SUMMARY
A micro-epidemic of hantavirus infections occurred in Lower Bavaria, South-East Germany, starting in April 2004. While only three cases were registered from 2001 to 2003, a dramatically increased number of clinically apparent human hantavirus infections (n=38) was observed in 2004, plus seven additional cases by June 2005. To determine the reservoir responsible for the infections, a total of 43 rodents were trapped in Lower Bavaria. Serological and genetic investigations revealed that Puumala virus (PUUV) is dominant in the local population of bank voles. Partial PUUV S segment nucleotide sequences originating from bank voles at four different trapping sites in Lower Bavaria showed a low divergence (up to 3·1%). This is contrasted by a nucleotide sequence divergence of 14–16% to PUUV strains detected in Belgium, France, Slovakia or North-Western Germany. PUUV sequences from bank voles in Lower Bavaria represent a new PUUV subtype which seems to be responsible for the observed increase of human hantavirus infections in 2004–2005.
INTRODUCTION
Hantaviruses, family Bunyaviridae, are carried by persistently infected rodents and are transmitted to humans by aerosolized excretions. The close association of hantavirus species and respective rodent hosts is explained by a co-evolution hypothesis. The geographical distribution of viruses is associated with the occurrence of reservoir hosts. Hantaviruses in Europe and Asia are carried by representatives of the rodent subfamilies Murinae and Arvicolinae. Certain hantavirus species associated with these rodent subfamilies can cause haemorrhagic fever with renal syndrome (HFRS), or its milder form Nephropathia epidemica (NE), in humans [1, 2]. The type species of the genus Hantavirus is the Hantaan virus (HTNV) carried by the striped field mouse, Apodemus agrarius [3].
Due to the presence of different hantaviruses and their rodent hosts, Germany is a melting pot of different strains with a high potential for hantavirus evolution. In Germany, human infections are mainly caused by Puumala virus (PUUV) carried by bank voles, Clethrionomys glareolus [4–7]. Although serological data suggest the presence of Dobrava virus (DOBV) in striped field, yellow-necked and wood mice (R. Ulrich, unpublished observations), thus far the molecular evidence to confirm these observations is lacking. This is in line with serological results of patients with HFRS-like disease which for a long time have indicated the presence of Murinae-associated hantaviruses, i.e. DOBV [8, 9]. Recently an A. agrarius-associated DOBV lineage (DOBV-Aa) was demonstrated to be the causative agent for HFRS in a patient from Northern Germany [10]. Further, Tula virus (TULV) carried by common voles (Microtus arvalis) occurs throughout Germany [11] but rarely causes human infections [11–13]. Detection of hantavirus-specific antibodies in other species, e.g. muskrat and water vole may reflect spillover infections [11].
The overall seroprevalence in humans in Germany is 1–2% [8, 11]. However, in the endemic region of Baden-Wuerttemberg the seroprevalence is remarkably higher [8, 12, 14]. Since becoming a notifiable disease in 2001, 140–240 cases of hantavirus infections in Germany have been reported annually [15–18]. The proportion of asymptomatic or mild non-diagnosed hantavirus infections in Germany is unclear, as data of representative seroepidemiological studies are missing.
In Bavaria, between 2001 and 2004, 125 human NE cases were reported, 65 originating from Under Franconia. In Lower Bavaria, only three cases were registered from 2001 to 2003. However, from April 2004 a dramatic increase in the number of NE cases (n=38 in 2004, n=7 until June 2005) was observed in Lower Bavaria in Freyung-Grafenau, Regen and Passau districts ([19], Fig. 1). The main symptoms were fever, lumbar/muscle pain, headache, nausea, vomiting and renal dysfunction. Thirty-one of the 38 (79%) patients were admitted to a hospital for clinical treatment, 25 (66%) of all patients showed signs of renal involvement, mainly polyuria and proteinuria. An infection with PUUV was serologically diagnosed for all these patients at the local diagnostic laboratories using commercially available immunofluorescence tests. Haemorrhagic manifestations were not reported.
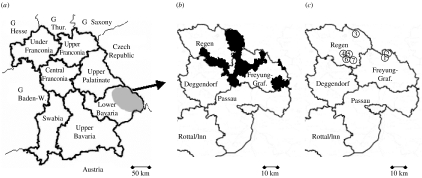
Fig. 1.
Geographic location of hantavirus micro-epidemic in South-East Germany (G). (a) Schematic map of Bavaria showing the districts. The grey ellipse marks the region in Lower Bavaria where 38 human hantavirus infections were recorded in 2004 (in 2005, seven cases until June). Baden-W., Baden-Wuerttemberg; Thur., Thuringia. (b) Geographical origin of 27 human infections in the two investigated districts, indicated by black areas. Freyung-Graf., Freyung-Grafenau. (c) Sites for rodent trapping (① Raimundsreuth; ② Glashütte; ③ Falkenstein; ④ Hangenleithen; ⑤ Mutzenwinkel; ⑥ Langfurth; ⑦ Schöfweg). Locations ④ and ⑥ were private houses, garage and garden from patients (i.e. rodents were directly trapped at the suspected sites of infection).
Around 78% of the patients were male; 66% were between 30 and 49 years old. Thirty-five of the 38 infections occurred in the districts Regen, Freyung-Grafenau and Passau, representing an area of above 4000 km2 ([19], Fig. 1a, b).
The objective of this study was to detect and characterize the hantavirus strain responsible for the increased number of NE cases in Lower Bavaria. Because the chances for virus or viral nucleic acid detection in humans are sparse, we decided to investigate the rodent reservoir population surrounding the observed clinical cases.
