SUMMARY
We characterized eleven H9N2 influenza A viruses isolated from chicken products imported from China. Genetically they were classified into six distinct genotypes, including five already known genotypes and one novel genotype. This suggested that such multiple genotypes of the H9N2 virus have possibly already become widespread and endemic in China. Two isolates have amino-acid substitutions that confer resistance to amantadine in the M2 region, and this supported the evidence that this mutation might be a result of the wide application of amantadine for avian influenza treatment in China. These findings emphasize the importance of surveillance for avian influenza virus in this region, and of quarantining imported chicken products as potential sources for the introduction of influenza virus.
INTRODUCTION
H9N2 influenza A viruses have been distributing widely from East Asia to points in the Middle East, Europe, and Southern Africa [1–3]. H9N2 influenza A viruses were also isolated from humans in 1999 and 2003 [4, 5]. The H9N2 viruses isolated in 1999 shared common internal gene components with the H5N1 influenza A viruses from 1997 that were lethal to humans in Hong Kong [6]. These H5N1 viruses were considered to be genetic reassortants generated from two or three viruses, including the H9N2 virus [7]. Of importance is the fact that recognition of human-like receptor specificity by recent H9N2 viruses in Asia could have facilitated direct transmission to humans [8]. Thus, H9N2 viruses along with H5N1 viruses are high on the list of candidates that could potentially cause a future human influenza pandemic.
However, since its first detection in southern China (i.e. A/goose/Guandong/1/96), the H5N1 influenza A virus has spread in Asian countries, where it is now enzootic and has caused multiple outbreaks in poultry [9, 10]. Among them, the isolation of an H5N1 virus from duck meat imported from China to Korea in 2001 raises the possibility of the introduction of viruses and transmission to humans from novel sources [11]. Given this possibility, the Japanese government decided in 2001 to halt the import of duck meat and to survey for the presence of the viruses in imported chicken products from China at the Animal Quarantine Service (AQS). During this process, H9N2 viruses and Newcastle disease viruses were isolated from chicken products such as meat and bone marrow.
In this study, we performed a genetic, antigenic, and pathogenic analysis of H9N2 viruses isolated from chicken products at the AQS in Japan in order to compare them with other influenza A viruses.
METHODS
Sampling and virus isolation
At the AQS in Japan, a total of 473 samples of frozen chicken meat and bone marrow collected between June 2001 and July 2002 were used for virus isolation. Briefly, applications were submitted to the AQS to import lots of chicken products (500–3000 cartons/lot under a single import application). One carton from each imported lot was randomly selected and tested for the presence of the influenza virus by sampling 1 g meat from each of 10 packages per carton, which was pooled and made into a 10% homogenate in phosphate buffered saline (PBS). The homogenates were then centrifuged at low speed (3000 g for 10 min) and the supernatants filtered through a sterile 0·45 μm membrane filter before inoculation into the allantoic cavity of embryonated specific pathogen-free (SPF) eggs. All isolates were identified as influenza A viruses of the H9N2 subtype by conventional haemagglutination inhibition (HI) and neuraminidase activity inhibition (NI) tests. Additionally, the H9N2 isolate, which was isolated in surveillance for imported chicken products at the AQS in 1997 (A/chicken/Osaka/aq48/97), was also used in this study (Table 1). The H9 prototype A/turkey/Wisconcin/66 strain (Ty/WI/66) and our previous isolates from imported parakeets, A/parakeet/Chiba/1/97 (Pa/Chiba/1/97) and A/parakeet/Narita/92A/98 (Pa/Narita/92A/98) [12] were also used in this study.
Table 1.
H9N2 influenza A viruses isolated from chicken products imported from China at AQS
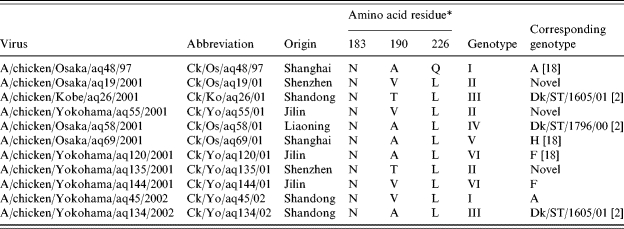
H3 numbering.
Antigenic characterization
All chicken antisera against Ty/WI/66, Pa/Chiba/1/97, A/chicken/Osaka/aq48/97 (Ck/Os/aq48/97) and A/chicken/Yokohama/aq55/2001 (Ck/Yo/aq55/01) were prepared in our laboratory according to a previously described method [7]. Briefly, 3-week-old SPF chickens were inoculated intranasally and orally with 0·1 ml allantoic fluid [∼106 50% egg infectious dose (EID50)]. The chickens were bled 3 weeks post-infection and boosted intravenously with 0·1 ml of infectious virus. HI and NI tests were performed according to standard procedures [13].
