SUMMARY
Clostridium difficile (C. diff) is a major nosocomial problem. Epidemiological surveillance of the disease can be accomplished by microbiological or administrative data. Microbiological tracking is problematic since it does not always translate into clinical disease, and it is not always available. Tracking by administrative data is attractive, but ICD-9 code accuracy for C. diff is unknown. By using a large administrative database of hospitalized patients with C. diff (by ICD-9 code or cytotoxic assay), this study found that the sensitivity, specificity, positive, and negative predictive values of ICD-9 coding were 71%, 99%, 87%, and 96% respectively (using micro data as the gold standard). When only using symptomatic patients the sensitivity increased to 82% and when only using symptomatic patients whose test results were available at discharge, the sensitivity increased to 88%. C. diff ICD-9 codes closely approximate true C. diff infection, especially in symptomatic patients whose test results are available at the time of discharge, and can therefore be used as a reasonable alternative to microbiological data for tracking purposes.
Clostridium difficile (C. diff) infection is a major worldwide nosocomial problem, affecting 3 million in-patients a year in the United States [1]. Both the severity and the incidence of the disease are increasing. According to the US National Nosocomial Infections Surveillance System (NNIS), from 1987 to 2001, rates of C. diff infection increased in ICU patients in hospitals with >500 beds and in non-ICU patients in hospitals with <250 beds [2]. Rates of C. diff infection also doubled in non-federal acute care hospitals from 1996 to 2003 [3]. A retrospective review of all cases of C. diff from a hospital in Quebec, Canada from 1991 to 2003 showed a greater than four-fold rise in incidence (eight-fold in those aged >65 years), a greater than two-fold rise in complications, and a three-fold rise in death [4]. A similar report from Pittsburgh, Pennsylvania, USA showed a doubling of the incidence in the disease from 1990–1999 to 2000–2001 [5]. Most recently, studies have reported epidemics of toxin gene–variant strains of C. diff that are associated with high morbidity and mortality [6, 7]. Given this documented increase in the severity and incidence of the disease, the need for easy and accurate epidemiological tracking is crucial.
Epidemiological surveillance of the disease can be accomplished by microbiological or administrative data. The gold standard for microbiological diagnosis is a C. diff cytotoxic assay; however, a positive test does not always translate into clinical disease. In one prospective study, McFarland et al. found that in a general medical population of patients who were C. diff-culture positive and asymptomatic, 16% were C. diff-toxin positive [8]. Kyne et al. found that in a general medical population of patients on antibiotics who acquired C. diff colonization in the hospital and remained asymptomatic, 79% of them were C. diff-toxin positive [9]. The prevalence of asymptomatic C. diff-toxin-positive patients is therefore variable, and dependent on the method of study and the risk of the patient population. Additionally, microbiological data are not always available from large administrative datasets, whereas billing codes are routinely available. The accuracy of ICD-9 coding is highly variable among diagnoses, and the accuracy of ICD-9 coding for C. diff infection is not well documented [10]. This study sought to determine the accuracy of ICD-9 coding for C. diff using a positive cytotoxic assay as the gold standard.
This was a retrospective cohort of all patients hospitalized at Brigham and Women's Hospital in 2004 who either had an ICD-9 diagnosis for C. diff (008·45) or a C. diff cytotoxic assay performed. Brigham and Women's Hospital is a 747-bed non-profit teaching affiliate of Harvard Medical School and a founding member of Partners HealthCare System located in Boston, Massachusetts, USA. The patients for the analysis were identified through the Partners HealthCare System Research Patient Database Repository (RPDR). The RPDR is an inclusive Partners Health System administrative database that contains over 2·5 million patients and 550 million records from patient encounters, laboratory results, and other medical care. Data from the Partners Health patient billing system is directly downloaded into RPDR and is 100% complete and accurate. IRB approval was granted through the Partners HealthCare system.
All in-patients for which there was either an ICD-9 code for C. diff or a C. diff cytotoxic assay performed in 2004 were included. Patients were categorized into four groups depending on their C. diff toxin results and ICD-9 coding: ICD-9+/toxin+, ICD9+/toxin−, ICD-9−/toxin+, and ICD-9−/toxin− and a 2×2 table was constructed. Chart review was undertaken on all ICD-9+/toxin+ patients and all discordant patients (ICD-9+/toxin− and ICD-9−/toxin+) to determine if the patient had symptoms and to determine why (or why not) an ICD-9 code was recorded. A patient was considered to have symptoms if the discharge summary recorded diarrhoea, abdominal pain/cramping, or radiological/colonoscopic evidence of colitis. Brigham and Women's Hospital utilizes a standard C. diff toxin A/B cytotoxic neutralization assay. This tissue culture-based test has a reported sensitivity of 94–100% and specificity of 99%.
