SUMMARY
Underreporting of hepatitis A infection in England may be high and a number of outbreaks have occurred undetected by routine surveillance. We evaluated surveillance of hepatitis A cases by employing capture–recapture analysis on data from two distinct outbreaks of hepatitis A. The overall reporting of cases of hepatitis A was 81·7% (95% CI 55·3–95) in the first outbreak in North East England and reporting through Lab Base was 65·7% (95% CI 42·8–76·4). In the second outbreak in the East Midlands the overall reporting of hepatitis A cases was 27·8% (95% CI 19–38·7) and through Lab Base 16·6% (95% CI 11·4–23·1). Underreporting of hepatitis A cases is high. Public health interventions exist to prevent and control outbreaks of hepatitis A. The lack of reliable data on incidence and prevalence hampers effective public health management of this disease.
INTRODUCTION
Hepatitis A is a viral infection that can be transmitted faeco-orally amongst vulnerable populations. An effective vaccine is available but it is not part of the routine immunization schedule in the United Kingdom. A large proportion of the UK population is susceptible to hepatitis A infection. A large national epidemic of hepatitis A peaked in 2002. The peak consisted of a series of outbreaks across the United Kingdom [1, 2]. Most of these outbreaks were not detected through the routine surveillance systems in place, raising concerns about data quality and the effectiveness of public health surveillance [3]. We aimed to quantify the degree of underreporting of cases of hepatitis A by the national laboratory reporting system (Lab Base) by applying capture–recapture techniques to data from two different outbreaks of this infection in England.
METHODS
The first outbreak occurred in the North East of England in 2002. Three data sources were available here. The local Health Protection Unit (HPU) supplied us with a list of cases. A pilot project carried out by the then PHLS (Public Health Laboratory Service) investigating genotyping of hepatitis A cases [4] provided us with another dataset. These two were compared to the Lab Base dataset. Comparison of cases was carried out using an Access database.
The second outbreak we considered occurred in the East Midlands in 2003. The local HPU supplied us with a dataset and this was compared with the Lab Base dataset. No other dataset was available for this outbreak. Comparison of cases was carried out using an Access database.
Capture–recapture analyses for both outbreaks used stata version 8.2 (Stata Corp., College Station, TX, USA). Capture–recapture techniques were applied to these data sources to allow for underreporting in the different data sources for the two distinct outbreaks [5].
Log-linear modelling was used to estimate the number of cases in each area. A saturated model was used for each dataset. For three data sources, the saturated model included two-way interactions, all of which we believed to be meaningful. The Akaike Information Criteria (AIC) was used to check that the interactions were reflected in the data. Profile Poisson likelihood intervals were used to calculate 95% confidence intervals (CI).
RESULTS
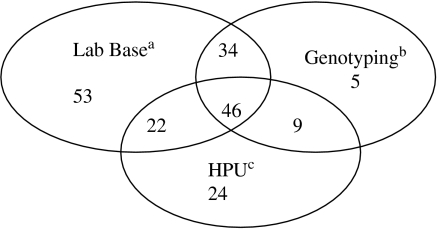
In the first outbreak Lab Base recorded 155 cases. The data source from the local HPU recorded 101 cases. The dataset derived from the genotyping project recorded 94 cases. In total 350 cases were detected. Cases were matched between all three sources using surnames, date of birth and sample date variables. After matching we concluded that the 350 entries represented 193 different cases (Fig. 1). The number of additional cases was estimated to be 43 (95% CI 10–169) based on the three-way interaction being zero. Knowing something of the administrative procedures which gave rise to the lists, we expected statistical dependency between pairs of lists, especially between the Lab Base data and the genotyping data, and an additional but smaller degree of dependency between all three lists.
Fig. 1.
Diagram of the North East England outbreak, 2002 showing the three data sources used in the analysis and the number of cases in each list and how many were common between all the lists. a Data from the national laboratory reporting system; b data derived from a project carried out by the national Public Health Laboratory on hepatitis A genotyping; c data from the local Health Protection Unit.
A saturated model (three main effects and three two-way interactions) was chosen to account for possible dependency between sources. Our estimate of 43 unlisted cases can be realistically increased to 300 or more by estimating the impact of varying visibility on the size of the three-way interaction. If we assume that each case has a visibility on the scale 0–1, maximal visibility where v=1 is associated with a zero three-way interaction. If the average visibility in the population is 0·8, 0·5 or 0·2 the three-way interaction is greater than 0 and increases the estimated number of missing cases to 103, 279 or 937 respectively. We assessed how sensitive the prediction is to matching errors. The data were adjusted in six different ways to represent the effect of a single erroneous match, and the model was refitted each time. The estimate of 43 increased by −4, 0, 5, 5, 12 and 18 in the six cases, and it can be assumed that roughly opposite effects would be produced by the converse errors.
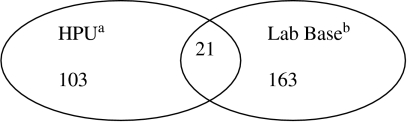
For the second outbreak, we only had two lists. The first was derived from Lab Base (184 cases) and the second a database from the HPU that had been enhanced through active case finding by one member of staff (124 cases). Matching was carried out using date of birth and sample dates. In total 308 cases were detected. After matching we concluded that the 308 entries represented 287 different cases (Fig. 2). Using a saturated model the number of additional cases was estimated to be 799 (95% CI 488–1310). As there were only two lists this is an independence model. The confidence interval was calculated from a standard error on the log scale. We adjusted the data to represent the effect of a single matching error in either direction. The point estimate of the number of missed cases became 751 or 853 instead of 799. The Table summarizes the estimates of potential underreporting derived from the capture–recapture analysis.