Genetic and phylogenetic analysis
RNA extraction, RT–PCR, and sequencing of the PCR products were carried out as described previously [12]. PCR amplification of the coding regions of the viral gene segments was performed with gene-specific primer sets (sequences of the primers available upon request). The nucleotide sequences were analysed using version 12.0 of the sequence analysis software package GENETYX-MAC (Software Development, Tokyo, Japan). The nucleotide sequences obtained from this study are available from GenBank under accession numbers AB256663–AB256750. Multiple alignments were conducted by the Cluster X program [14], and phylogenetic trees were constructed by the neighbour-joining (NJ) method as previously described [15]. Based on each phylogenetic analysis of each gene segment, the genotypes of the H9N2 viruses isolated from chicken products were compared with previously reported genotypes [2, 7, 16–18].
Pathogenicity to chickens
Three isolates, Ck/Os/aq19/01, Ck/Ko/aq26/01, and Ck/Yo/aq55/01, were chosen for the pathogenicity test. For pathogenicity testing according to the guidelines established by the World Organization for Animal Health (Office International des Epizooties; OIE) [19], 6-week-old SPF chickens were used in this study. Eight chickens were inoculated intravenously with 0·2 ml of a 1:10 dilution of infected allantoic fluid, and clinical signs were observed daily for 10 days. In addition, we performed an intracerebral inoculation test according to that for the Newcastle disease virus based on the guidelines established by the OIE [19]. Ten chickens were inoculated intracerebrally with 0·05 ml of a 1:10 dilution of infected allantoic fluid and clinical signs were observed daily for 8 days.
RESULTS
Genetic and phylogenetic analysis
The HA genes of the H9N2 viruses isolated from chicken products belonged to the A/chicken/Beijing/1/94 (Ck/BJ/1/94) sub-lineage [Fig. (a); phylogenetic trees of genes of influenza A viruses can be seen in the Figure on the Journal’s website] as described by Li et al. [18]. The amino-acid sequence at the HA cleavage site of H9N2 isolates from chicken products possessed a PARSSR-GLF motif, which corresponded to low-pathogenicity viruses [1, 16, 20]. The amino acids at the receptor binding site of HA proteins are associated with differences in the receptor binding specificity [21]. Table 1 shows the amino-acid positions 183, 190, and 226 (numbering according to H3 HA). All H9N2 viruses isolated from chicken products possessed N at amino-acid position 183. Except for one strain (Ck/Os/aq48/97), all H9N2 viruses isolated from chicken products possessed L at amino-acid position 226. Five potential glycosylation sites in HA1 (11, 123, 200, 280, and 287) were conserved in all H9N2 viruses isolated from chicken products (data not shown).
The M genes of the H9N2 viruses isolated from chicken products also belonged to the Ck/BJ/1/94 sub-lineage [Fig. (b)] [18]. Resistance to the two types of influenza antiviral compounds (M2 ion channel blockers, e.g. amantadine and rimantadine, and NA inhibitors, e.g. oseltamivir and zanamivir [22]) is associated with particular mutations. Viruses become resistant to amantadine through a single amino-acid substitution at positions 26, 27, 30, 31, or 34 in the transmembrane region of the M2 protein [23]. Two isolates, Ck/Os/19/01 and Ck/Yo/45/02, have the amino-acid substitution at position 30 (A to T) and 31 (S to N), and these substitutions confer resistance to amantadine in the M2 protein, respectively.
The NA genes of the H9N2 viruses isolated from the chicken products also divided into two sub-lineages (Ck/BJ/1/94-like and Ck/HK/G9/97-like) [18] [Fig. (c)]. Six isolates clustered with Ck/BJ/1/94-like viruses, which contain a three (at amino-acid positions 63–65) amino-acid deletion in the stalk region. The other five isolates clustered with Ck/HK/G9/97-like viruses, which do not contain a stalk deletion. Through a single amino-acid substitution at positions 119, 152, 274, 292 or 294 in the NA active centre [24, 25], viruses can become resistant to oseltamivir. None of the amino-acid substitutions that were resistant to oseltamivir in the NA protein were found in the H9N2 viruses isolated from chicken products.