METHODS
Trapping of small rodents
Rodents were collected from 12 to 15 October 2004, using Sherman live traps at seven locations in Lower Bavaria including forests, fields, blackberry bushes and farms. Sampling areas were chosen according to the occurrence of hantavirus-induced NE cases as reported by the patients. Patients were interviewed to discover possible contacts with rodents in the nearby area, but also to analyse other possible contacts with rodents due to travelling or outdoor activities in other locations (Fig. 1a–c; for precise locations of trapping sites see http://www.maporama.de). Animals were trapped after decoying using 0·5–l plastic cups with apple pieces as bait located every 5–10 m, with 2–3×20 bait lines per location. Trapping (5–9 traps/area, 33 traps in total) was performed on sites with signs of rodent activities. Traps were controlled twice a day. Animals were sacrificed by exposure to CO2 and killed humanely. Location, trapping site, species, sex, reproductive and physical conditions were recorded for each rodent. Blood samples were taken by cardial puncture from 29 mice, as 14 rodents were found dead in their traps or the amount of taken blood drawn was not sufficient to gain serum. Rodents were placed into separate plastic bags and transported to the laboratory on dry ice.
Preparation of tissue samples
Rodents were necropsied under BSL-2 conditions. Tissue samples (heart, lung, liver, spleen, kidney, gonads) were aseptically removed. Transudates were collected from 15 rodents (12 C. glareolus, one Apodemus flavicollis, one Apodemus sylvaticus, one Mus musculus) by rinsing liver necropsies with 300 μl phosphate-buffered saline (PBS). Tissues were homogenized in MEM+10% FCS+antibiotics with the Fast Prep BIO101 matrix (Qbiogene, Heidelberg, Germany).
Preparation of viral RNA and rodent mitochondrial (mt) DNA
Nucleic acids were extracted from tissue homogenates using the RNeasy kit (Qiagen, Hilden, Germany) according the manufacturer's instructions.
Controls
Plasmid pCR2 derivatives (Invitrogen, Karlsruhe, Germany) containing the S segment of PUUV CG18-20 or DOBV were provided by Dr Weidmann (University of Freiburg, Germany). After linearizing plasmid DNA with HindIII, 1 μg DNA was in vitro-transcribed using T7 polymerase and subsequently purified with the PanRNA kit (PanBiotech, Aidenbach, Germany) according to the manufacturer's instructions.
Enzyme-linked immunosorbent assay (ELISA)
Twenty-nine rodent sera and 15 transudates were tested in an indirect IgG screening ELISA using yeast-expressed hexahistidine (His)-tagged recombinant nucleocapsid (rN) proteins of PUUV strain Vranica/Hällnäs (PUUV-Vra) and DOBV strain Slovenia (DOBV-Slo) [20, 21] according to Schmidt et al. [22]. Microtitre plates (Polysorp; Nunc, Roskilde, Denmark) were coated with 0·2 μg rN protein/well from PUUV-Vra or DOBV-Slo diluted in carbonate buffer (1 h, 37°C) and blocked with 1% bovine serum albumin (BSA)/0·05% Tween-20 in PBS. Sera or transudates diluted 1/200 in 0·5% BSA/0·05% Tween-20 were added. Bound antibodies were detected with horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (1/3000, 1 h, Bio-Rad, Hercules, CA, USA). O-phenylenediamine (OPD) substrate (Sigma, Deisenhofen, Germany) was added and the reaction stopped by addition of 50 μl 1 m H2SO4 (10 min). The optical density (OD) was measured at 492 nm (reference 620 nm). As a control, sera of C57BL/6 mice were used that had been immunized with hepatitis B virus core particles (HBcd) or HBcd carrying the amino-terminal 120 amino acids (aa) of the nucleocapsid protein of the hantaviruses DOBV-Slo and PUUV-Vra. The final OD value for each serum sample was calculated as the difference of the OD values measured in antigen-containing and antigen-free wells. These final OD values for serum dilutions of 1/200 were regarded as positive if the mean OD exceeded the cut-off value of 0·120, which was determined as three times the OD of the negative control sample. The threefold OD value was obtained with the negative control sample (HBcd) [23]. End-point titres were determined by successive twofold dilutions and defined as the highest serum dilution where the OD value was higher than the cut-off value.
SDS–polyacrylamide gel electrophoresis (SDS–PAGE) and Western blot (WB)
SDS–PAGE and WB analysis were performed as previously described [22]. Aliquots of purified rN protein of PUUV-Vra or DOBV-Slo were run in a SDS polyacrylamide gel and blotted onto nitrocellulose membranes. After blocking (1% BSA/0·05% Tween-20), membranes were incubated with rodent sera or transudates [1/200, 1 h, room temperature (RT)]. Filters were washed three times and incubated with HRP-conjugated anti-mouse IgG (1/250, 1 h, RT; Sigma). Strips were stained with 4-chloro-1-naphthol (Sigma) and washed with deionized water after colour development was complete.
Immunfluorescence assay (IFA)
Multi-well glass slides spotted with PUUV- or HTNV-infected Vero E6 cells were used according to the manufacturer's recommendations (PUUV and HTNV Hantavirus Antibody IF Test, Progen, Heidelberg, Germany). Rodent sera or transudates were diluted 1/16 or 1/32 in PBS and fluorescein isothiocyanate (FITC)-conjugated anti-mouse immunoglobulin (H+L) goat IgG (1/40, Sigma) was used to detect bound antibodies.
