SUMMARY
The stability of IS6110 restriction fragment length polymorphism (RFLP) pattern was determined in 31 isolates from patients with multidrug-resistant tuberculosis (MDR-TB). These patients were in actual chains of transmission and they referred to the National Institute of Tuberculosis and Lung Diseases, Tehran, Iran. Susceptibility testing against first- and second-line drugs were performed by the proportional method on Lowenstein–Jensen culture media. Thereafter, DNA fingerprinting by IS6110 with direct repeat (DR) region as a probe was performed by standard protocols. The rate of IS6110 changes was 16%, although, no variation was found in the DR region, in a time-span of 1–63 months. The strains with unstable IS6110 patterns were resistant to all drugs tested, and the majority of them (60%) were collected from HIV-positive patients. The results demonstrated that for a reliable interpretation of strain typing, it is better to use an additional marker along with IS6110 RFLP.
INTRODUCTION
The restriction fragment length polymorphism (RFLP) with insertion sequence element (IS6110) as a probe is the standard technique for the comparison of Mycobacterium tuberculosis isolates on the molecular level [1, 2]. IS6110 is a genetic marker and is found in different numbers and locations within the chromosomal region of the M. tuberculosis complex [3]. During the last 10 years, RFLP data have become the primary tool for study of the tuberculosis (TB) epidemic in different settings [4–6]. The analyses is based on the observation that the polymorphism of IS6110 RFLP among unrelated clinical isolates is very high, whereas, epidemiologically related strains show identical or similar fingerprint patterns [5, 6]. This scenario, while convenient, ignores the possibility of bacterial multiplication that may give rise to clonal variants of itself, characterized by minor changes in the IS6110-based RFLP banding patterns. This is supported by studies in which M. tuberculosis isolates were serially passaged in vitro or in vivo for >2–3 years [7, 8]. In a similar context, other studies demonstrated that the degree of IS6110 RFLP patterns might vary for different families of M. tuberculosis strains or for strains that are isolated in different geographical region [9, 10]. Recently, Warren et al. [7] demonstrated that M. tuberculosis populations prior to or after anti-TB treatment (early or late phase of disease) have different rates of IS6110 instability. Similar results were seen in data from The Netherlands [10]. Thereby, for correct interpretation of molecular typing in the epidemiology of TB, it is essential to estimate the rate at which the IS6110 RFLP pattern might change. The information is particularly critical for long-term population-based studies. For this reason, we attempted to evaluate the stability of RFLP patterns among epidemiologically related multidrug-resistant tuberculosis (MDR-TB) patients that were collected from the heavily populated city of Tehran over a 6-year period. The various risk factor(s) that might be associated with the change of IS6110 RFLP patterns were also investigated using classical epidemiology. Furthermore, since composition of the direct repeat (DR) region appears to be more stable than the IS6110-associated RFLP, the polymorphism in this region was determined by spoligotyping. In addition, the possible relation between IS6110 instability and different families of M. tuberculosis was investigated.
MATERIALS AND METHODS
Data collection
The study was conducted from 18 January 1999 to 27 November 2005. Clinical and epidemiological information was collected by trained technicians, using standard questionnaires. For each case information was obtained on gender (female/male), age, contact (family contact/close contact), duration of contact (6 months, <6 months, >6 months), previous TB history (yes, no, not known), present address, associated medical data such as HIV infection (yes, no, not known), tuberculin skin test (+, −, equivocal) and chest radiography finding (bilateral pulmonary involvement with cavity, bilateral pulmonary involvement without cavity, cavitary lesions and non-cavitary non-bilateral pulmonary involvement). The patients with similar or highly similar IS6110 fingerprint patterns were interviewed together. Although, in this study, the majority of patients (64·5%) were family members and we could easily establish a transmission link among them. The study was approved by the Institutional Review Board at the National Research Institute of Tuberculosis and Lung Diseases (NRITLD) in Tehran, Iran.
Bacterial strains
Primary isolation and culture of Mycobacterium isolates were followed in accordance to established procedures [11]. Drug susceptibility testing was performed against isoniazid (INH), rifampicin (RIF), streptomycin (SM), ethambutol (ETB), pyrazinamide (PZA), kanamycin (KM), amikacin (AM), capreomycin (CM), ciprofloxacin (CIP), d-cycloserine (CS), ethionamid (ETH) and P-amino salicylic acid (PAS) [11]. All the drugs were purchased from Sigma Chemical Co. (St. Louis, MO, USA). The MDR-TB patients that were resistant to all second-line drugs tested, were denoted as super MDR-TB cases.