Fig. 2.
Diagram of the East Midlands outbreak, 2003 showing the two data sources used in the analysis and the number of cases in each list and how many were common to both lists. a Data from the local Health Protection Unit; b data from the national laboratory reporting system.
Table.
Reporting of cases of hepatitis A through the surveillance system in two distinct outbreaks in England

CI, Confidence interval.
Varying catchability of cases can be allowed for by stratifying the analysis by variables thought to be related to capture (e.g. risk factor information, ethnicity, post code, etc.), and then performing separate capture–recapture analyses for each stratum. Unfortunately our data did not contain enough additional information to allow further stratification analysis to allow for varying catchability.
DISCUSSION
Capture–recapture techniques have historically been used to estimate the size of wildlife populations. When applying this technique to human epidemiology there are some limitations to consider. Capture in one list is unlikely to be independent of capture in another list. If a case appears on a HPU database, that person has most probably sought medical advice and been tested and so is more likely to appear on a laboratory database. Moreover, the probability of capture (catchability) is not necessarily homogeneous across all cases of hepatitis A. We concluded that the total effect of any errors we may have made in matching was small compared with the uncertainty represented by the confidence limits. This infection affects marginalized groups in society including injecting drug users (IDUs) who may be less visible to health services than other vulnerable groups such as returning travellers. The analysis of data from the 2003 East Midlands outbreak showed lower levels of reporting of cases within Lab Base than for the 2002 North East England outbreak. The active case finding that was carried out by the HPU in the East Midlands enhanced the HPU dataset identifying additional cases that had not been notified to the HPU. This enhanced dataset from the HPU may have contributed to a much lower level of Lab Base reporting being noted in the analysis. The apparent better reporting of cases within Lab Base in the 2002 outbreak in the North East may have reflected better reporting in that particular area or may be spurious. Cases may have been missed from all three datasets because they may not have been reported to the HPU and the laboratory may have not reported this on Lab Base.
Our analysis of two outbreaks taking place at different times and in different regions shows that levels of underreporting of hepatitis A cases in Lab Base may be high. National surveillance relies on Lab Base data. As effective interventions to prevent further spread of hepatitis A are available, underreporting matters because it prevents prompt and effective public health action to protect immediate contacts of cases and their communities [6]. Most of the UK population is susceptible to hepatitis A [7], therefore cases and outbreaks that start in high-risk groups may spread to the general population. The regions with the highest rates of infection in IDUs in recent years also have the highest rates of infection in children [3] which may be due to spread of the infection to the general population or to socio-economic factors that increase the risk of hepatitis A both in children and IDUs. Lack of detailed risk factor information, ethnicity or postcode information in Lab Base data hampered our ability to carry out a stratified analysis. At a national level geographical analysis of outbreaks has to be done by laboratory rather than patient location. Some patients may live a long way from the laboratory where the blood test was processed. This hampers accurate geographical analysis of outbreaks. Without detailed risk factor information it is difficult to address inequity of access to appropriate care and control measures for vulnerable groups and difficult to develop and implement appropriate policy to contain the spread of this infection. Local health protection units may hold valuable information about outbreaks of hepatitis A including additional risk factor information and patient postcodes. Incorporation of this information with Lab Base data needs to be improved in order to enhance national surveillance of hepatitis A.
ACKNOWLEDGEMENTS
We thank Dr Wendy Phillips (Doncaster HPU) and Ms Susie Singleton (East Midlands HPU) who provided Health Protection unit data on cases. We also thank Anjna Mistry (HPA, CFI) who provided Lab Base data and Dr Siew Lin Ngui (HPA, CFI) who provided genotyping data.
DECLARATION OF INTEREST
None.
REFERENCES
- 1.CDR http://www.hpa.org.uk/cdr/PDFfiles/2004/cdr3504.pdf. http://www.hpa.org.uk/cdr/PDFfiles/2004/cdr3504.pdf . CDR weekly Laboratory Reports of hepatitis A in England and Wales: 2003. ( ). Accessed 24 July 2005.
- 2.Health Protection Agency. http://www.hpa.org.uk/cdr/PDFfiles/2004/cdr4204.pdf. http://www.hpa.org.uk/cdr/PDFfiles/2004/cdr4204.pdf ). Accessed 24 July 2005.
- 3.Perrett K et al. Changing epidemiology of hepatitis A: should we be doing more to vaccinate injecting drug users? Communicable Disease and Public Health. 2003;6:97–100. [PubMed] [Google Scholar]
- 4.Ngui SL p. 106. . Molecular epidemiology of hepatitis A virus infection during 2002. Health Protection Agency First Scientific Conference. University of Warwick, September 2003, p.
- 5.International Working Group for Disease Monitoring and Forecasting. Capture–recapture and multiple-record systems estimation II: applications in human diseases. American Journal of Epidemiology. 1995;142:1059–1068. [PubMed] [Google Scholar]
- 6.Crowcroft NS et al. Guidelines for the control of hepatitis A virus infection. Communicable Disease and Public Health. 2001;4:213–227. [PubMed] [Google Scholar]
- 7.Morris MC et al. The changing epidemiological pattern of hepatitis A in England and Wales. Epidemiology and Infection. 2002;128:457–463. doi: 10.1017/s095026880200701x. [DOI] [PMC free article] [PubMed] [Google Scholar]