RT–PCR amplification and nucleotide sequencing
PUUV-specific amplification was performed using primers targeting the S segment nucleotides, nt, 342–1102 of strain PUUV CG18-20) – PUUV342: 5′-TAT GGT AAT GTC CTT GAT GT-3′ and cPUUV 1102: 5′-GCC ATD ATD GTR TTY CTC AT-3′ (modified following refs [24, 25]). Briefly, 5 μl of RNA were amplified using 20 pmol of primers and the SuperScriptII One-step RT–PCR system with Platinum Taq DNA polymerase (Invitrogen) in a final volume of 50 μl according to the manufacturer's instructions. Following reverse transcription at 50°C for 30 min, denaturation at 94°C for 2 min, DNA was amplified in 40 cycles for 15 s at 94°C, 30 s at 54°C, 1 min at 68°C. A final extension for 10 min at 68°C was added. For RNA samples without detectable amplicons in the agarose gel following the first PCR, a nested PCR (nPCR, nt 390–721 of strain PUUV CG18-20) was set up using 5 μl of the first reaction and primers PUUV390 (5′-GGN CAR ACA GCR GAT TGG T-3′) and cPUUV721 (5′-ACH CCC ATN ACW GGR CTY AT-3′) and amplification and extension conditions as previously descibed [24, 25]. A RT–PCR specific for DOBV was performed using primers DOBV-M6 (5′-AGY CCW GTN ATG RGW GTR ATT GG-3′) and DOBV-M8 (5′-GAK GCC ATR ATN GTR TTY CKC ATR TCC TG-3′) as described previously [26].
Morphological species determination of the 10 PUUV-PCR-positive bank voles was confirmed using a PCR specific for the mt cytochrome B (cyt B) gene [27]. For one A. flavicollis (no. 5/04) this gene was amplified using primers H16 (5′-CWG GTT GRC CTC CRA TTC AWG T-3′) and L14648 (5′-TGA ATY TGA GGR GGA TTC TCA GTA-3′). Denaturation and extension were performed as described above, annealing was modified by running five cycles reduced from 60 to 50°C each for 30 s, and 40 cycles at 50°C for 30 s.
Amplified products were run in agarose gels, visualized with ethidium bromide and UV illumination. For sequencing, amplicons were purified using the QIAquick PCR purification kit according to the manufacturer's instructions (Qiagen). Direct sequencing of the purified PCR products was done using S segment-specific and cyt B-specific primers respectively. Mitochondrial cyt B sequences were also submitted to GenBank (Accession numbers: DQ090751–DQ090761).
Alignments and phylogenetic analyses
Alignments of nt and deduced amino-acid (aa) sequences were carried out using Clustal W ([28], BioEdit version 7.0.2). Phylogenetic analyses were performed using PHYLIP version 3.5c and maximum parsimony [29]. Sequences were deposited in GenBank. For comparative purposes, sequences of hantaviruses and rodent cyt B were obtained from GenBank (Fig. 2).
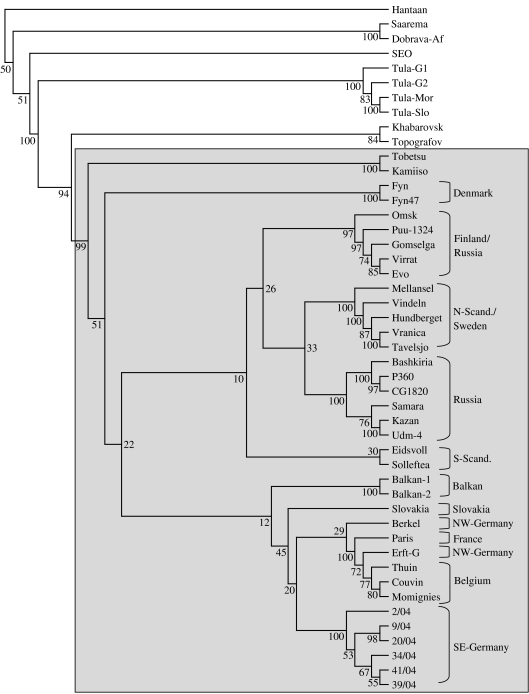
Fig. 2.
Phylogenetic tree of PUUV sequences from the investigated rodents from South-East Germany in comparison to other Eurasian hantaviruses. The phylogenetic tree is based on a 712 nt fragment of the viral S segment (nt 364–1076). Figures on each branch represent the percentage of bootstrap support for maximum parsimony (100×bootstrap). Bootstrap values >50 are indicated. Puumala viruses are shaded. Abbreviations: N-Scand., North Scandinavia; S-Scand., South Scandinavia (Norway, Sweden), NW-Germany, North-West Germany; SE-Germany, South-East Germany. Abbreviations for strains: Hantaan, Hantaan virus strain 76–118, M14626; Saaremaa, Apodemus agrarius-associated Saaremaa/160V virus, AJ009773; Dobrava-Af, Dobrava virus strain associated to Apodemus flavicollis, L41916; SEO, Seoul virus strain SR-11, M34881; Tula-G1, Tula virus strain Germany 1, AF164093; Tula-G2, Tula virus strain Germany 2, AF164094; Tula-Mor, Tula virus strain Moravia/5302Ma/94, Z49915; Tula-Slo, Tula virus strain Slovakia, AJ223600; Khabarovsk, Khabarovsk virus strain MF-43, U35255; Topografov, Topografov virus strain Ls136V, AJ011646; Tobetsu, PUUV strain Tobetsu 60Cr/93, AB010731; Kamiiso, PUUV strain Kamiiso 8Cr/95, AB010730; Fyn, PUUV strain Fyn, AJ238791; Fyn47, PUUV strain Fyn 47, AJ278092; Omsk, PUUV strain Omsk/CG144/199-2000, AF367064; Puu-1324, PUUV strain 1324CG/79, Z46942; Gomselga, PUUV strain Karelia/Gomselga, AJ238790; Virrat, PUUV strain Virrat/25Cg/95, Z69985; Evo, PUUV strain Evo/12Cg/93, Z30702; Mellansel, PUUV strain Mellansel/Cg47/94, AJ223374; Vindeln, PUUV strain Vindeln/L20Cg/83, Z48586; Hundberget, PUUV strain Hundberget/Cg36/95, AJ223371; Vranica, PUUV strain Vranica/Hällnäs, U14137; Tavelsjo, PUUV strain Tavelsjo Cg81/94, AJ223380; Bashkiria, PUUV strain Bashkortostan/2001/CG17B, AF442613; P360, PUUV strain P360, L11347; CG18-20, PUUV strain CG18-20/Ufa83/Häallnaäs B1, M32750; Samara, PUUV strain Samara/FS-808 AF411446; Kazan, PUUV strain Kazan, Z84204; Udm-4, PUUV strain Udmurtia 444Cg/88, Z30706; Eidsvoll, PUUV strain Eidsvoll 1124v, AJ223368; Solleftea, PUUV strain Puu/Solleftea/Cg3/95, AJ223376; Balkan-1, PUUV strain Balkan 1, AJ314600; Balkan-2, PUUV strain Balkan 2, AJ314601; Slovakia, PUUV virus strain Opina916-Slovakia, AF294652; Berkel, PUUV strain Berkel, L36943; Paris, PUUV strain Paris 90-13 CG13891, U22423; Erft-G, PUUV strain Erft Germany, AJ238779; Thuin, PUUV strain Thuin AJ277030; Couvin, PUUV strain Couvin, AJ277034; Momignies, PUUV strain Momignies, AJ277032.