DNA fingerprinting
Extraction of bacterial DNA and DNA fingerprinting with RFLP using IS6110 as a probe, was performed by standard protocols. PvuII enzyme (Boehringer Mannheim, Mannheim, Germany) digested the DNA of the M. tuberculosis 14 323 reference strains, and was also used in each Southern blot experiment, as an external size standard for quality control of IS6110 experiments [5].
Spoligotyping
Spoligotyping was performed as described by Kamerbeek et al. [12] with a commercially available kit (Isogen Bioscience B.V., Maarsen, The Netherlands). In brief, the DR region was amplified by PCR using primers derived from the DR sequence. The amplified DNA was hybridized to a set of 43 immobilized oligonucleotides derived from the spacer sequence of M. tuberculosis H37Rv and M. bovis BCG P3 by reverse line blotting.
Computer-assisted analyses of fingerprints
The autoradiograph of IS6110 RFLP was scanned with a Snap Scan 1236 Scanner. Bionumerics Software version 2.5 (Applied Maths, Kortrijk, Belgium) was used to analyse the molecular patterns generated by IS6110 RFLP. The similarity matrices were constructed using the Jaccard index with a linear error tolerance of 1–3% proportional to the size of the bands. The dendograms were generated by the hierarchic unweighted-pair group method analyses (UPGMA). Strains were classified as a ‘cluster’ if the isolates were different by a single number of the band. However, only patients with confirmed classical epidemiology data were included in this study.
Statistical analyses
The continuous variables were expressed as group means±standard deviation (s.d.). The primary variable was instability of IS6110 in MDR-TB patients. Secondary variables included gender, age, drug abuse, HIV status, family/close contact, pattern of drug resistance, PPD (protein purified derivatives) test, duration of contact and M. tuberculosis superfamilies. The null hypothesis was that IS6110 is stable in the real chain of transmission. Statistical analyses were used as appropriate. All P values were two-tailed. A P value of <0·05 was considered statistically significant. Findings were analysed using SAS software version 9.01 (SAS Institute, Cary, NC, USA).
RESULTS
In total five out of 31 (16%), MDR-TB patients had altered IS6110 patterns. The change in IS6110 banding pattern at the 95% confidence interval (CI) was 0·05–0·34 and with an exact binomial test, the null hypothesis had a P value of 0·000. Thereby, both 95% CI and exact binomial statistical tests rejected the stability of the IS6110 marker. HIV positivity (60%), drug abuse (60%), bilateral pulmonary with cavity (60%), super MDR resistance (100%), and close contact (60%), were more common in patients with unstable IS6110 patterns (see Table). However, only HIV positivity and super MDR resistance (100%) were statistically significant (P<0·05). The number of IS6110 copies per isolate varied from 6 to 14, with a mean age of nine bands.
Table.
Characteristics of each patient with stable or unstable IS6110-banding patterns
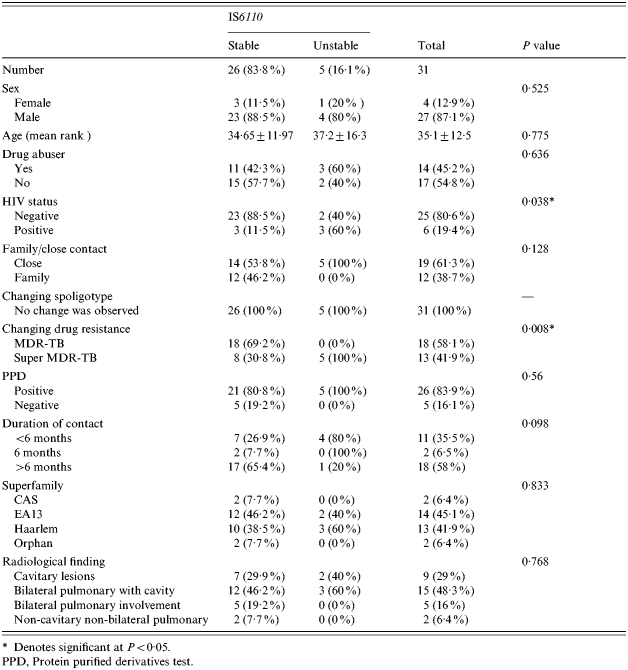
Denotes significant at P<0·05.
