Abstract
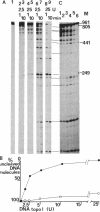
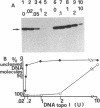
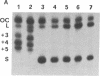
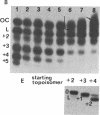
The effects of supercoiling on the cleavage reaction by eukaryotic DNA topoisomerases I (wheat germ, chicken erythrocyte, and calf thymus) have been analyzed on DNA fragments (0.96 and 2.3 kilobases) encompassing an immunoglobulin kappa light-chain promoter. In one topological condition of the substrate, the absolutely relaxed state, cleavage was found to be impeded. This finding defines the topology-dependent step of the eukaryotic DNA topoisomerase I reaction and shows that for the cleavage reaction topology is more critical than sequence effects. These findings suggest a simple model for the regulation of the DNA topoisomerase I reaction based on topological factors, which may explain the regulatory function of the enzyme in in vivo eukaryotic transcription.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baase W. A., Wang J. C. An omega protein from Drosophila melanogaster. Biochemistry. 1974 Oct 8;13(21):4299–4303. doi: 10.1021/bi00718a009. [DOI] [PubMed] [Google Scholar]
- Bonven B. J., Gocke E., Westergaard O. A high affinity topoisomerase I binding sequence is clustered at DNAase I hypersensitive sites in Tetrahymena R-chromatin. Cell. 1985 Jun;41(2):541–551. doi: 10.1016/s0092-8674(85)80027-1. [DOI] [PubMed] [Google Scholar]
- Brill S. J., DiNardo S., Voelkel-Meiman K., Sternglanz R. Need for DNA topoisomerase activity as a swivel for DNA replication for transcription of ribosomal RNA. 1987 Mar 26-Apr 1Nature. 326(6111):414–416. doi: 10.1038/326414a0. [DOI] [PubMed] [Google Scholar]
- Camilloni G., Della Seta F., Negri R., Grazia Ficca A., Di Mauro E. Structure of RNA polymerase II promoters. Conformational alterations and template properties of circularized Saccharomyces cerevisiae GAL1-GAL10 divergent promoters. EMBO J. 1986 Apr;5(4):763–771. doi: 10.1002/j.1460-2075.1986.tb04279.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camilloni G., Di Martino E., Caserta M., di Mauro E. Eukaryotic DNA topoisomerase I reaction is topology dependent. Nucleic Acids Res. 1988 Jul 25;16(14B):7071–7085. doi: 10.1093/nar/16.14.7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champoux J. J., Dulbecco R. An activity from mammalian cells that untwists superhelical DNA--a possible swivel for DNA replication (polyoma-ethidium bromide-mouse-embryo cells-dye binding assay). Proc Natl Acad Sci U S A. 1972 Jan;69(1):143–146. doi: 10.1073/pnas.69.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champoux J. J. Strand breakage by the DNA untwisting enzyme results in covalent attachment of the enzyme to DNA. Proc Natl Acad Sci U S A. 1977 Sep;74(9):3800–3804. doi: 10.1073/pnas.74.9.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depew D. E., Wang J. C. Conformational fluctuations of DNA helix. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4275–4279. doi: 10.1073/pnas.72.11.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards K. A., Halligan B. D., Davis J. L., Nivera N. L., Liu L. F. Recognition sites of eukaryotic DNA topoisomerase I: DNA nucleotide sequencing analysis of topo I cleavage sites on SV40 DNA. Nucleic Acids Res. 1982 Apr 24;10(8):2565–2576. doi: 10.1093/nar/10.8.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley D. E., Coleclough C., Perry R. P. Functional significance and evolutionary development of the 5'-terminal regions of immunoglobulin variable-region genes. Cell. 1982 Jun;29(2):681–689. doi: 10.1016/0092-8674(82)90184-2. [DOI] [PubMed] [Google Scholar]