Phylogenetic analysis of the remaining gene segments of the H9N2 isolates from chicken products suggested that they were divided into six genotypes (temporarily designated I–VI) [Fig. (d–h), Table 1]. These sequences were compared with representative H9N2 viruses, including Ck/BJ/1/94, A/quail/Hong Kong/G1/97 (Qa/HK/G1/97), A/chicken/Hong Kong/G9/97 (Ck/HK/G9/97), A/duck/Hong Kong/Y280/97 (Dk/HK/Y280/97), A/chicken/Shanghai/F/98 (Ck/SH/F/98), and some duck viruses isolated in Shantou, Guangdong Province in mainland China, reported previously by others [2, 7, 16–18]. Compared with the H9N2 genotypes that have been described (Table 1), genotype I seemed to correspond to genotype A defined by Li et al. [18]. Genotypes III and VI seemed to correspond to genotypes Dk/ST/1605/01 and Dk/ST/1796/00 defined by Li et al. [2] respectively. Genotype V seemed to correspond to genotype H defined by Li et al. [18]. Genotype VI seemed to correspond to genotype F defined by Li et al. [18]. The remaining genotype II seemed to be a novel genotype in which the PB1 belonged to the Ck/BJ/1/94 lineage, with the other seven segments corresponding to genotype I as defined by Li et al. [18].
Antigenic analysis
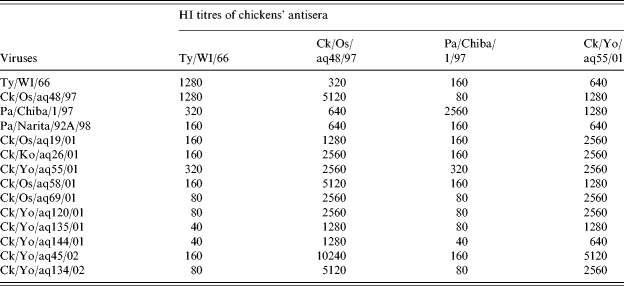
Antisera against the H9N2 isolates from chicken products imported from China reacted with all tested strains within the threefold dilution of the homologous HI titres (Table 2). The reactivity of the H9N2 isolates from chicken products to antisera against the H9 prototype Ty/WI/66 was diverse. However, antisera against Pa/Chiba/1/97, which belongs to the Qa/HK/G1/97 lineage [12], reacted with most tested strains except Ck/Yo/aq55/01, with the fourfold dilution lower than the homologous HI titres.
Table 2.
Cross haemaggulutination inhibition test among H9N2 viruses
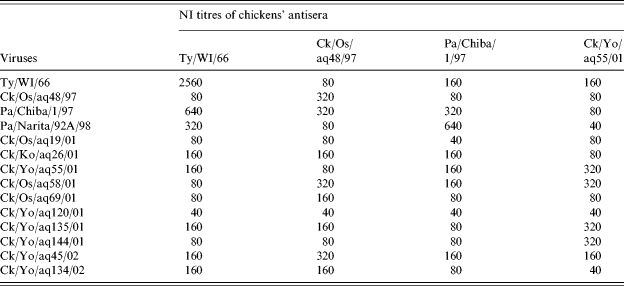
NI tests revealed that the antisera against the H9N2 isolates from the chicken products imported from China also reacted with most tested strains within the threefold dilution of the homologous NI titres (Table 3). Differing from the results of the HI tests, the antisera against Pa/Chiba/1/97 reacted with all tested strains within the threefold dilution of the homologous NI titres. The antisera against the H9 prototype Ty/WI/66 reacted with most H9N2 isolates from chicken products, with the fourfold dilution being lower than the homologous NI titres.
Table 3.
Cross neuraminidase activity inhibition test among H9N2 viruses
Pathogenicity tests
The three H9N2 isolates from chicken products were not judged as highly pathogenic for chickens by the OIE guidelines, since none of the chickens died and no clinical signs were observed in the inoculated chickens. In the intracerebral inoculation tests, only one chicken inoculated with Ck/Ko/aq26/01 died during the observation period. The range of values of the intracerebral pathogenicity index of the tested viruses was 0·00–0·2, which showed low pathogenicity to chickens.
DISCUSSION
We characterized the H9N2 isolates from chicken products imported from China. The HA of these H9N2 viruses seemed to be antigenically diverse, but the NA seemed to have a close relationship with one another. The amino acids of the receptor binding sites in HA are similar to those affecting humans. Almost all viruses possess L at position 226, which is identical to the human H2 and H3 strains [8]. L at position 226 binds to the Neu Acα2,6 Gal linkage, whereas that possessing Q 226 binds to the Neu Acα2,3 Gal linkage [26]. These results suggested that the recent H9N2 viruses might be candidates for a pandemic influenza outbreak in humans.