RESULTS
Rodent trapping
Fourty-three rodents were collected in seven places located close to putative infection sites of NE patients (see Fig. 1c; Falkenstein: 49° 4′ N, 13° 14′ E; Raimundsreuth: 48° 52′ N, 13° 32′ E; Glashütte: 48° 52′ N, 13° 33′ E; Hangenleithen: 48° 52′ N, 13° 10′ E; Schöfweg/Mutzenwinkel: 48° 0′ N, 13° 14′ E; Langfurth: 48° 49′ N, 13° 12′ E) within 99 trapping nights in October 2004. Trapped rodents belonged to four species: 29 bank voles (C. glareolus), 11 yellow-necked mice (A. flavicollis), two wood mice (A. sylvaticus), and one house mouse (Mus musculus). Thus, trapping index by apple decoying was 43%.
Serological investigations of trapped rodents
Initial serological screening of 29 rodent sera revealed seven ELISA-positive samples reacting with DOBV-Slo and/or PUUV-Vra, originating from six bank voles and one yellow-necked mouse (Table 1). PUUV reactivity was confirmed by WB and IFA. The DOBV ELISA reactivity of five sera was confirmed by WB. For 5 out of the 7 sera the ELISA demonstrated an at least two-fold higher end-point titre for PUUV than for DOBV. From 15 transudates (11 bank voles, 3 yellow-necked mice, 1 house mouse) five bank vole samples were PUUV-reactive in ELISA and WB (Table 1). Three out of these five transudates also reacted with DOBV rN protein. Investigation of the transudates confirmed the positive serology for nos. 20/04 and 34/04. For three additional bank voles (nos. 9/04, 39/04, 41/04), for which no serum was available, hantavirus-specific antibodies were detected. In summary, 10 samples (7 sera, 3 liver transudates, 23%), originating from nine bank voles and one yellow-necked mouse, trapped in four of the seven investigated locations were seroreactive.
Table 1.
Hantavirus-specific serology and RT–PCR results
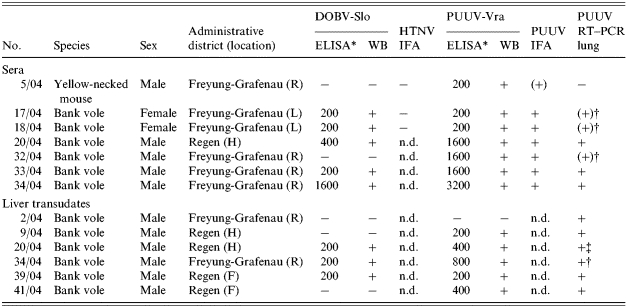
H, Hangenleiten; F, Falkenstein; R, Raimundsreuth; L, Langfurth; n.d., not determined.
End-point titres: −, negative; +, positive; (+), weakly positive. † (+), Nested RT–PCR positive; ‡ see upper part of the table.
RT–PCR analysis of rodent tissue samples
Lung, kidney and bladder samples of all 43 rodents were analysed for the presence of viral RNA by PUUV-specific RT–PCR. For seven of the 43 (24%) lung samples from bank voles an amplicon of the expected size (761 bp) was detected by PUUV-specific RT–PCR (Table 1). Three corresponding kidney samples (7%) and two bladder samples (5%) also yielded this fragment (Table 2). Interestingly, PUUV-specific RNA was detected in further tissues of some of these PUUV-positive rodents: in the hearts of five, livers of two, spleen of one and gonads of one other bank vole respectively (Table 2). For lung RNA without amplicon in the RT–PCR, sensitivity was enhanced using nPCR. Amplicons of the expected 331 bp were obtained for a further three bank voles (nos. 17/04, 18/04, 32/04; Table 1). In two of these three voles the nPCR was also positive using bladder, kidney and liver samples (Table 2). The DOBV-specific RT–PCR amplified the in vitro-transcribed DOBV S segment RNA, but was negative for all rodent lungs, kidneys and bladders. Tissue samples from the seropositive A. flavicollis (no. 5/04) were negative in PUUV- and DOBV-specific RT–PCRs (Table 1). Sequencing of the cyt B gene (1090 nt) of PUUV-positive bank voles revealed a 98–99% homology to published C. glareolus sequences [26]. The partial cyt B gene (514 nt) of A. flavicollis no. 5/04 showed 100% homology to that of one originating from Ukraine (GenBank AY158454).
Table 2.