- Kirkegaard K., Wang J. C. Mapping the topography of DNA wrapped around gyrase by nucleolytic and chemical probing of complexes of unique DNA sequences. Cell. 1981 Mar;23(3):721–729. doi: 10.1016/0092-8674(81)90435-9. [DOI] [PubMed] [Google Scholar]
- Le Bret M. Catastrophic variation of twist and writhing of circular DNAs with constraint? Biopolymers. 1979 Jul;18(7):1709–1725. doi: 10.1002/bip.1979.360180710. [DOI] [PubMed] [Google Scholar]
- Liu L. F., Wang J. C. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci U S A. 1987 Oct;84(20):7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller M. T. Quantitation of eukaryotic topoisomerase I reactivity with DNA. Preferential cleavage of supercoiled DNA. Biochim Biophys Acta. 1985 Mar 20;824(3):263–267. doi: 10.1016/0167-4781(85)90057-0. [DOI] [PubMed] [Google Scholar]
- Ness P. J., Parish R. W., Koller T. Mapping of endogenous nuclease-sensitive regions and of putative topoisomerase sites of action along the chromatin of Dictyostelium ribosomal RNA genes. J Mol Biol. 1986 Apr 5;188(3):287–300. doi: 10.1016/0022-2836(86)90155-5. [DOI] [PubMed] [Google Scholar]
- Pulleyblank D. E., Shure M., Tang D., Vinograd J., Vosberg H. P. Action of nicking-closing enzyme on supercoiled and nonsupercoiled closed circular DNA: formation of a Boltzmann distribution of topological isomers. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4280–4284. doi: 10.1073/pnas.72.11.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schon E., Evans T., Welsh J., Efstratiadis A. Conformation of promoter DNA: fine mapping of S1-hypersensitive sites. Cell. 1983 Dec;35(3 Pt 2):837–848. doi: 10.1016/0092-8674(83)90116-2. [DOI] [PubMed] [Google Scholar]
- Seidman J. G., Leder P. A mutant immunoglobulin light chain is formed by aberrant DNA- and RNA-splicing events. Nature. 1980 Aug 21;286(5775):779–783. doi: 10.1038/286779a0. [DOI] [PubMed] [Google Scholar]
- Shore D., Baldwin R. L. Energetics of DNA twisting. II. Topoisomer analysis. J Mol Biol. 1983 Nov 15;170(4):983–1007. doi: 10.1016/s0022-2836(83)80199-5. [DOI] [PubMed] [Google Scholar]
- Thomsen B., Mollerup S., Bonven B. J., Frank R., Blöcker H., Nielsen O. F., Westergaard O. Sequence specificity of DNA topoisomerase I in the presence and absence of camptothecin. EMBO J. 1987 Jun;6(6):1817–1823. doi: 10.1002/j.1460-2075.1987.tb02436.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venditti S., Caserta M., Di Mauro E., Camilloni G. DNA conformational variations in the in vitro torsionally strained Ig kappa light chain gene localize on consensus sequences. Biochim Biophys Acta. 1988 Nov 10;951(1):139–148. doi: 10.1016/0167-4781(88)90034-6. [DOI] [PubMed] [Google Scholar]
- Vinograd J., Lebowitz J., Watson R. Early and late helix-coil transitions in closed circular DNA. The number of superhelical turns in polyoma DNA. J Mol Biol. 1968 Apr 14;33(1):173–197. doi: 10.1016/0022-2836(68)90287-8. [DOI] [PubMed] [Google Scholar]
- Wang J. C. DNA topoisomerases. Annu Rev Biochem. 1985;54:665–697. doi: 10.1146/annurev.bi.54.070185.003313. [DOI] [PubMed] [Google Scholar]
- Weintraub H., Cheng P. F., Conrad K. Expression of transfected DNA depends on DNA topology. Cell. 1986 Jul 4;46(1):115–122. doi: 10.1016/0092-8674(86)90865-2. [DOI] [PubMed] [Google Scholar]
- Zhang H., Wang J. C., Liu L. F. Involvement of DNA topoisomerase I in transcription of human ribosomal RNA genes. Proc Natl Acad Sci U S A. 1988 Feb;85(4):1060–1064. doi: 10.1073/pnas.85.4.1060. [DOI] [PMC free article] [PubMed] [Google Scholar]