This surveillance of chicken products imported from China was conducted from 2001 to 2002, and the H9N2 viruses were isolated from 17 (2·7%) of 621 samples at a comparatively high frequency. This suggests that such contaminated chicken products could be a potential source for the introduction of avian influenza viruses into other countries. The pathogenicity index indicates that the H9N2 viruses isolated from chicken products were of low pathogenicity to chickens, and the reason for the frequent isolation of low-pathogenic H9N2 viruses from chicken products in this study was not completely understood. In outbreaks of the H9N2 virus in Iran, a mixed infection caused severe clinical features with bacteria or mycoplasma [27]. In addition, Kishida et al. revealed that co-infection with Streptococcus or Haemophilus increased the virulence of H9N2 viruses in chickens [28]. Hence, the same secondary pathogens may be related to severe pathogenicity and isolation of the virus from novel sources such as chicken products.
So far, the prevalence of multiple H9N2 genotypes in mainland China has been reported [2, 16–18]. Previously, Liu et al. showed that all eight segments of H9N2 viruses isolated in China between 1995 and 1999 genetically belonged to Ck/HK/G9/97 lineages [20]. Thereafter, multiple genotypes of the H9N2 viruses have been generated from complicated reassortment with various viruses [18]. In this study, the multiple genotypes of H9N2 viruses, which include the genotype isolated from only duck [2], were detected in chicken products imported from China. Interestingly, most genotypes of the poultry products originated from only one province; genotype III only in Shandong, IV from Liaoning, V only in Shanghai, and VI only in Jilin. In contrast, genotype I originated from Shanghai and Shandong and genotype II from Shenzhen or Jilin. This may suggest that there is a prevalent genotype in each province, and that it may be related to the emergence of novel genotype H5N1 viruses [2, 6].
The amino-acid substitutions, which confer resistance to amantadine in the M2 region [23], were detected in two isolates (Ck/Os/aq19/01 and Ck/Yo/aq45/02 isolated in Shenzhen and Shandong respectively). These isolates were obtained from chicken products with an unknown history of amantadine antiviral therapy. As previously suggested [18], this mutation may be the result of the application of amantadine for avian influenza treatment, and there may be a wide distribution of this resistant strain to these drugs in mainland China. Continuing surveillance of avian influenza viruses in this region is needed.
ACKNOWLEDGEMENTS
This work was partially supported by a Grant-in-Aid from the Zoonoses Control Project of the Ministry of Agriculture, Forestry and Fisheries, Japan.
DECLARATION OF INTEREST
None.
NOTE
Supplementary information accompanies this paper on the Journal’s website (http://journals.cambridge.org).
REFERENCES
- 1.Banks J et al. Phylogenetic analysis of influenza A viruses of H9 haemagglutinin subtype. Avian Pathology. 2000;29:353–360. doi: 10.1080/03079450050118485. [DOI] [PubMed] [Google Scholar]
- 2.Li KS et al. Characterization of H9 subtype influenza viruses from the ducks of southern China: a candidate for the next influenza pandemic in humans. Journal of Virology. 2003;77:6988–6994. doi: 10.1128/JVI.77.12.6988-6994.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Naeem K et al. Avian influenza A subtype H9N2 in poultry in Pakistan. Veterinary Record. 1999;145:560. doi: 10.1136/vr.145.19.560. [DOI] [PubMed] [Google Scholar]
- 4.Peiris M et al. Human infection with influenza H9N2. Lancet. 1999;354:916–917. doi: 10.1016/s0140-6736(99)03311-5. [DOI] [PubMed] [Google Scholar]
- 5.Butt KM et al. Human infection with an avian H9N2 influenza A virus in Hong Kong in 2003. Journal of Clinical Microbiology. 2005;43:5760–5767. doi: 10.1128/JCM.43.11.5760-5767.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin YP et al. Avian-to-human transmission of H9N2 subtype influenza A viruses: relationship between H9N2 and H5N1 human isolates. Proceedings of the National Academy of Sciences USA. 2000;97:9654–9658. doi: 10.1073/pnas.160270697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guan Y et al. Molecular characterization of H9N2 influenza viruses: were they donors of the ‘internal’ genes of H5N1 viruses in Hongkong. Proceedings of the National Academy of Sciences USA. 1999;96:9363–9367. doi: 10.1073/pnas.96.16.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matrosovich MN, Krauss S, Webster RG. H9N2 influenza A viruses from poultry in Asia have human virus-like receptor specificity. Virology. 2001;281:156–162. doi: 10.1006/viro.2000.0799. [DOI] [PubMed] [Google Scholar]
