SUMMARY
Enterohaemorrhagic Escherichia coli O157 (O157) is infectious to humans, particularly children, at very low doses and causes not only haemorrhagic colitis but also other serious symptoms. To investigate an association between intestinal bacterial flora and resistance to such infections, we screened faecal samples for the presence of enteric bacteria that are able to suppress the growth of O157. Samples from 303 individuals, 35 children (aged ⩽6 years) and 268 adults (aged 20–59 years), were examined. Colonies with different appearances on sorbitol MacConkey agar medium were screened for the production of bacteriocins inhibitory for O157 in an overlay agar plate assay. O157-inhibiting strains were isolated from 52 individuals. The prevalence of these bacteria tended to rise with age, and was significantly higher among 40- to 59-year-old adults (23/101, 22·8%) than among children (3/35, 8·6%; P<0·05). To test the hypothesis that these bacteriocin-producing strains contribute to resistance against O157 in human adults, we examined faecal samples of 25 healthy O157 carriers. Inhibitory bacteria were more prevalent among the latter (9/25, 36·0%) than among age-matched subjects who did not carry O157 (49/268, 18·3%). It appears, therefore, that inhibitory bacteria in the human gut may play a role in inhibiting propagation of O157 and/or suppressing expression of virulence factors by this pathogen.
INTRODUCTION
Enterohaemorrhagic Escherichia coli O157 (O157) are infectious to humans at very low doses and cause not only haemorrhagic colitis but also other serious symptoms, such as haemolytic–uraemic syndrome (HUS) and neurological abnormalities. However, only about 30% of 30- to 49-year old adults become symptomatic after colonization, while the remainder become healthy carriers of the organism [1–3]. In contrast, most children and elderly people fall ill due to the infection. Mechanisms of the difference of host susceptibility between adults and children have not as yet been elucidated but reduced gastric secretion of elderly people may account in part for their lower resistance [4].
Immunity is thought to contribute to the resistance of adults to O157. However, serum antibodies specific to the O157 lipopolysaccharide (LPS) antigens were not common even among adults, except those who had occupational contact with farm animals [5, 6], and no age-related increase in seropositivity against the shiga toxin was noted [4]. Children who recovered from HUS also did not develop a long-lasting humoral immune response to the O157 antigen [7]. Further, the serological responses did not correlate with elimination of infection in calves or protection of calves against reinfection with the same strain, although antibodies to shiga toxin 1 and O157 LPS increased in calves during experimental infections with this organism [8]. The presence of healthy children carrying O157 prompted us to consider other factors not involved with immunological defence [2, 9–13].
Normal gastrointestinal flora is composed of autochthonous and allochthonous microorganisms which have marked effects on the morphological, physiological, and immunological development of the host [14]. The bacterial flora play an important role in the prevention and/or the amelioration of intestinal infections through a variety of mechanisms such as (i) direct inhibition of pathogens by bacteriocins and/or metabolites, (ii) competition for receptors and nutrients, and (iii) stimulation of innate or acquired immunity of the host. Some strains have been used as probiotic agents based on the principle of competitive exclusion or the displacement of diarrhoeagenic bacteria with non-pathogenic strains [15].
Bacteriocins are bacterial proteins which are lethal for other members of the same or closely related species. The bacteriocins produced by and active against E. coli and related bacteria are called colicins. Indigenous bacteria producing colicins are assumed to have a competitive advantage over non-producing strains since the toxin acts directly against susceptible bacteria. Colicinogenic E. coli have been reported to be effective in inhibiting diarrhoeagenic E. coli including serotype O157 in vitro [16–19]. Zhao et al. [20] suggested that competitive exclusion of O157 could occur in cattle by administration of colicinogenic E. coli strains. However, it is not clear whether colicinogenic bacteria are beneficial to humans in vivo in preventing infections due to diarrhoeagenic E. coli.
The purpose of this investigation was to obtain epidemiological clues as to whether intestinal colicinogenic flora explained the resistance of adults to O157 infections. We focused specifically on the detection of enteric colicinogenic bacteria that could suppress propagation of O157. The carriage rate of these bacteria was analysed in an age-stratified manner, and the rate among healthy carriers of O157 was compared to that among persons who did not carry this organism.
