubMed] [Google Scholar]
- 7.Korhonen TK et al. P-antigen-recognizing fimbriae from human uropathogenic Escherichia coli strains. Infection and Immunity. 1982;37:286–291. doi: 10.1128/iai.37.1.286-291.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Russo TA, Johnson JR. A proposal for a new inclusive designation for extraintestinal pathogenic isolates of Escherichia coli: ExPEC. Journal of Infectious Diseases. 2000;181:1753–1754. doi: 10.1086/315418. [DOI] [PubMed] [Google Scholar]
- 9.Johanson IM et al. Pap, papG and prsG DNA sequences in Escherichia coli from the fecal flora and the urinary tract. Microbial Pathogenesis. 1993;15:121–129. doi: 10.1006/mpat.1993.1062. [DOI] [PubMed] [Google Scholar]
- 10.Johnson JR. Virulence factors in Escherichia coli urinary tract infection. Clinical Microbiology Review. 1991;4:80–128. doi: 10.1128/cmr.4.1.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donnenberg MS, Welch RA, Mobley HL, Warren JW. Urinary Tract Infection. Washington DC: ASM Press; 1996. Virulence determinants in uropathogenic Escherichia coli; pp. 135–174. : pp. [Google Scholar]
- 12.Johnson JR. Microbial virulence determinants and the pathogenesis of urinary tract infection. Infectious Diseases Clinics North America. 2003;17:261–278. doi: 10.1016/s0891-5520(03)00027-8. [DOI] [PubMed] [Google Scholar]
- 13.Johnson DE et al. The role of cytotoxic necrotizing factor-1 in colonization and tissue injury in a murine model of urinary tract infection. FEMS Immunology and Medical Microbiology. 2000;28:37–41. doi: 10.1111/j.1574-695X.2000.tb01454.x. [DOI] [PubMed] [Google Scholar]
- 14.Mobley HL et al. Isogenic P-fimbrial deletion mutants of pyelonephritogenic Escherichia coli: the role of alpha Gal (1-4) beta Gal binding in virulence of a wild-type strain. Molecular Microbiology. 1993;10:143–155. doi: 10.1111/j.1365-2958.1993.tb00911.x. [DOI] [PubMed] [Google Scholar]
- 15.Russo T et al. The O4 specific antigen moiety of lipopolysaccharide but not the K54 group 2 capsule is important for urovirulence of an extraintestinal isolate of Escherichia coli. Infection and Immunity. 1996;64:2343–2348. doi: 10.1128/iai.64.6.2343-2348.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Russo TA et al. Loss of the O4 antigen moiety from the lipopolysaccharide of an extraintestinal isolate of Escherichia coli has only minor effects on serum sensitivity and virulence in vivo. Infection and Immunity. 1995;63:1263–1269. doi: 10.1128/iai.63.4.1263-1269.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson JR et al. Identification of urovirulence traits in Escherichia coli by comparison of urinary and rectal E. coli isolates from dogs with urinary tract infection. Journal of Clinical Microbiology. 2003;41:337–345. doi: 10.1128/JCM.41.1.337-345.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.London N et al. Comparison of virulence factors of urinary and faecal Escherichia coli isolated from the same patient. Canadian Journal of Infectious Diseases. 1995;6:392. [Google Scholar]
- 19.Murray PR. Manual of Clinical Microbiology. 8th edn. Washington, DC: ASM Press; 2003. [Google Scholar]
- 20.Lidin-Janson G et al. The homogeneity of the faecal coliform flora of normal school-girls, characterized by serological and biochemical properties. Medical Microbiology and Immunology (Berlin) 1978;164:247–253. doi: 10.1007/BF02125493. [DOI] [PubMed] [Google Scholar]
- 21.Clermont O, Bonacorsi S, Bingen E. Rapid and simple determination of the Escherichia coli phylogenetic group. Applied and Environmental Microbiology. 2000;66:4555–4558. doi: 10.1128/aem.66.10.4555-4558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson JR, Stell AL. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. Journal of Infectious Diseases. 2000;181:261–272. doi: 10.1086/315217. [DOI] [PubMed] [Google Scholar]
- 23.Johnson JR, Stell AL, Delavari P. Canine feces as a reservoir of extraintestinal pathogenic Escherichia coli. Infection and Immunity. 2001;69:1306–1314. doi: 10.1128/IAI.69.3.1306-1314.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson JR, Kuskowski M. Clonal origin, virulence factors, and virulence. Infection and Immunity. 2000;68:424–425. doi: 10.1128/iai.68.1.424-425.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Picard B et al. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infection and Immunity. 1999;67:546–553. doi: 10.1128/iai.67.2.546-553.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Versalovic J et al. Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods in Molecular and Cellular Biology. 1994;5:25–40. [Google Scholar]
- 27.Terai A et al. Molecular epidemiological evidence for ascending urethral infection in acute bacterial prostatitis. Journal of Urology. 2000;164:1945–1947. [PubMed] [Google Scholar]
- 28.Johnson JR et al. Colonization with and acquisition of uropathogenic Escherichia coli as revealed by polymerase chain reaction-based detection. Journal of Infectious Diseases. 1998;177:1120–1124. doi: 10.1086/517409. [DOI] [PubMed] [Google Scholar]
- 29.Stapleton A et al. Effect of secretor status on vaginal and rectal colonization with fimbriated Escherichia coli in women with and without recurrent urinary tract infection. Journal of Infectious Diseases. 1995;171:717–720. doi: 10.1093/infdis/171.3.717. [DOI] [PubMed] [Google Scholar]
- 30.Foxman B et al. Transmission of uropathogens between sex partners. Journal of Infectious Diseases. 1997;175:989–992. doi: 10.1086/514007. [DOI] [PubMed] [Google Scholar]
- 31.Schubert S et al. Yersinia high-pathogenicity island contributes to virulence in Escherichia coli causing extraintestinal infections. Infection and Immunity. 2002;70:5335–5337. doi: 10.1128/IAI.70.9.5335-5337.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goullet P, Picard B. Electrophoretic type B2 of carboxylesterase B for characterisation of highly pathogenic Escherichia coli strains from extra-intestinal infections. Journal of Medical Microbiology. 1990;33:11–16. doi: 10.1099/00222615-33-1-11. [DOI] [PubMed] [Google Scholar]
- 33.Zhang L, Foxman B, Marrs C. Both urinary and rectal Escherichia coli isolates are dominated by strains of phylogenetic group B2. Journal of Clinical Microbiology. 2002;40:3951–3955. doi: 10.1128/JCM.40.11.3951-3955.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schlager TA et al. Clonal diversity of Escherichia coli colonizing stools and urinary tracts of young girls. Infection and Immunity. 2002;70:1225–1229. doi: 10.1128/IAI.70.3.1225-1229.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schlager TA et al. Variation in frequency of the virulence-factor gene in Escherichia coli clones colonizing the stools and urinary tracts of healthy prepubertal girls. Journal of Infectious Diseases. 2003;188:1059–1064. doi: 10.1086/377643. [DOI] [PubMed] [Google Scholar]