The sensitivity, specificity, positive predictive value and negative predictive values of ICD-9 codes were calculated in a standard fashion. Age, gender, race, length of stay, and month of admission for ICD-9+/toxin+ patients, ICD-9+/toxin− patients, and ICD-9−/toxin+ patients were compared by χ2 (categorical variables) and ANOVA (continuous variables). P values <0·05 were considered significant.
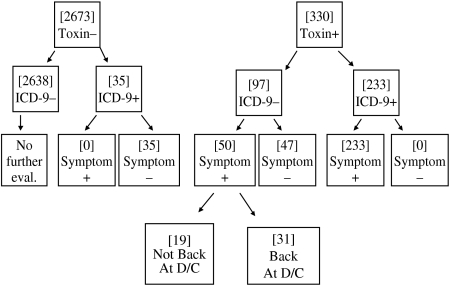
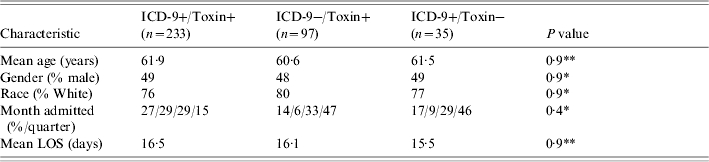
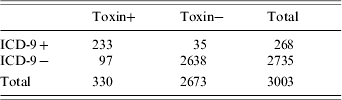
Baseline patient characteristics are outlined in Table 1. There were no statistically significant differences between the groups with respect to age, gender, race, length of stay, or month of admission. There were a total of 5173 C. diff cytotoxic assays corresponding to 3003 patients (due to repeat testing during the same hospital stay). Of these 3003 patients, 330 had a positive cytotoxic assay (11%) (Table 2 and Fig.). There were 97 ICD-9−/toxin+ patients, of whom 50 (52%) were symptomatic and 47 (48%) who were asymptomatic. There were 233 ICD9+/toxin+ patients, all of whom were symptomatic. There were 35 ICD-9+/toxin− patients, of whom none were symptomatic. When using all patients with positive cytotoxic assays as the gold standard, the sensitivity, specificity, positive, and negative predictive values of ICD-9 coding were 71%, 99%, 87%, and 96% respectively (Table 2). In determining why an ICD-9 code was assigned to the 35 ICD-9+/toxin− patients, all had a past history of C. diff outlined in the discharge summary (in the past medical history) but it was not an active issue during the hospital stay. In determining why an ICD-9 code was not assigned to the 97 ICD-9−/toxin+ patients, 47 of them did not have any chart evidence of symptoms and the diagnosis of C. diff was not recorded anywhere in the chart. An additional 19 patients did not have the test results back at the time of discharge so the diagnosis was not known at the time of discharge from the hospital. If limiting the analysis to only include patients with symptoms, the sensitivity increased to 82%, and if limiting to patients with symptoms and a positive toxin at the time of discharge, the sensitivity increased to 88%.
Table 1.
Baseline demographics for three groups of patients
LOS, Length of stay.
χ2, ** ANOVA.
Table 2.
Sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) of ICD-9 codes for C. difficile infection
Sensitivity, 71%; Specificity, 99%; PPV, 87%; NPV, 96%.
Fig.
Patients according to toxin, ICD-9, and symptoms.
The primary benefit of epidemiological tracking of C. diff through microbiological data is the high sensitivity; however, it is not specific for symptomatic disease. This can be problematic since the prevalence of asymptomatic toxin+ patients varies in the literature, dependent on the study methodology and the risk of the patient population [8, 9]. In this study, 47 out of 330 toxin-positive patients (14%) were asymptomatic, which may be an underestimation since it was a retrospective chart review where underestimation of symptoms is not unexpected.
The primary benefit of epidemiological tracking of C. diff through administrative data is the high specificity. In this study, there were only 35 ICD-9+/toxin− patients that accounted for a small relative number and did not significantly affect the overall specificity. Upon chart review, all of them had a prior history of C. diff infection in the past medical history for which they were incorrectly given an ICD-9 code. This small number should not significantly affect the accuracy of epidemiological tracking.