MATERIALS AND METHODS
Bacterial strains
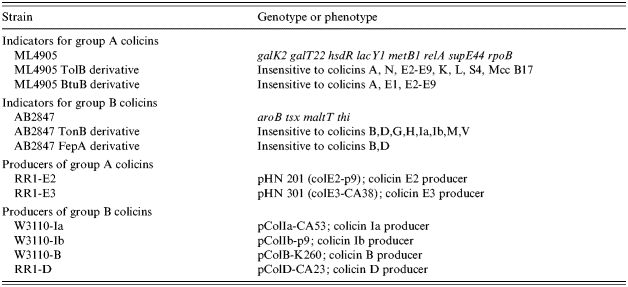
E. coli O157:H7 strain 96-98-83, which was isolated from a patient during a large outbreak of O157 infections that occurred in Sakai City, Japan, 1996 [21, 22], was used as an indicator to detect bacteria that could kill or inhibit growth of O157. Standard strains carrying colicin plasmids were used for identification of the colicins produced by isolated bacteria (Table 1). To investigate whether colicinogenic organisms from healthy carriers of O157 could inhibit the O157 clone isolated from each carrier, the O157 strain of each carrier was used as an additional indicator of bacteriocin activity.
Table 1.
E. coli strains used for colicin typing
Isolation of indigenous bacteria
A total of 303 stool specimens were taken from healthy personnel for routine monitoring of asymptomatic carriers of O157 between October and December 2000. Samples were streaked on sorbitol MacConkey agar plates (Nissui Pharmaceutical, Tokyo, Japan) and incubated overnight at 37°C. Three to five sorbitol non-fermenting colonies were picked as possible O157 isolates, and other morphologically distinguishable colonies were also transferred to triple-sugar-iron (TSI) and lysine-indole-motility (LIM) media (Nissui Pharmaceutical), and identified by standard biochemical tests [23]. Twenty-five carriers and five patients suffering from O157 infection were also examined for organisms inhibitory for O157.
Detection of colicinogenic bacteria active against E. coli O157
Overlay tests were used to detect faecal bacteria inhibitory for O157 [24]. Each isolate from faecal specimens was cultured in tryptone soya broth; TSB (Oxoid, Basingstoke, Hampshire, UK) at 37°C. A 2 μl sample of each overnight culture was spotted onto a tryptone soya agar; TSA (Oxoid) plate (20 isolates/plate) and incubated overnight at 37°C. A second TSA plate containing 0·25 μg/ml of mitomycin C (Sigma, St. Louis, MO, USA) was also inoculated with the panel of isolates. Both plates were then overlaid with 10 ml of TSB containing 1% agar and 105–106 c.f.u./ml of the indicator strain E. coli O157:H7 strain 96-98-83 and incubated overnight. The mitomycin C plate overlay also contained 0·25 μg/ml of mitomycin C. Colonies showing zones of inhibition of the indicator strain were considered to be putative O157 inhibitors.
Determination of the proteinaceous nature of the inhibitor
Putative colicinogenic strains were grown as colonies on agar plates as described above. A 20 μl volume of α-chymotrypsin (Sigma) or protease (Sigma) each at 20 mg/ml was poured into a small hole cut adjacent to the colony; sterile distilled water served as a negative control for protease activity. The plate was incubated at 30°C for 30 min to allow diffusion of the enzyme and then overlaid with agar containing the indicator strain as above. The absence of a zone of inhibition in the region of the protease confirmed the proteinaceous nature of the inhibitory substance produced by a strain.
Typing of colicins
Preliminary identification of the colicins produced by the O157 inhibitors was performed phenotypically according to the method of Pugsley & Oudega [25]. Colicins were classified into group A or group B using strains in which the gene coding the colicin receptor was mutated (Table 1); group A colicins are inactive against TolB or BtuB mutants, and group B are inactive against TonB or FepA mutants. Colicinogenic strains were assigned to four categories (i) group A producer, (ii) group B producer, (iii) simultaneous producer of groups A and B, and (iv) non-responder. E. coli K12 strains harbouring different known colicin plasmids were also used as indicators instead of the reference O157 strain (Table 1). These strains are immune to the colicin that they produce so an O157 inhibitor producing inhibition of all but one of the K12 strains was considered to produce the same colicin as that produced by the unaffected control strain.