- 9.Li K et al. Genesis of a highly pathogenic and potentially pandemic H5N1 influenza virus in eastern Asia. Nature. 2004;430:209–213. doi: 10.1038/nature02746. [DOI] [PubMed] [Google Scholar]
- 10.World Health Organization Global Influenza Program Surveillance Network. Evolution of H5N1 avian influenza viruses in Asia. Emerging Infectious Diseases. 2005;11:1515–1521. doi: 10.3201/eid1110.050644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tumpey TM et al. Characterization of a highly pathogenic H5N1 avian influenza A virus isolated from duck meat. Journal of Virology. 2002;76:6344–6355. doi: 10.1128/JVI.76.12.6344-6355.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mase M et al. Imported parakeets harbor H9N2 influenza A viruses that are genetically closely related to those transmitted to humans in Hong Kong. Journal of Virology. 2001;75:3490–3494. doi: 10.1128/JVI.75.7.3490-3494.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Madeley CR, Allan WH, Kendal AP. Studies with avian influenza A viruses: serological relations of the haemagglutinin and neuraminidase antigens of ten virus isolates. Journal of General Virology. 1971;12:69–78. doi: 10.1099/0022-1317-12-2-69. [DOI] [PubMed] [Google Scholar]
- 14.Thompson JD et al. The CLUSTAL-X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Research. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 16.Guo YJ et al. Characterization of the pathogenicity of members of the newly established H9N2 influenza virus lineages in Asia. Virology. 2000;267:279–288. doi: 10.1006/viro.1999.0115. [DOI] [PubMed] [Google Scholar]
- 17.Choi YK et al. Continuing evolution of H9N2 influenza viruses in Southeastern China. Journal of Virology. 2004;78:8609–8614. doi: 10.1128/JVI.78.16.8609-8614.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li C et al. Evolution of H9N2 influenza viruses from domestic poultry in Mainland China. Virology. 2005;340:70–83. doi: 10.1016/j.virol.2005.06.025. [DOI] [PubMed] [Google Scholar]
- 19.Office International des EpizootiesNewcastle Disease, chapter 2.1.15Manual of Diagnostic Tests and Vaccines for Terrestrial Animals 5th ednParis, France: Office International des Epizooties; 2004http://www.oie.int [Google Scholar]
- 20.Liu J et al. H9N2 influenza viruses prevalent in poultry in China are phylogenetically distinct from A/quail/Hong Kong/G1/97 presumed to be the donor of the internal protein genes of the H5N1 Hong Kong/97 virus. Avian Pathology. 2003;32:551–560. doi: 10.1080/0307-9450310001596728. [DOI] [PubMed] [Google Scholar]
- 21.Rogers GN et al. Single amino acid substitutions in influenza haemagglutinin change receptor binding specificity. Nature. 1983;304:76–78. doi: 10.1038/304076a0. [DOI] [PubMed] [Google Scholar]
- 22.Monto AS. The role of antivirals in the control of influenza. Vaccine. 2003;21:1796–1800. doi: 10.1016/s0264-410x(03)00075-6. [DOI] [PubMed] [Google Scholar]
- 23.Crumpacker C, Knipe DM, Howley PM.Antiviral therapyFields Virology 4th ednPhiladelphia: Lippincott Williams & Wilkins; 2001393–433. [Google Scholar]
- 24.Gubareva LV, Webster RG, Hayden FG. Detection of influenza virus resistance to neuraminidase inhibitors by an enzyme inhibition assay. Antiviral Research. 2002;53:47–61. doi: 10.1016/s0166-3542(01)00192-9. [DOI] [PubMed] [Google Scholar]
- 25.Kiso M et al. Resistant influenza A viruses in children treated with oseltamivir: descriptive study. Lancet. 2004;364:759–765. doi: 10.1016/S0140-6736(04)16934-1. [DOI] [PubMed] [Google Scholar]
- 26.Weis W et al. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature. 1988;333:426–431. doi: 10.1038/333426a0. [DOI] [PubMed] [Google Scholar]
- 27.Nill H, Asasi K. Natural cases and an experimental study of H9N2 avian influenza in commercial broiler chickens of Iran. Avian Pathology. 2002;31:247–252. doi: 10.1080/03079450220136567. [DOI] [PubMed] [Google Scholar]
- 28.Kishida N et al. Co-infection of Staphylococcus aureus or Haemophilus paragallinarum exacerbates H9N2 influenza A virus infection in chickens. Archives of Virology. 2004;149:2095–2104. doi: 10.1007/s00705-004-0372-1. [DOI] [PubMed] [Google Scholar]