PPD, Protein purified derivatives test.
Diversity by RFLP
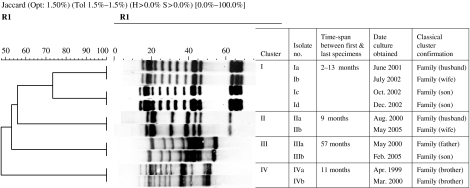
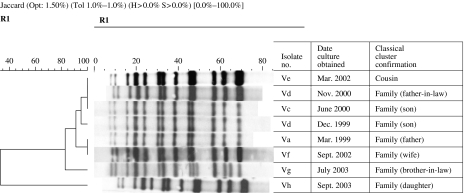
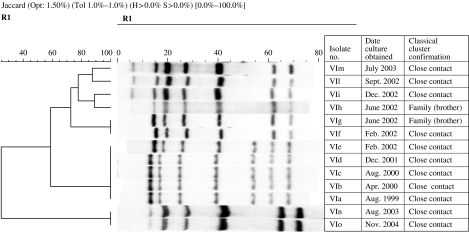
Based on the IS6110 RFLP banding patterns, strains isolated from 31 MDR-TB patients were grouped in six to clusters (I, II, III, IV, V, and VI). Ten patients in four clusters (I, II, III, IV) had stable IS6110 and the time-span between collection of first and last specimen ranged from 2 to 57 months (Fig. 1). The remaining two clusters (V, VI) contained 21 patients, of whom five had altered IS6110 patterns. Cluster V represented family outbreaks (Fig. 2), whereas cluster VI represented family and community outbreaks combined. The first strain in cluster VI, denoted as ‘VIa’ was collected from a HIV-positive patient in late 1999 (Fig. 3). Within 8–30 months, we detected five more patients that were identified as close contacts with the VIa index case. Four of them (VIb, VIc, VId, VIe) had identical RFLP patterns to the VIa patient, and one of them had similar but not identical IS6110 banding patterns (VIf). Later, the patient with the VIf RFLP pattern transmitted the diseases to three more individuals; two were his family members (VIg, VIh) and one was his close contact (VIi). During 2003–2004, another four patients were referred to the NRITLD for MDR treatment. Retrospective analyses showed that these patients (VIl, VIm, VIn, and VIo) were also in close contact with patients with VIi and VIc banding patterns. Overall, in this cluster, four different RFLP-banding patterns were identified (VIa, VIf, VIi, and VIn).
Fig. 1.
Ten patients in four clusters representing the family outbreaks. The IS6110 patterns are stable in the second outbreaks strains. The average time between the first and last specimen collection is 2–57 months.
Fig. 2.
In this cluster, three RFLP-banding patterns were observed; Va (original one), Vd (size differences) and Vg (addition of two bands). The time-span between first and last specimen collection was 6–54 months.
Fig. 3.
The single cluster representing the family and community outbreaks combined. The time-span between the first and last specimens was 1–63 months. In this cluster, four different RFLP-banding patterns were identified (VIa, VIf, VIi, VIn).
M. tuberculosis superfamilies by spoligotyping
When the spoligotyes from the isolated TB strains were compared to earlier published spoligopatterns [13], our isolates could be identified as members of the following superfamilies, East-African Indian (EAI3, 45%), Central Asian family (CAS, 2%), Haarlem (41·9%) and two were Orphan (the pattern which had not been reported before and seen for the first time). All of the strains had stable DR regions throughout the study periods.