PCR
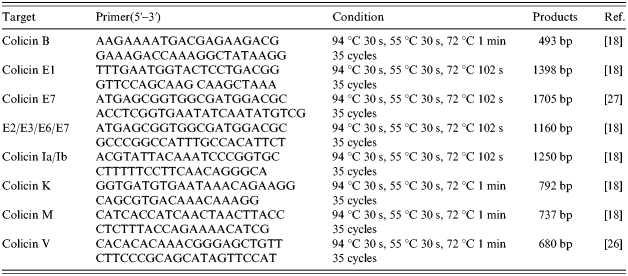
PCR was used as an alternative method to identify the colicin plasmids of O157 inhibitors [18, 26, 27]. The primer sets and amplification conditions used are shown in Table 2. Amplicons were detected and sized against molecular-weight markers by electrophoresis in 2% agarose gel. The gels were stained with ethidium bromide and amplicons visualized under UV light.
Table 2.
Oligonucleotide primers and PCR conditions for amplification of colicin genes
Statistics
χ2 for independence test and Fisher's exact probability test were used for statistical analysis.
RESULTS
Prevalence of O157 inhibitors among humans
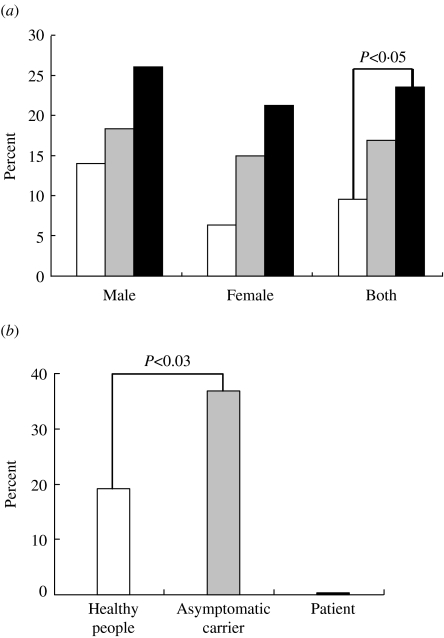
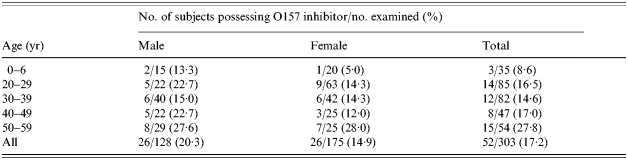
A total of 303 individuals comprising 35 infants (aged ⩽6 years) and 268 adults (aged 20–59 years) were examined. Bacteria showing inhibition of the O157 indicator strain were isolated from 52 individuals (Fig. 1). The prevalence of O157 inhibitors increased with age, and was significantly higher among 40- to 59-year-old adults (23/101, 22·8%; P<0·05) than among children (3/35, 8·6%) (Fig. 2 a, Table 3). Inhibitory bacteria were also significantly more prevalent among O157 healthy carriers (9/25, 36·0%) than among non-carriers (49/268, 18·3%; P<0·03) (Fig. 2 b). Five patients failed to yield O157 inhibitors from their faecal flora.
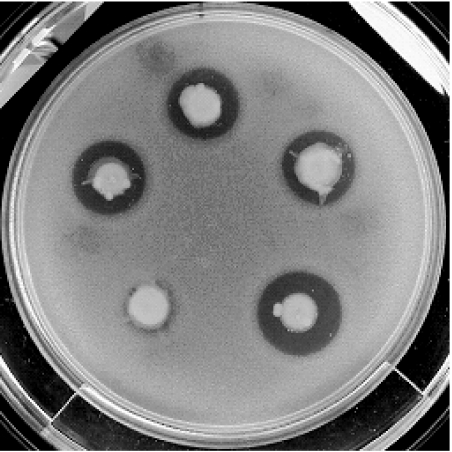
Fig. 1.
Inhibition of E. coli O157 in agar overlay by colicinogenic bacteria.
