Abstract
We have developed a quantitative method for examining the removal of N-methylpurines from specific genes to investigate their possible differential repair throughout the genome. Chinese hamster ovary cells were exposed to dimethyl sulfate, and the isolated DNA was treated with an appropriate restriction endonuclease. The DNA was heated to convert remaining N-methylpurines to apurinic sites to render them alkaline-labile. Duplicate samples heated in the presence of methoxyamine to protect the apurinic sites from alkaline hydrolysis provided controls to assess total DNA. After alkaline hydrolysis, agarose gel electrophoresis, Southern transfer, and probing for the fragment of interest, the ratios of band intensities of the test DNA sample to its methoxyamine-treated control counterpart were calculated to yield the percentage of fragments containing no alkaline-labile sites. The frequency of N-methylpurines was measured at different times after dimethyl sulfate treatment to study repair. We found no differences between the rates of repair of N-methylpurines in the active dihydrofolate reductase gene and a nontranscribed region located downstream from it in treated cells. Also, similar rates of repair were observed in the transcribed and nontranscribed strands of the gene, in contrast to previous results for the removal of cyclobutane pyrimidine dimers. Thus, there does not appear to be a coupling of N-methylpurine repair to transcription in Chinese hamster ovary cells. However, the repair in the dihydrofolate reductase domain appears to be somewhat more efficient than that in the genome overall. Our method permits the quantifying at the defined gene level of abasic sites or of any DNA adduct that can be converted to them.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abanobi S. E., Columbano A., Mulivor R. A., Rajalakshmi S., Sarma D. S. In vivo replication of hepatic deoxyribonucleic acid of rats treated with dimethylnitrosamine: presence of dimethylnitrosamine-induced O6-methylguanine, N7-methylguanine, and N3-methyladenine in the replicated hybrid deoxyribonucleic acid. Biochemistry. 1980 Apr 1;19(7):1382–1387. doi: 10.1021/bi00548a018. [DOI] [PubMed] [Google Scholar]
- Bohr V. A., Hanawalt P. C. Enhanced repair of pyrimidine dimers in coding and non-coding genomic sequences in CHO cells expressing a prokaryotic DNA repair gene. Carcinogenesis. 1987 Sep;8(9):1333–1336. doi: 10.1093/carcin/8.9.1333. [DOI] [PubMed] [Google Scholar]
- Bohr V. A., Okumoto D. S., Ho L., Hanawalt P. C. Characterization of a DNA repair domain containing the dihydrofolate reductase gene in Chinese hamster ovary cells. J Biol Chem. 1986 Dec 15;261(35):16666–16672. [PubMed] [Google Scholar]
- Bohr V. A., Phillips D. H., Hanawalt P. C. Heterogeneous DNA damage and repair in the mammalian genome. Cancer Res. 1987 Dec 15;47(24 Pt 1):6426–6436. [PubMed] [Google Scholar]
- Brent T. P. 3-Methyladenine-DNA glycosylase: a probe for determining alkylation damage and repair in human fibroblasts. Biochem Biophys Res Commun. 1979 Dec 14;91(3):795–802. doi: 10.1016/0006-291x(79)91950-8. [DOI] [PubMed] [Google Scholar]