DISCUSSION
In a recent study by Alito et al. [9] it was found that the rate of change of IS6110 RFLP patterns, in particular MDR-TB strains, may run too fast for a reliable interpretation of strain typing. These authors suggested that the instability of IS6110 RFLP in the second outbreak strain may be somehow related to the selective pressure of a combination of drugs, as these drug-resistant patients were difficult to treat. Similarly, the IS6110 typing of isolates after a large outbreak of the MDR W strain in the United States over a 3-year period showed a significant divergence of IS6110 RFLP [14], indicating that within a few years a significant fraction of the M. tuberculosis progeny may change. In our study, 16% of epidemiologically related MDR-TB strains had altered DNA genotypes. These isolates were all super MDR-TB cases, and the majority of them (60%) were found to be positive by HIV testing. Therefore, our results showed that the rate of mobility of IS6110 in super MDR-TB cases is faster than MDR-TB patients. However, it is unclear whether these changes were due to bacterial characteristics or of the patients involved. In contrast to our observations, other researchers have not found a relation between instability of IS6110 banding pattern and susceptibility or patients’ HIV serum status [10, 15]. However, in most of previously reported cases, the M. tuberculosis cultures were obtained from serial patient isolates [10, 15, 16] and perhaps the mobility of IS6110 in M. tuberculosis strains remaining in the body of the patient may differ from that in strains that undergo the actual chains of transmission [16]. It is generally accepted that the instability of IS6110 RFLP is associated with duration of infection and size of bacterial population [7, 10]. However, to clarify such matters, well-described human population studies are mandatory [17, 18]. For example we found different results in relation to instability of IS6110 and duration of contact. As shown in Figure 3, two brothers (VIg, VIh) developed diseases from a common source of infection (VIf). Both had the same time of exposure (4 months) and at a similar time their smear and culture became positive. The IS6110 DNA fingerprinting was identical to original strain. In contrast, another two patients (VIe, VIf) with the same source (VIa) and duration of contact (18 months), produced RFLP patterns with a single-band variation. It is possible that the IS6110 elements in the second outbreak strains may be inserted in a particular genomic promoter region that resulted in a higher transposition pressure. Therefore, it is possible to say that the factor(s) influencing the transposition frequency may be strain dependent [10]. Warren et al. [7] demonstrated that IS6110 evaluation is dependent on the number of IS6110 elements. They showed that strains with fewer than five IS6110 elements were genotypically invariant. In this study, all the collected strains had more than five IS6110 hybridizing bands and we could not compare the instability of IS6110 in the strain with low and high copy numbers. Overall, our results justify the use of the second typing method for detecting recent transmission. Today, spoligotyping has been useful in determining more distant relationships among M. tuberculosis isolates [13, 19]. The method is based on the detection of various non-repetitive spacer sequences located between small DR units in the DR locus of M. tuberculosis complex strains. In our study, the strains with altered IS6110 patterns had identical spoligopatterns; therefore, the DR is a more stable marker than IS6110 RFLP. Recently, based on spoligopatterns, nine potential superfamilies or clades of Mycobacterium–TB complex have been identified worldwide (M. African, Beijing, M. bovis, EAI, CAS, T, Haarlem, X and LAM families) [13]. In the present study, from five patients with unstable IS6110, three (60%) belonged to the Haarlem family. Recently, Moss et al. [20] demonstrated that, eight of 129 W-type MDR-TB strains exhibited IS6110 RFLP patterns slightly different from those of the predominant type. Hence, these authors suggested that the transposition rate may be genotype-dependent. However, in our study since the number of isolates was limited to 31, no possible relation between different superfamilies of M. tuberculosis and instability of IS6110 could be suggested. Therefore, the study needs further investigation in serial patients or in patients with real chains of transmission. In conclusion, the rate of changes of any molecular marker(s) that are used for epidemiological studies of TB must be low enough to ensure that most cases linked through recent transmission have identical fingerprints. The data presented in this study, demonstrated that the super MDR-TB strains have higher mutational frequency. Therefore, we have to consider this observation while we investigate the recent transmission within the country.
ACKNOWLEDGEMENTS
This work was sponsored by a grant from NRITLD-WHO/11/005. We gratefully thank Centre National de Reference des Mycobacteries, Institute Pasteur, Paris (France) for technical instruction and training. Finally, we thank all the TB patients and their families who patiently helped us to obtain the required information.
DECLARATION OF INTEREST
None.