Fig. 2.
Isolation rates of E. coli O157 inhibitors. (a) Isolation rates in each group classified on the basis of age and sex. □, Age 0–6 years;  , 20–39 years; ■, 40–59 years. (b) Comparison of healthy adults and asymptomatic carriers.
, 20–39 years; ■, 40–59 years. (b) Comparison of healthy adults and asymptomatic carriers.
Table 3.
Distribution of E. coli O157 inhibitors in each age group of healthy persons
Sensitivity of O157 organisms to colicins
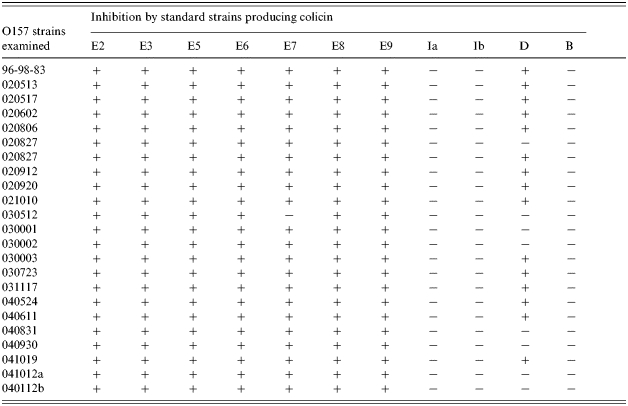
All of 23 E. coli O157 strains examined were sensitive to colicins E2/E3/E5/E6/E8/E9 (Table 4); a strain that showed resistance to colicin E7 proved to be an E7 producer. A further 15 strains were also inhibited by the colicin D-producing strain. Patch tests were performed to examine the sensitivity of O157 to colicins [23] but strains that showed inhibition of O157 indicator in the overlay test failed in the patch tests, although they were able to suppress the growth of colicin-sensitive standard strains (data not shown).
Table 4.
Sensitivity of E. coli O157 strains to colicinogenic standard strains
Identification of O157 inhibitors and their colicin
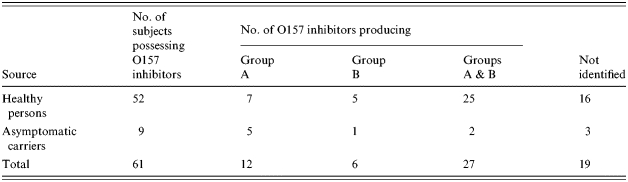
A total of 53 strains of O157 inhibitors were isolated from 52 healthy persons, and 11 strains were from nine asymptomatic carriers of O157. All strains were identified as E. coli except for a strain of Klebsiella oxytoca. Colicins of 64 strains of O157 inhibitors were sensitive to protease and 34 showed resistance to α-chymotrypsin. Addition of mitomycin C to the test medium was effective in inducing colicins in 43 strains, as evidenced by an enlarged zone or initiation of inhibition. The colicins of nine strains were not influenced by mitomycin C but the inhibitory activity of 12 strains was suppressed by this compound. The properties of the colicins were not associated with their source of isolation.
Table 5 shows that bacteria classified as group A producers (12 strains) or groups A and B producers (27 strains) were more prevalent than those producing only group B (six strains). The remainder (19 strains) were untypable. Forty-six of the 64 strains did not yield amplicons with the PCR used. However, primers for genes coding for colicins E (E2, E3, E6, E7), M, V, B, and K generated amplicons in 7, 7, 4, 4, and 4 strains of O157 inhibitors, respectively.
Table 5.