Detection of PUUV by RT–PCR (S-segment, nt 342–1122) and nested RT–PCR (nt 390–721) using RNA isolated from different bank vole tissues
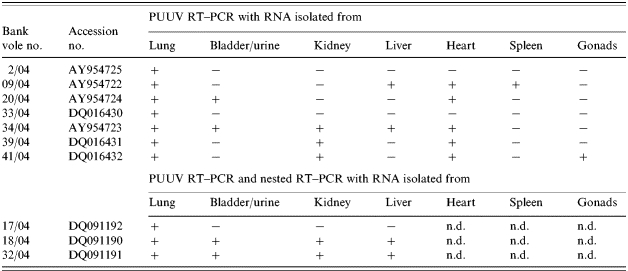
n.d., Not determined; −, negative; +, positive.
Sequence and phylogenetic analysis
Nucleotide sequences of lung-derived PCR products were compared to corresponding partial S segments of PUUV strains from Europe and Asia. Nucleotide sequences of PCR products from different tissues of bank vole nos. 9/04, 34/04 and 32/04 were identical to that of lungs of the corresponding rodent (data not shown). The seven PUUV nucleotide sequences (size 712 nt without primers) obtained from Bavarian bank voles showed up to 3·1% divergence (Table 3). The three additional nucleotide sequences obtained by nRT–PCR did not enhance this level of divergence (nos. 17/04 and 18/04 identical and 98·6% homologous to no. 32/04). Nucleotide sequences of two bank voles (nos. 2/04, 33/04) trapped at the same site were identical for the analysed region (data not shown). The level of divergence to the corresponding S-segment region of previously described PUUV strains from Slovakia, Germany, France and Belgium was 14·0–16·6%. Divergence from partial S segments of PUUV strains of Scandinavia and Balkan origin was 16·6–17·6%, and from Russian PUUV, Danish PUUV and Tobetsu-Kamiiso clusters it was 18–20%. In a 611-bp fragment of the S segment PUUV sequences originating from Austrian bank voles [24] were 81% identical to the Bavarian PUUV sequences (data not shown). The difference at the nt level was 29% for the North-American Isla Vista and Prospect Hill viruses and 35–37% for the El Moro Canyon virus group, 31–32% for TULV, 41–42% for DOBV, Seoul Virus (SEOV) and the outgroup HTNV. The deduced sequences of the 237 aa partial nucleocapsid (N) protein were identical for three bank voles (nos. 34/04, 39/04, 41/04). The other partial N proteins from Bavarian PUUV shared 98·7–99·5% aa similarity, the shorter polypeptides (109 aa) of nos. 17/04, 18/04 and 32/04 were identical. The polypeptide divergence to PUUV strains of other geographical origin was 0·9–4·7%, to the Tobetsu-Kamiiso cluster 4·7–5·1%, Topografov-Khabarovsk viruses 14–14·8%, TULV 24·5–25·8%, DOBV and SEOV 41·8–45·6% and to the outgroup HTNV 57%. Phylogenetic analysis of the partial S segments demonstrated a separate PUUV clade for Asia and several European PUUV clades geographically localized in Denmark, Finland/Russia, Northern Scandinavia/Sweden, Russia, Southern Scandinavia, Balkans and Central Europe (Fig. 2). When analysing the phylogeny of the Central European PUUV clade, three major sublineages were found: Slovakia, North-Western Germany/France/Belgium, and a new Central-European sublineage in South-East Germany.
Table 3.
Comparison of nucleotide (nt) identity, and amino acid (aa) similarity, of partial PUUV S segments and deduced nucleocapsid proteins of the South-East German strains and of other selected hantaviruses. Gaps and stop codons are included in the calculation (the nucleotide sequences of 02/04 and 33/04 are identical)
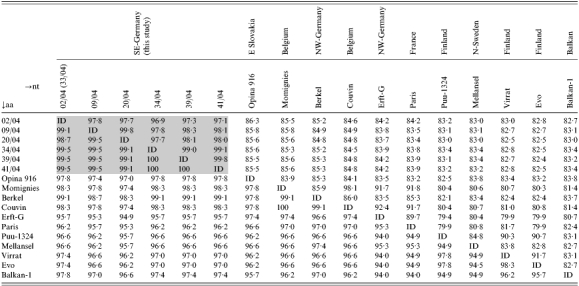
Opina916, Puumala virus strain Opina916-Slovakia, AF294652; Momignies, Puumala virus strain Momignies, AJ277032; Berkel, PUUV strain Berkel, L36943; Couvin, Puumala virus strain Couvin, AJ277034; Erft-G, PUUV strain Erft, Germany, AJ238779; Paris, PUUV strain Paris 90-13 CG13891, U22423; Puu-1324, PUUV strain 1324CG/79, Z46942; Mellansel, PUUV strain Mellansel/Cg47/94, AJ223374; Virrat, PUUV strain Virrat/25Cg/95, Z69985; Evo, PUUV strain Evo/12Cg/93, Z30702; Balkan-1, Puumala virus strain Balkan-1, AJ314600.
ID, 100% identity.
DISCUSSION
PUUV is endemic in Central Europe and in some parts of Germany, e.g. Suebian Alb in Baden-Wuerttemberg and Under Franconia, with sporadic epidemic outbreaks as well as isolated cases. From 2001 to 2003 only three infections were diagnosed in Lower Bavaria [19]. An increased incidence of 38 NE cases was observed in this area in 2004 (45 cases up to June 2005) when around 31 of the 38 (82%) patients in 2004 were referred to a hospital. For all patients reported PUUV infections were diagnosed [19]. Interestingly, similar reports came from neighbouring countries: in Austria and Czech Republic in 2004 an increase of human hantavirus infections was notified (S. Aberle and M. Pejcoch, personal communication). Whereas last year no increase in the numbers of clinical cases was found in certain parts of Germany, Belgium and France, a large number of infections was apparent in the 20 first weeks of 2005 (P. Heyman et al., personal communication).