- Cathcart R., Goldthwait D. A. Enzymatic excision of 3-methyladenine and 7-methylguanine by a rat liver nuclear fraction. Biochemistry. 1981 Jan 20;20(2):273–280. doi: 10.1021/bi00505a007. [DOI] [PubMed] [Google Scholar]
- Galbraith A. I., Itzhaki R. F., Craig A. W., Margison G. P. Distribution and repair of O6-methylguanine in different fractions of rat liver DNA after dimethylnitrosamine administration. Chem Biol Interact. 1980 Mar;29(3):347–355. doi: 10.1016/0009-2797(80)90153-2. [DOI] [PubMed] [Google Scholar]
- Gallagher P. E., Brent T. P. Partial purification and characterization of 3-methyladenine-DNA glycosylase from human placenta. Biochemistry. 1982 Dec 7;21(25):6404–6409. doi: 10.1021/bi00268a013. [DOI] [PubMed] [Google Scholar]
- Hanawalt P. C., Cooper P. K., Ganesan A. K., Smith C. A. DNA repair in bacteria and mammalian cells. Annu Rev Biochem. 1979;48:783–836. doi: 10.1146/annurev.bi.48.070179.004031. [DOI] [PubMed] [Google Scholar]
- Hoffmann G. R. Genetic effects of dimethyl sulfate, diethyl sulfate, and related compounds. Mutat Res. 1980 Jan;75(1):63–129. doi: 10.1016/0165-1110(80)90028-7. [DOI] [PubMed] [Google Scholar]
- Leadon S. A. Differential repair of DNA damage in specific nucleotide sequences in monkey cells. Nucleic Acids Res. 1986 Nov 25;14(22):8979–8995. doi: 10.1093/nar/14.22.8979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leadon S. A., Hanawalt P. C. Cell-cycle-dependent repair of damage in alpha and bulk DNA of monkey cells. Mutat Res. 1986 Jul;166(1):71–77. doi: 10.1016/0167-8817(86)90042-8. [DOI] [PubMed] [Google Scholar]
- Leadon S. A., Zolan M. E., Hanawalt P. C. Restricted repair of aflatoxin B1 induced damage in alpha DNA of monkey cells. Nucleic Acids Res. 1983 Aug 25;11(16):5675–5689. doi: 10.1093/nar/11.16.5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb L. A. Apurinic sites as mutagenic intermediates. Cell. 1985 Mar;40(3):483–484. doi: 10.1016/0092-8674(85)90191-6. [DOI] [PubMed] [Google Scholar]
- Lunn G., Sansone E. B. Validation of techniques for the destruction of dimethyl sulfate. Am Ind Hyg Assoc J. 1985 Mar;46(3):111–114. doi: 10.1080/15298668591394518. [DOI] [PubMed] [Google Scholar]
- Male R., Nes I. F., Kleppe K. Purification and properties of 3-methyladenine-DNA glycosylase from L-cells. Eur J Biochem. 1981 Dec;121(1):243–248. doi: 10.1111/j.1432-1033.1981.tb06455.x. [DOI] [PubMed] [Google Scholar]
- Margison G. P., Margison J. M., Montesano R. Methylated purines in the deoxyribonucleic acid of various Syrian-golden-hamster tissues after administration of a hepatocarcinogenic dose of dimethylnitrosamine. Biochem J. 1976 Sep 1;157(3):627–634. doi: 10.1042/bj1570627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margison G. P., O'Connor P. J. Biological implications of the instability of the N-glycosidic bone of 3-methyldeoxyadenosine in DNA. Biochim Biophys Acta. 1973 Dec 21;331(3):349–356. doi: 10.1016/0005-2787(73)90021-x. [DOI] [PubMed] [Google Scholar]