Distribution of colicin types of E. coli O157 inhibitors in healthy persons
DISCUSSION
We have explored the hypothesis that colicinogenic bacteria in the human gut may protect individuals from infectious diseases caused by the highly pathogenic E. coli O157 by suppressing the growth of the latter in vivo. We set out to establish whether normal bowel flora produced bacteriocins active on O157 and, given the observed association of O157 infections in the young and the elderly, whether the prevalence of carriage of colicinogenic bacteria differed across age groups. We were able to demonstrate that O157 inhibitors were more prevalent among the middle-aged than among children (Table 3, Fig. 1) and these bacteria were also more prevalent among asymptomatic O157 carriers than among non-carriers (Fig. 1); five symptomatic patients did not have O157 inhibitors. These results suggest that the presence of colicinogenic bacteria with activity towards O157 strains might explain, at least in part, the resistance of adults to O157 infections. Presumably these bacteria limit the propagation of O157 and/or hinder expression of virulence factors. The reason why possession of colicinogenic bacteria increased with age is not clear. One might speculate that only organisms that have advantageous properties over other bacteria might be able better to remain in the intestine of older persons following repeated conflicts with colicin-susceptible bacteria competing for the same niche in vivo. Indeed, the prevalence of O157 inhibitors among asymptomatic carriers of this E. coli serotype was significantly different (P=0·03) over non-carriers but this result is tempered by the size of the sample because of the difficulties of finding and obtaining specimens from asymptomatic carriers. In order to evaluate our hypothesis further we are currently collecting stool samples from O157 symptomatic patients to establish the prevalence of O157 inhibitory bacteria in their intestines.
Most children and elderly people fall ill due to O157 infection while we have shown asymptomatic carriage of O157 is common among adults. We assumed that if colicinogeny was responsible for suppression of O157 strains then this property would be expressed by bacteria common among the intestinal flora and taxonomically close to E. coli. For these reasons we concentrated on detection of colicinogenic Enterobacteriaceae. Lactic acid bacteria are known to be effective probiotics because they are generally recognized as safe. However, inhibition of Gram-negative bacteria by lactic acid bacteria has not been widely reported although Brashears et al. [28] recently showed that the presence of lactic acid bacteria were inhibitory to O157 in the bovine rumen. Nevertheless, all O157-inhibiting strains proved on subsequent testing to be E. coli with a single exception and were isolated from 18·3% of healthy adults. This suggests that the natural E. coli flora may contribute to the resistance to O157 infections in adults.
Twenty-three strains of O157 were sensitive to E2/E3/E5/E6/E8/E9 but insensitive to B, Ia, and Ib (Table 4), a result similar to that of Bradley et al. [29]. We attempted to screen for O157 inhibitors using the patch test which is an alternative method to detect bacteriocins but were unsuccessful. Compared to the agar overlay method, the amount of colicins released into the medium in the patch tests was insufficient for inhibition of the O157 indicators but the method was adequate for the colicin-sensitive standard strains. Bradley et al. [29] considered that O157 strains were most likely resistant due to masking of the bacteriocin receptor by O-antigen side-chains of LPS. It was noted that O157 strains were sensitive to colicins E, K, and N of group A [16, 17] and to colicins G and H of group B [16, 29]. Although most of our O157 inhibitors produce group A colicins or both groups A and B, further work is necessary to identify the colicins involved fully.
It is not known whether indigenous colicinogenic bacteria are beneficial to prevent clinical manifestation of infection caused by O157. A possibility is that colicins may ameliorate the disease process not only by inhibition of growth but also by suppressing the expression of virulence factors. There is no direct evidence available to support this but there are experimental data which suggest that colicinogenic E. coli could be effective in inhibiting diarrhoeagenic E. coli including serotype O157 in vitro [16–19] and in cattle [20]. However, O157 strains may also have obtained a selective advantage by acquisition of the ability to produce colicins [29]. Scotland et al. [30] indicated that some O157 strains could produce colicin D, and these strains were isolated more readily from bloody diarrhoea rather than ordinary diarrhoea. Bradley et al. [29] speculated that colicin-producing O157 might tend to cause infectious disease because all non-colicinogenic O157 in their study were from asymptomatic carriers; this was not confirmed in the present study as three of 15 O157 strains from carriers produced colicins.
In conclusion, colicinogenic bacteria inhibitory for E. coli O157 are more prevalent among the faecal flora of adult humans than among children. These bacteria may contribute to the resistance to O157 infection in the former group. Further, O157 inhibitors may contribute to amelioration of the effects of infections caused by E. coli O157 as they tended to be more prevalent among asymptomatic carriers than non-carriers. Although current results support an active role of colicinogenic bacteria in the control of enteric infections due to O157 further studies into the aetiological and ecological significance of O157 inhibitors are warranted.