We investigated potential causal links between rodent populations, the hantaviruses they carry and hantavirus emergence in Lower Bavaria. Patients living in the affected districts were reported to have cleaned houses, garages, or other buildings, or had direct contact with rodents or their faeces. Additionally, a recent serological study of approximately 600 samples from persons living in the affected area revealed a higher percentage of PUUV-reactive sera than expected from the average hantavirus prevalence (R. Wölfel et al., unpublished observations). Investigation of 43 rodents trapped close to the potential exposure sites of some patients revealed 23·3% hantavirus-positive rodents. The detection of 31% PUUV-seroreactive and 34·5% RT–PCR-positive bank voles suggested PUUV as the causative agent for this outbreak. With roughly every third mouse carrying PUUV, it seems likely that the infected local C. glareolus population was the source of human infections. The failure to detect DOBV sequences in rodents confirms the suggestion that PUUV is responsible for the outbreak.
We analysed rodent samples by serological and molecular assays. All PUUV-seroreactive voles were (n)RT–PCR-positive, while nine of 10 PCR-positive were seroreactive. The bank vole (no. 34/04) with the highest end-point titre in PUUV ELISA showed amplicons in the highest number (n=5) of different tissues. Samples from vole nos. 17/04 and 18/04 with a low PUUV ELISA titre were positive exclusively in the nRT–PCR. Differences observed in the end-point titres for PUUV and DOBV ELISA, with lacking reactivity of a few samples in DOBV ELISA, were in line with serological findings for human sera. This demonstrates the necessity of the use of a homologous antigen for highly sensitive detection of hantavirus infections [22, 30, 31].
Comparing RT–PCR data of different tissues demonstrated that the lung represents the most useful sample for detection of hantavirus RNA as evidenced by other investigations [24, 27, 32, 33]. However, if only other tissues (heart, kidney) are available, these can be used for assays, but because of lower detection rates this may not allow conclusions on hantavirus prevalence. Accordingly, screening of kidneys of A. flavicollis from Greece [34] and A. agrarius from Hungary [26] resulted in the detection of DOBV-related sequences.
This study was conducted to characterize a PUUV strain found in Lower Bavaria, which is genetically distinct from other known PUUV strains and, therefore, represents a new subtype of PUUV. Although in Germany up to 240 hantavirus infections are reported annually, few data exist on characterization of virus species and their reservoirs. So far, two PUUV sequences have been published: the Berkel strain, found in a patient [5], and the Clethrionomys-derived PUUV Erft which is 14% divergent to the Berkel strain, while both originated from the same region in North-West Germany [35]. We present the first collection of sequences obtained from an outbreak in Germany and from South-East Germany in particular. The observed PUUV divergence of 0·2–3·1% mirrors the natural range of diversity of S segments as observed for PUUV strains from Denmark (5% divergence, [33]), TULV strains from Slovakia (4%, [36]) and DOBV strains in Slovenia (up to 5%, [37]). However, the obtained PUUV nucleotide sequences were 14·7–16·3% divergent to that from other regions in Germany indicating a new PUUV sublineage in Lower Bavaria. Phylogenetic analysis supported considerable genetic variation among PUUV in Germany. Membership to a genetic sublineage was described by a nucleotide sequence diversity of 8%, diversity of >14% outgrouped a sequence [35].
Rodent population density may also influence the increase of human NE cases in Lower Bavaria. Several studies showed the correlation of NE infections with the population of their rodent reservoirs [38, 39]. Serological examinations of bank voles in Belgium, over a period of 5 years, indicated an up to eightfold difference in animals carrying PUUV antibody, depending on the respective population density. A pullulation of bank voles in 1999 coincided with an increase of PUUV-seropositive animals [40]. The high trapping efficiency may reflect an increased population of rodents in Lower Bavaria. Unfortunately, in contrast to Scandinavian countries [41], there is no official surveillance of rodent populations in Germany so far. However, patients living in a rural environment and forestry offices observed a high prevalence of tree fruits (e.g. a proliferation of beech-nut, acorns) and consequently, with the higher availability of sources of nutrition, a massive increase of the rodent population in Bavaria in 2004. Further studies in collaboration with local forest authorities will gain more data on population dynamics. Our trapping by decoying and use of Sherman traps was highly efficient. Usually, captures effectively depend on exogenous variables such as habitat, weather, water and food availability, behaviour of target species, etc. [42, 43].
The high prevalence of PUUV observed in C. glareolus may be responsible for the observed spillover infection to a A. flavicollis (no. 5/04). Spillover was previously described for PUUV from C. glareolus to Microtus arvalis and A. sylvaticus [44], DOBV from A. flavicollis to A. sylvaticus and Mus musculus [45] and TULV from Microtus arvalis to Pitymys subterraneus [46].
Additional studies are in need to understand the reasons for this outbreak, the role of rodent population density as well as PUUV prevalence in rodents. Future trapping activities will monitor the rodent populations in the affected part of Germany. Likewise, the characterization of the Bavarian subtype will be continued.
ACKNOWLEDGEMENTS
The authors gratefully acknowledge the excellent help in rodent trapping from Dr Mirko Köhler, Aileene Lorber, Andreas Knoche, and Florian Goldberg. Thanks are due to Harald Weber, Peter Klein, Gudrun Zöller, Aileene Lorber and Roswitha Mattis for their excellent technical assistance. The authors also thank Prof. Dr Stephan Aberle (Vienna), Dr Herve Zeller (Lyon), Dr Milan Pejcoch (Prague), Paul Heyman (Brussels), Dr Jerrold J. Scharninghausen (Würzburg), Dr Graf (Freyung) for communication of unpublished data, and Dr Böer, Local Health Office Regen, Dr Schraml, Local Health Office Freyung-Grafenau, Dr Wifling, Local Health Office Passau, and Dr Schoder, Schöfweg, for information about patients and infection sites. Prof. Dr Bäumler, Technical University Munich, supported the project with Sherman traps.