The primary limitation of epidemiological tracking of C. diff through administrative data is the sensitivity, which in this study was 71%. This is similar to a recent study that reported an ICD-9 code sensitivity of 78% compared to C. diff toxin in hospitalized patients [11]. That study, however, did not undertake a chart review to determine if the patients were symptomatic or if the test result was back at the time of discharge. In our study, the sensitivity increased to 82% when only evaluating symptomatic patients and further increased to 88% when only evaluating symptomatic patients with a test result at the time of hospital discharge. This is an important distinction since the patient population that researchers would be most interested in tracking would be symptomatic patients whose diagnosis was determined by the time of hospital discharge. Therefore, by using ICD-9 codes for C. diff tracking purposes, a 12% under-reporting rate would be expected if trying to capture symptomatic C. diff toxin+ patients whose diagnosis is known at the time of discharge.
The primary limitation of this study is its single institution design, potentially limiting its generalizability, and that it was retrospective, so symptoms may not have been appropriately documented in the chart. However, the strengths include the complete analysis of 5173 toxin assays on 3003 patients, the direct examination of the discharge summary of all toxin-positive and discordant patients, and the use of an objective gold standard.
In conclusion, C. diff ICD-9 codes closely approximate true C. diff infection, especially in symptomatic patients whose test results are available at the time of discharge, and can be used as a reasonable alternative for epidemiological tracking when microbiological data in unavailable.
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Kyne L et al. Health care costs and mortality associated with nosocomial diarrhoea due to Clostridium difficile. Clinical Infectious Diseases. 2002;34:346–353. doi: 10.1086/338260. [DOI] [PubMed] [Google Scholar]
- 2.Archibald LK, Banerjee SN, Jarvis WR. Secular trends in hospital-acquired Clostridium difficile disease in the United States, 1987–2001. Journal of Infectious Diseases. 2004;189:1585–1589. doi: 10.1086/383045. [DOI] [PubMed] [Google Scholar]
- 3.McDonald LC, Owings M, Jernigan DB. Clostridium difficile infection in patients discharged from US short-stay hospitals, 1996–2003. Emerging Infectious Diseases. 2006;12:409–415. doi: 10.3201/eid1203.051064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pepin J et al. Clostridium difficile-associated diarrhoea in a region of Quebec from 1991 to 2003: a changing pattern of disease severity. Canadian Medical Association Journal. 2004;171:466–472. doi: 10.1503/cmaj.1041104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muto CA et al. A large outbreak of Clostridium difficile-associated disease with an unexpected proportion of deaths and colectomies at a teaching hospital following increased fluoroquinolone use. Infection Control and Hospital Epidemiology. 2005;26:273–280. doi: 10.1086/502539. [DOI] [PubMed] [Google Scholar]
- 6.Loo VG et al. A predominantly clonal multi-institutional outbreak of Clostridium difficile-associated diarrhoea with high morbidity and mortality. New England Journal of Medicine. 2005;353:2442–2449. doi: 10.1056/NEJMoa051639. [DOI] [PubMed] [Google Scholar]
- 7.McDonald LC et al. An epidemic, toxin gene–variant strain. Clostridium difficile New England Journal of Medicine. 2005;353:2433–2441. doi: 10.1056/NEJMoa051590. of . [DOI] [PubMed] [Google Scholar]
- 8.McFarland LV et al. Nosocomial acquisition of Clostridium difficile infection. New England Journal of Medicine. 1989;320:204–210. doi: 10.1056/NEJM198901263200402. [DOI] [PubMed] [Google Scholar]
- 9.Kyne L et al. Asymptomatic carriage of Clostridium difficile and serum levels of IgG antibody against toxin A. New England Journal of Medicine. 2000;342:390–397. doi: 10.1056/NEJM200002103420604. [DOI] [PubMed] [Google Scholar]
- 10.Berthelson CL. Evaluation of coding data quality of the HCUP National Inpatient Sample. Top Health Information Management. 2000;21:10–23. [PubMed] [Google Scholar]
- 11.Dubberke ERet al. ICD-9 codes and surveillance for Clostridium difficile-associated disease. Emerging Infectious Diseases [serial on the Internet] (http://www.cdc.gov/ncidod/EID/vol12no10/06-0016.htm). Accessed 4 October 2006 [DOI] [PMC free article] [PubMed]