ACKNOWLEDGEMENTS
This research was partly supported by a grant (no. 13680154) from the Japan Society for the Promotion of Science.
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Watanabe H et al. Molecular analysis of enterohemorrhagic Escherichia coli isolates in Japan and its application to epidemiological investigation. Pediatrics International. 1999;41:202–208. doi: 10.1046/j.1442-200x.1999.4121043.x. [DOI] [PubMed] [Google Scholar]
- 2.Ludwig K et al. Shiga toxin-producing Escherichia coli infection and antibodies against Stx2 and Stx1 in household contacts of children with enteropathic hemolytic-uremic syndrome. Journal of Clinical Microbiology. 2002;40:1773–1782. doi: 10.1128/JCM.40.5.1773-1782.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Terajima J et al. Shiga toxin-producing Escherichia coli O157:H7 in Japan. Emerging Infectious Diseases. 1999;5:301–302. doi: 10.3201/eid0502.990222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karmali MA. Infection by verocytotoxin-producing Escherichia coli. Clinical Microbiology Reviews. 1989;2:15–38. doi: 10.1128/cmr.2.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reymond D et al. Neutralizing antibodies to Escherichia coli Vero cytotoxin 1 and antibodies to O157 lipopolysaccharide in healthy farm family members and urban residents. Journal of Clinical Microbiology. 1996;34:2053–2057. doi: 10.1128/jcm.34.9.2053-2057.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evans J et al. Evidence of persisting serum antibodies to Escherichia coli O157 lipopolysaccharide and Verocytotoxin in members of rural communities in England. European Journal of Epidemiology. 2000;16:885–889. doi: 10.1023/a:1011072907877. [DOI] [PubMed] [Google Scholar]
- 7.Ludwig K et al. Escherichia coli O157 fails to induce a long-lasting lipopolysaccharide-specific, measurable humoral immune response in children with hemolytic-uremic syndrome. Journal of Infectious Diseases. 2002;186:566–569. doi: 10.1086/341781. [DOI] [PubMed] [Google Scholar]
- 8.Johnson RP, Cray WC, Jr., Johnson ST. Serum antibody responses of cattle following experimental infection with Escherichia coli O157:H7. Infection and Immunity. 1996;64:1879–1883. doi: 10.1128/iai.64.5.1879-1883.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Donnell JM et al. Outbreak of Vero cytotoxin-producing Escherichia coli O157 in a child day care facility. Communicable Disease and Public Health. 2002;5:54–58. [PubMed] [Google Scholar]
- 10.Bielaszewska M et al. Human Escherichia coli O157:H7 infection associated with the consumption of unpasteurized goat's milk. Epidemiology and Infection. 1997;119:299–305. doi: 10.1017/s0950268897008297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Akashi S et al. A severe outbreak of haemorrhagic colitis and haemolytic uraemic syndrome associated with Escherichia coli O157:H7 in Japan. European Journal of Pediatrics. 1994;153:650–655. doi: 10.1007/BF02190685. [DOI] [PubMed] [Google Scholar]
- 12.Brewster DH et al. An outbreak of Escherichia coli O157 associated with a children's paddling pool. Epidemiology and Infection. 1994;112:441–447. doi: 10.1017/s0950268800051141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lerman Y et al. A cluster of cases of Escherichia coli O157 infection in a day-care center in a communal settlement (Kibbutz) in Israel. Journal of Clinical Microbiology. 1992;30:520–521. doi: 10.1128/jcm.30.2.520-521.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berg RD. The indigenous gastrointestinal microflora. Trends in Microbiology. 1996;4:430–435. doi: 10.1016/0966-842x(96)10057-3. [DOI] [PubMed] [Google Scholar]
- 15.Nurmi E, Nuotio L, Schneitz C. The competitive exclusion concept: development and future. International Journal of Food Microbiology. 1992;15:237–240. doi: 10.1016/0168-1605(92)90054-7. [DOI] [PubMed] [Google Scholar]