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Plyusnin A, Krüger DH, Lundkvist A. Hantavirus infections in Europe. Advances in Virus Research. 2001;57:105–136. doi: 10.1016/s0065-3527(01)57002-5. [DOI] [PubMed] [Google Scholar]
- 2.Schmaljohn CS, Hjelle B. Hantaviruses. A global disease problem. Emerging Infectious Diseases. 1997;3:95–104. doi: 10.3201/eid0302.970202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee HW, Lee PW, Johnson KM. Isolation of the etiologic agent of Korean hemorrhagic fever. Journal of Infectious Diseases. 1978;137:298–308. doi: 10.1093/infdis/137.3.298. [DOI] [PubMed] [Google Scholar]
- 4.Pilaski J et al. Haemorrhagic fever with renal syndrome in Germany. Lancet. 1991;337:111. doi: 10.1016/0140-6736(91)90765-h. [DOI] [PubMed] [Google Scholar]
- 5.Pilaski J et al. Genetic identification of a new Puumala virus strain causing severe haemorrhagic fever with renal syndrome in Germany. Journal of Infectious Diseases. 1994;170:1456–1462. doi: 10.1093/infdis/170.6.1456. [DOI] [PubMed] [Google Scholar]
- 6.Kunz A et al. Epidemic nephropathy. An important differential diagnosis of acute renal failure in Reutlingen, an endemic area [in German] Deutsche Medizinische Wochenschrift. 2002;127:1685–1689. doi: 10.1055/s-2002-33374. [DOI] [PubMed] [Google Scholar]
- 7.Rasche FM et al. Thrombocytopenia is a predictor for acute renal failure in Puumala hantavirus infections. Emerging Infectious Diseases. 2004;10:1420–1425. doi: 10.3201/eid1008.031069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zöller L et al. Seroprevalence of hantavirus antibodies in Germany as determined by a new recombinant enzyme immunoassay. European Journal of Clinical Microbiology and Infectious Diseases. 1995;14:303–315. doi: 10.1007/BF02116523. [DOI] [PubMed] [Google Scholar]
- 9.Meisel H et al. First case of infection with hantavirus Dobrava in Germany. European Journal of Clinical Microbiology and Infectious Diseases. 1998;17:884–885. doi: 10.1007/s100960050214. [DOI] [PubMed] [Google Scholar]
- 10.Klempa B et al. First molecular identification of human Dobrava virus infection in Central Europe. Journal of Clinical Microbiology. 2004;42:1322–1325. doi: 10.1128/JCM.42.3.1322-1325.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ulrich R et al. Prevalence of hantavirus infections in Germany [in German] Bundesgesundheitsblatt. 2004;47:661–670. doi: 10.1007/s00103-004-0858-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kimmig P et al. Epidemiology of hantaviruses in Germany [in German] Gesundheitswesen. 2001;63:107–112. doi: 10.1055/s-2001-10961. [DOI] [PubMed] [Google Scholar]
- 13.Klempa B et al. Occurrence of renal and pulmonary syndrome in a region of North-East Germany where Tula hantavirus circulates. Journal of Clinical Microbiology. 2003;41:4894–4897. doi: 10.1128/JCM.41.10.4894-4897.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kimmig P, Wagner-Wiening C, Hassler D. Hantaviruses – ‘exotic’ viruses in Germany [in German] Deutsche Medizinische Wochenschrift. 2002;127:2369–2370. [PubMed] [Google Scholar]
- 15.Anon Berlin: Robert Koch-Institut; 2002. pp. 61–63. . Infektionsepidemiologisches Jahrbuch meldepflichtiger Krankheiten für 2001. [Google Scholar]
- 16.Anon Berlin: Robert Koch-Institut; 2003. pp. 65–68. . Infektionsepidemiologisches Jahrbuch meldepflichtiger Krankheiten für 2002. [Google Scholar]
- 17.Anon Berlin: Robert Koch-Institut; 2004. pp. 73–77. . Infektionsepidemiologisches Jahrbuch meldepflichtiger Krankheiten für 2003. [Google Scholar]
- 18.Anon Berlin: Robert Koch-Institut; 2005. pp. 79–83. . Infektionsepidemiologisches Jahrbuch meldepflichtiger Krankheiten für 2004. [Google Scholar]
- 19.Hautmann W, Essbauer S, Ulrich R. Cumulative appearance of clinical apparent hantavirus infections in Lower Bavaria in 2004 [in German] Epidemiological Bulletin. 2005;10:85–86. [Google Scholar]
- 20.Dargeviciute A et al. Yeast-expressed Puumala hantavirus nucleocapsid protein induces protection in a bank vole model. Vaccine. 2002;4:3523–3531. doi: 10.1016/s0264-410x(02)00341-9. [DOI] [PubMed] [Google Scholar]
- 21.Razanskiene A et al. High yields of stable and highly pure nucleocapsid proteins of different hantaviruses can be generated in the yeast Saccharomyces cerevisiae. Journal of Biotechnology. 2004;111:319–333. doi: 10.1016/j.jbiotec.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 22.Schmidt J et al. Development and evaluation of serological assays for the detection of human hantavirus infections caused by Sin Nombre virus. Journal of Clinical Virology. 2005;33:247–253. doi: 10.1016/j.jcv.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 23.Geldmacher A et al. A hantavirus nucleocapsid protein segment exposed on hepatitis B virus core particles is highly immunogenic in mice when applied without adjuvants or in the presence of pre-existing anti-core antibodies. Vaccine. 2005;23:3973–3983. doi: 10.1016/j.vaccine.2005.02.025. [DOI] [PubMed] [Google Scholar]
- 24.Bowen MD et al. Puumala virus and two genetic variants of Tula virus are present in Austrian rodents. Journal of Medical Virology. 1997;53:174–181. doi: 10.1002/(sici)1096-9071(199710)53:2<174::aid-jmv11>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 25.Scharninghausen JJ et al. Genetic evidence for Tula virus in Microtus arvalis and Microtus agrestis populations in Croatia. Vector-Borne and Zoonotic Disease. 2002;2:19–27. doi: 10.1089/153036602760260742. [DOI] [PubMed] [Google Scholar]