REFERENCES
- Van Embden JDA et al. Strain identification of M. tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. Journal of Clinical Microbiology. 1993;31:406–409. doi: 10.1128/jcm.31.2.406-409.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes PF et al. Patterns of tuberculosis transmission in central Los Angeles. Journal of the American Medical Association. 1997;278:1159–1163. [PubMed] [Google Scholar]
- Kremer K et al. Use of IS6110 DNA fingerprinting in tracing man to man transmission of M. tuberculosis in the Czech Republic. Central European Journal of Public Health. 1996;4:3–6. [PubMed] [Google Scholar]
- Small PM et al. The epidemiology of tuberculosis in San Francisco. A population-based study using conventional and molecular methods. New England Journal of Medicine. 1994;330:1703–1709. doi: 10.1056/NEJM199406163302402. [DOI] [PubMed] [Google Scholar]
- van Soolingen D et al. DNA fingerprinting of M. tuberculosis. Methods in Enzymology. 1994;253:196–205. doi: 10.1016/0076-6879(94)35141-4. [DOI] [PubMed] [Google Scholar]
- Alland D et al. Transmission of tuberculosis in New York City. An analysis by DNA fingerprinting and conventional epidemiologic methods. New England Journal of Medicine. 1994;330:1710–1716. doi: 10.1056/NEJM199406163302403. [DOI] [PubMed] [Google Scholar]
- Warren RM et al. Calculation of the stability of the IS6110 banding pattern in patients with persistent M. tuberculosis disease. Journal of Clinical Microbiology. 2002;40:1705–1708. doi: 10.1128/JCM.40.5.1705-1708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh RW et al. Stability of M. tuberculosis DNA genotypes. Journal of Infectious Diseases. 1998;177:1107–1111. doi: 10.1086/517406. [DOI] [PubMed] [Google Scholar]
- Alito A et al. The IS6110 restriction fragment length polymorphism in particular multidrug-resistant M. tuberculosis strains may evolve too fast for reliable use in outbreaks investigation. Journal of Clinical Microbiology. 1999;37:788–791. doi: 10.1128/jcm.37.3.788-791.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boer AS et al. Analysis of rate of change of IS6110 RFLP patterns of M. tuberculosis based on serial patient isolates. Journal of Infectious Diseases. 1999;180:1238–1244. doi: 10.1086/314979. [DOI] [PubMed] [Google Scholar]
- Kent PT, Kubica GP. Public health mycobacteriology: a guide for level III laboratory. Atlanta: Centers for Diseases Control; 1985. pp. 12–39. , pp. [Google Scholar]
- Kamerbeek J et al. Simultaneous detection and strain differentiation of M. tuberculosis for diagnosis and epidemiology. Journal of Clinical Microbiology. 1997;35:907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filliol I et al. Global distribution of M. tuberculosis spoligotypes. Emerging Infectious Diseases. 2002;8:1347–1349. doi: 10.3201/eid0811.020125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bifani PJ et al. Origin and interstate spread of a New York City multidrug-resistant M. tuberculosis clone family. Journal of the American Medical Association. 1996;275:452–457. [PubMed] [Google Scholar]
- Ritacco VM et al. Nosocomial spread of human immunodeficiency virus-related multidrug-resistant tuberculosis in Buenos Aires. Journal of Infectious Diseases. 1997;176:637–642. doi: 10.1086/514084. [DOI] [PubMed] [Google Scholar]
- Niemann S, Richter E, Rusch Gerdes S. Stability of M. tuberculosis IS6110 restriction fragment length polymorphism patterns and spoligotypes determined by analyzing serial isolates from patients with drug resistant tuberculosis. Journal of Clinical Microbiology. 1999;37:409–412. doi: 10.1128/jcm.37.2.409-412.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cave MD et al. Stability of DNA fingerprint pattern produced with IS6110 in strains of M. tuberculosis. Journal of Clinical Microbiology. 1994;32:262–266. doi: 10.1128/jcm.32.1.262-266.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niemann S et al. Stability of IS6110 restriction fragment length polymorphism patterns of M. tuberculosis strains in actual chains of transmission. Journal of Clinical Microbiology. 2000;38:2563–2567. doi: 10.1128/jcm.38.7.2563-2567.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filliol I et al. Snapshot of moving and expanding clones of M. tuberculosis and their global distribution assessed by spoligotyping in an international study. Journal of Clinical Microbiology. 2003;41:1963–1970. doi: 10.1128/JCM.41.5.1963-1970.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss AR et al. A city-wide outbreak of a multiple-drug-resistant strain of M. tuberculosis in New York city. International Journal of Tuberculosis and Lung Diseases. 1997;1:115–121. [PubMed] [Google Scholar]