- 16.Murinda SE, Roberts RF, Wilson RA. Evaluation of colicins for inhibitory activity against diarrheagenic Escherichia coli strains, including serotype O157:H7. Applied Environmental Microbiology. 1996;62:3196–3202. doi: 10.1128/aem.62.9.3196-3202.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jordi BJ et al. Sensitivity of Shiga toxin-producing Escherichia coli (STEC) strains for colicins under different experimental conditions. FEMS Microbiology Letters. 2001;204:329–334. doi: 10.1111/j.1574-6968.2001.tb10906.x. [DOI] [PubMed] [Google Scholar]
- 18.Schamberger GP, Diez-Gonzalez F. Characterization of colicinogenic Escherichia coli strains inhibitory to enterohemorrhagic Escherichia coli. Journal of Food Protection. 2004;67:486–492. doi: 10.4315/0362-028x-67.3.486. [DOI] [PubMed] [Google Scholar]
- 19.Schamberger GP, Diez-Gonzalez F. Selection of recently isolated colicinogenic Escherichia coli strains inhibitory to Escherichia coli O157:H7. Journal of Food Protection. 2002;65:1381–1387. doi: 10.4315/0362-028x-65.9.1381. [DOI] [PubMed] [Google Scholar]
- 20.Zhao T et al. Reduction of carriage of enterohemorrhagic Escherichia coli O157:H7 in cattle by inoculation with probiotic bacteria. Journal of Clinical Microbiology. 1998;36:641–647. doi: 10.1128/jcm.36.3.641-647.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watanabe H, Wada A, Inagaki Y. Outbreaks of enterohaemorrhagic Escherichia coli O157:H7 infection by two different genotype strains in Japan, 1996. Lancet. 1996;348:831–832. doi: 10.1016/s0140-6736(05)65257-9. [DOI] [PubMed] [Google Scholar]
- 22.Nishikawa Y et al. Phage Typing and DNA-based comparison of strains of enterohemorrhagic Escherichia coli O157 from apparently sporadic infections in Osaka City, Japan 1996. Japan Journal of Infectious Diseases. 2001;54:140–143. [PubMed] [Google Scholar]
- 23.Holt JG. Bergey's Manual of Determinative Bacteriology. 9th ed. Philadelphia: Lippincott Williams & Wilkins; 2000. pp. 175–289. , pp. [Google Scholar]
- 24.De Martinis EC, Franco BD. Inhibition of Listeria monocytogenes in a pork product by a Lactobacillus sake strain. International Journal of Food Microbiology. 1998;42:119–126. doi: 10.1016/s0168-1605(98)00059-2. [DOI] [PubMed] [Google Scholar]
- 25.Pugsley AP, Oudega B, Hardy KG. Plasmids: a practical approach. Oxford: IRL Press; 1987. Methods for studying colicins and their plasmids; pp. 105–161. , pp. [Google Scholar]
- 26.Johnson JR, Stell AL. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. Journal of Infectious Diseases. 2000;181:261–272. doi: 10.1086/315217. [DOI] [PubMed] [Google Scholar]
- 27.Nandiwada LS et al. Characterization of an E2-type colicin and its application to treat alfalfa seeds to reduce Escherichia coli O157:H7. International Journal of Food Microbiology. 2004;93:267–279. doi: 10.1016/j.ijfoodmicro.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 28.Brashears MM, Jaroni D, Trimble J. Isolation, selection, and characterization of lactic acid bacteria for a competitive exclusion product to reduce shedding of Escherichia coli O157:H7 in cattle. Journal of Food Protection. 2003;66:355–363. doi: 10.4315/0362-028x-66.3.355. [DOI] [PubMed] [Google Scholar]
- 29.Bradley DE, Howard SP, Lior H. Colicinogeny of O157:H7 enterohemorrhagic Escherichia coli and the shielding of colicin and phage receptors by their O-antigenic side chains. Canadian Journal of Microbiology. 1991;37:97–104. doi: 10.1139/m91-014. [DOI] [PubMed] [Google Scholar]
- 30.Scotland SM et al. Properties of strains of Escherichia coli belonging to serogroup O157 with special reference to production of Vero cytotoxins VT1 and VT2. Epidemiology and Infection. 1987;99:613–624. doi: 10.1017/s0950268800066462. [DOI] [PMC free article] [PubMed] [Google Scholar]