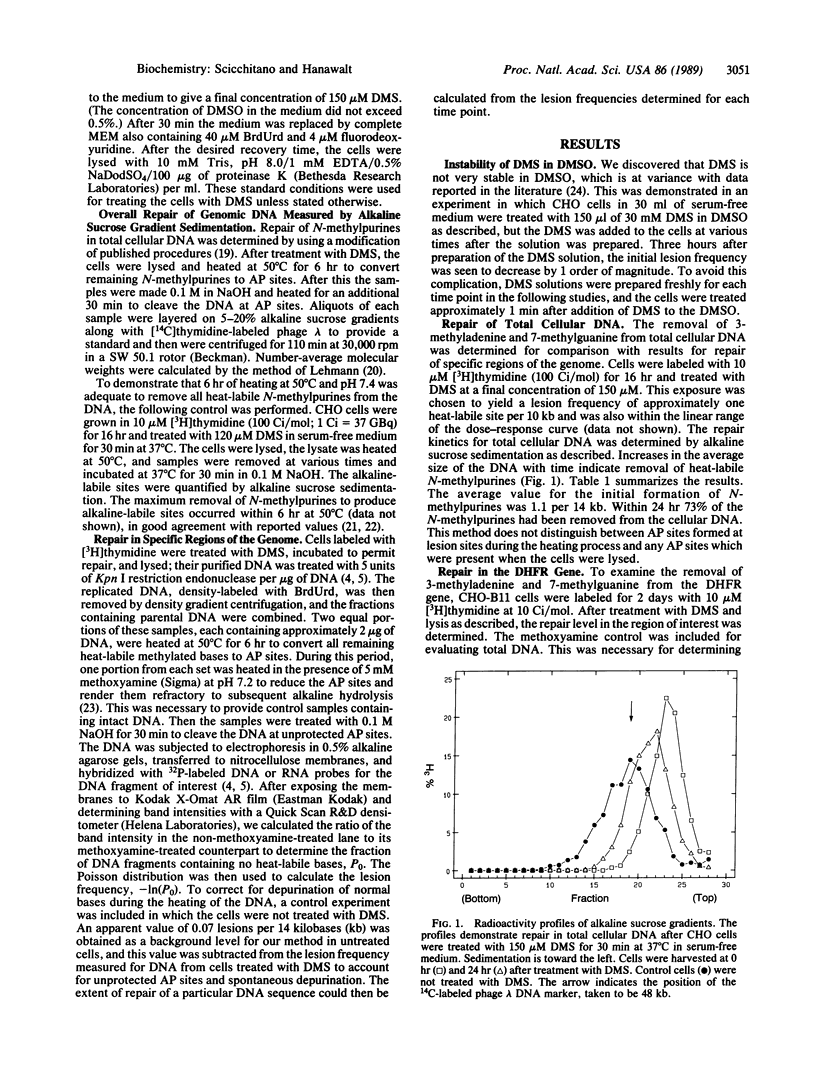
- Mellon I., Spivak G., Hanawalt P. C. Selective removal of transcription-blocking DNA damage from the transcribed strand of the mammalian DHFR gene. Cell. 1987 Oct 23;51(2):241–249. doi: 10.1016/0092-8674(87)90151-6. [DOI] [PubMed] [Google Scholar]
- Nakabeppu Y., Yamashita K., Sekiguchi M. Purification and characterization of normal and mutant forms of T4 endonuclease V. J Biol Chem. 1982 Mar 10;257(5):2556–2562. [PubMed] [Google Scholar]
- Nose K., Nikaido O. Transcriptionally active and inactive genes are similarly modified by chemical carcinogens or X-ray in normal human fibroblasts. Biochim Biophys Acta. 1984 Apr 5;781(3):273–278. doi: 10.1016/0167-4781(84)90093-9. [DOI] [PubMed] [Google Scholar]
- Paquette Y., Crine P., Verly W. G. Properties of the endonuclease for depurinated DNA from Escherichia coli. Can J Biochem. 1972 Nov;50(11):1199–1209. doi: 10.1139/o72-163. [DOI] [PubMed] [Google Scholar]
- Price J. A., Heller E., Goldthwait D. A. The release of 3-methyladenine from nucleosomal DNA by a 3-methyladenine DNA glycosylase. Carcinogenesis. 1983;4(2):145–152. doi: 10.1093/carcin/4.2.145. [DOI] [PubMed] [Google Scholar]
- Ryan A. J., Billett M. A., O'Connor P. J. Selective repair of methylated purines in regions of chromatin DNA. Carcinogenesis. 1986 Sep;7(9):1497–1503. doi: 10.1093/carcin/7.9.1497. [DOI] [PubMed] [Google Scholar]
- Shackleton J., Warren W., Roberts J. J. The excision of N-methyl-N-nitrosourea-induced lesions from the DNA of Chinese hamster cells as measured by the loss of sites sensitive to an enzyme extract that excises 3-methylpurines but not O6-methylguanine. Eur J Biochem. 1979 Jul;97(2):425–433. doi: 10.1111/j.1432-1033.1979.tb13130.x. [DOI] [PubMed] [Google Scholar]
- Smith C. A. DNA repair in specific sequences in mammalian cells. J Cell Sci Suppl. 1987;6:225–241. doi: 10.1242/jcs.1984.supplement_6.16. [DOI] [PubMed] [Google Scholar]
- Sweet J. M., Carda B., Small G. D. Repair of 3-methyladenine and 7-methylguanine in nuclear DNA of Chlamydomonas: requirement for protein synthesis. Mutat Res. 1981 Nov;84(1):73–82. doi: 10.1016/0027-5107(81)90050-6. [DOI] [PubMed] [Google Scholar]
- Vos J. M., Hanawalt P. C. Processing of psoralen adducts in an active human gene: repair and replication of DNA containing monoadducts and interstrand cross-links. Cell. 1987 Aug 28;50(5):789–799. doi: 10.1016/0092-8674(87)90337-0. [DOI] [PubMed] [Google Scholar]
- Walker I. G., Ewart D. F. The nature of single-strand breaks in DNA following treatment of L-cells with methylating agents. Mutat Res. 1973 Sep;19(3):331–341. doi: 10.1016/0027-5107(73)90234-0. [DOI] [PubMed] [Google Scholar]
- Warren W., Crathorn A. R., Shooter K. V. The stability of methylated purines and of methylphosphotriesters in the DNA of V79 cells after treatment with N-methyl-N-nitrosourea. Biochim Biophys Acta. 1979 Jun 20;563(1):82–88. doi: 10.1016/0005-2787(79)90009-1. [DOI] [PubMed] [Google Scholar]