- 26.Scharninghausen JJ et al. Genetic evidence for Dobrava virus in Apodemus agrarius in Hungary. Emerging Infectious Diseases. 1999;5:468–470. doi: 10.3201/eid0503.990324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dekonenko A et al. Genetic similarity of Puumala viruses found in Finland and Western Siberia and of the mitochondrial DNA of their rodent hosts suggests a common evolutionary origin. Infection, Genetics and Evolution. 2003;3:245–257. doi: 10.1016/s1567-1348(03)00088-1. [DOI] [PubMed] [Google Scholar]
- 28.Hall TA. BioEdit. a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symposium Series. 1999;41:95–98. [Google Scholar]
- 29.Felsenstein J. PHYLIP – phylogeny interference package. Cladistics. 1989;5:164–166. [Google Scholar]
- 30.Sjölander KB, Lundkvist Å. Dobrava virus infection: serological diagnosis and cross-reactions to other hantaviruses. Journal of Virological Methods. 1999;80:137–143. doi: 10.1016/s0166-0934(99)00037-3. [DOI] [PubMed] [Google Scholar]
- 31.Schmidt J et al. Serological assays for the detection of human Andes hantavirus infections based on its yeast-expressed nucleocapsid protein. Intervirology. 2006;49:173–184. doi: 10.1159/000089378. [DOI] [PubMed] [Google Scholar]
- 32.Lundkvist Å et al. Isolation and characterization of Puumala hantavirus from Norway. Evidence for a distinct phylogenetic sublineage. Journal of General Virology. 1998;79:2603–2614. doi: 10.1099/0022-1317-79-11-2603. [DOI] [PubMed] [Google Scholar]
- 33.Sironen T et al. Distribution of Puumala hantavirus in Denmark. Analysis of bank voles (Clethrionomys glareolus) from Fyn and Jutland. Vector-Borne and Zoonotic Disease. 2002;2:37–45. doi: 10.1089/153036602760260760. [DOI] [PubMed] [Google Scholar]
- 34.Papa A et al. Preliminary characterization and natural history of hantaviruses in rodents in northern Greece. Emerging Infectious Diseases. 2000;6:645–655. doi: 10.3201/eid0606.000618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heiske A et al. A new Clethrionomys-derived hantavirus from Germany: evidence for the distinct genetic sublineages of Puumala viruses in Western Europe. Virus Research. 1999;61:101–112. doi: 10.1016/s0168-1702(99)00024-6. [DOI] [PubMed] [Google Scholar]
- 36.Sibold C et al. Recombination in Tula hantavirus. Analysis of genetic lineages from Slovakia. Journal of Virology. 1999;73:667–675. doi: 10.1128/jvi.73.1.667-675.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Avsic-Zupanc T et al. Genetic analysis of wild-type Dobrava hantavirus in Slovenia: co-existence of two distinct genetic lineages within the same natural focus. Journal of General Virology. 2000;81:1747–1755. doi: 10.1099/0022-1317-81-7-1747. [DOI] [PubMed] [Google Scholar]
- 38.Hjelle B, Glass GE. Outbreak of hantavirus infection in the Four Corners region of the United States in the wake of the 1997–1998 El Nino-southern oscillation. Journal of Infectious Diseases. 2000;181:1569–1573. doi: 10.1086/315467. [DOI] [PubMed] [Google Scholar]
- 39.Calisher CH et al. Do unusual site-specific population dynamics of rodent reservoirs provide clues to the natural history of hantaviruses? Journal of Wildlife Diseases. 2001;37:280–288. doi: 10.7589/0090-3558-37.2.280. [DOI] [PubMed] [Google Scholar]
- 40.Escutenaire S et al. Spatial and temporal dynamics of Puumala hantavirus infection in red bank vole (Clethrionomys glareolus) populations in Belgium. Virus Research. 2000;67:91–107. doi: 10.1016/s0168-1702(00)00136-2. [DOI] [PubMed] [Google Scholar]
- 41.Korpimaki E et al. Predator-induced synchrony in population oszillations of coexisting small animal species. Proceedings of the Royal Society of London. Series B: Biological Sciences. 2005;272:193–202. doi: 10.1098/rspb.2004.2860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Call MS., Cooperrider AY, Boyd RJ, Stuart HR. Inventory and Monitoring of Wildlife Habitat. Denver, CO: US Department of the Interior, Bureau of Management Service Center; 1986. Rodents and insectivores; pp. 429–452. , pp. [Google Scholar]
- 43.Margaletic J, Glavas M, Bäumler W. The development of mice and voles in an oak forest with a surplus of acorns. Journal of Pesticide Science. 2002;75:95–98. [Google Scholar]
- 44.Klingström J et al. Rodent host specificity of European hantaviruses: evidence of Puumala virus interspecific spillover. Journal of Medical Virology. 2002;68:581–588. doi: 10.1002/jmv.10232. [DOI] [PubMed] [Google Scholar]
- 45.Weidmann M et al. Identification of genetic evidence for Dobrava virus spillover in rodents by nested reverse transcription (RT)-PCR and TaqMan RT-PCR. Journal of Clinical Microbiology. 2005;43:808–812. doi: 10.1128/JCM.43.2.808-812.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Song JW, Gligic A, Yangihara R. Identification of Tula hantavirus in Pitymys subterraneus captured in the Cacak region of Serbia-Yugoslavia. International Journal of Infectious Diseases. 2002;6:31–36. doi: 10.1016/s1201-9712(02)90133-5. [DOI] [PubMed] [Google Scholar]