Abstract
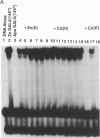
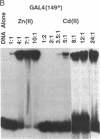
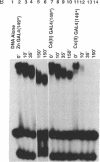
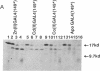
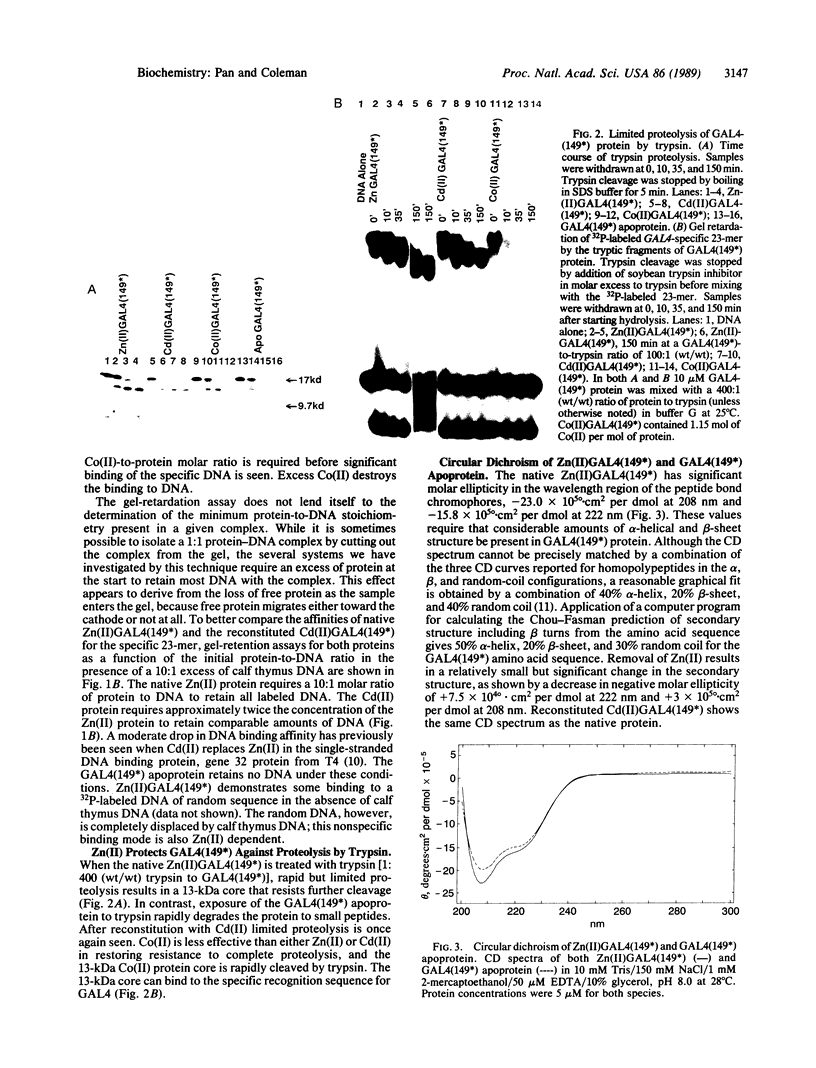
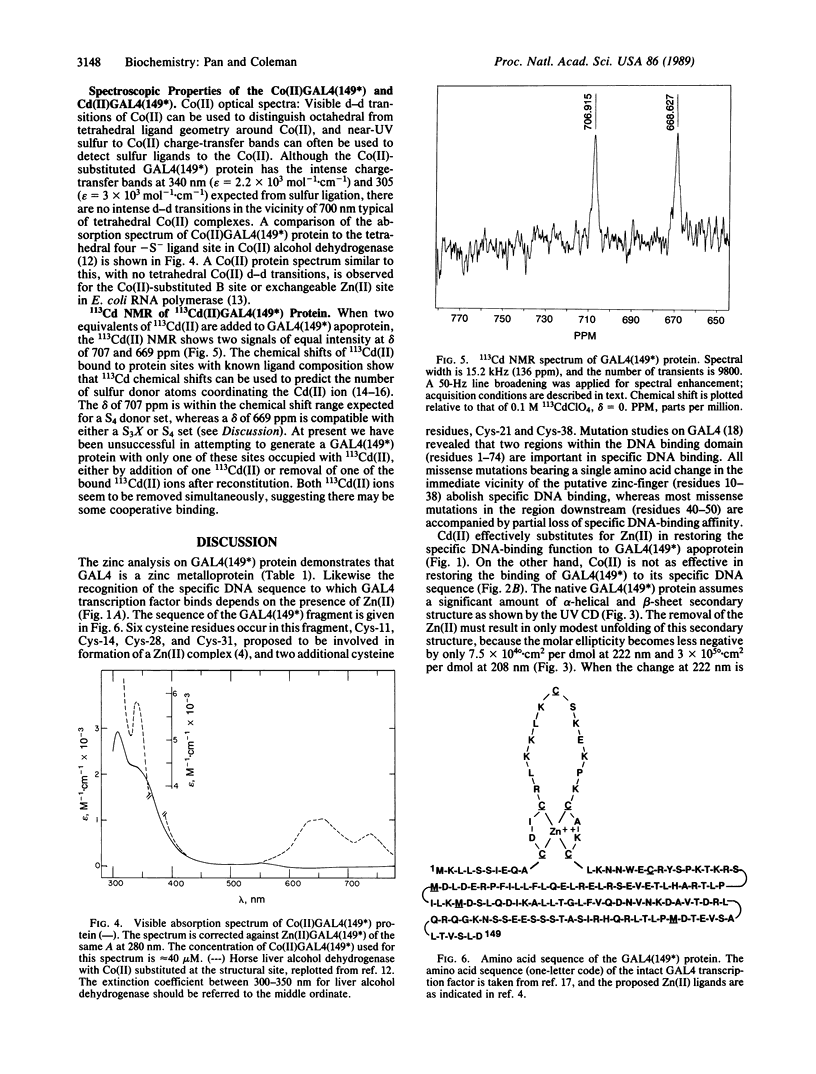
The transcription factor GAL4 from Saccharomyces cerevisiae contains a "zinc-finger"-like motif, Cys-Xaa2-Cys-Xaa6-Cys-Xaa6-Cys-Xaa2-Cys-Xaa6+ ++-Cys, within its DNA-binding domain. A GAL4 fragment consisting of residues 1-147 plus two additional residues from the cloning vector [denoted GAL4(149*)] has been cloned and overexpressed in Escherichia coli. This fragment includes the entire DNA-binding domain (residues 1-74). The homogeneous GAL4-(149*) protein contains 1-1.5 moles of Zn(II) per mole of protein. The GAL4(149*) protein binds tightly to the specific 17-base-pair palindromic DNA sequence found at GAL4 binding sites as shown by gel-retention assays using a 32P-labeled 23-mer containing this sequence. Removal of the intrinsic Zn(II) by EDTA at low pH abolishes binding to the 23-mer. The GAL4(149*) apoprotein can be reconstituted with Zn(II), Cd(II), or Co(II) with restoration of specific DNA binding. Titration of GAL4(149*) apoprotein with 113Cd(II) shows two 113Cd(II) binding sites on the molecule, one with delta of 707 ppm, suggesting coordination to four sulfur atoms, and one with delta of 669 ppm, suggesting coordination to three or four sulfur atoms. Because GAL4(149*) protein contains only six cysteine residues within its DNA-binding domain, the precise coordination of the two Cd(II) ions cannot be stated with certainty; one or more shared -S- ligands could exist. GAL4(149*) protein contains approximately 40% alpha-helix and approximately 20% beta-sheet, estimated from circular dichroism. Removal of the native Zn(II) ion causes limited unfolding of secondary structure, but less than one turn of alpha-helix. The binding of Zn(II), Cd(II), and, to a lesser extent, Co(II) to GAL4(149*) apoprotein protects the protein from proteolysis by trypsin, which produces a 13-kDa DNA-binding core.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Diakun G. P., Fairall L., Klug A. EXAFS study of the zinc-binding sites in the protein transcription factor IIIA. Nature. 1986 Dec 18;324(6098):698–699. doi: 10.1038/324698a0. [DOI] [PubMed] [Google Scholar]
- Engeseth H. R., McMillin D. R., Otvos J. D. Comparative Cd-113 nuclear magnetic resonance studies of Cd(II)-substituted blue copper proteins. J Biol Chem. 1984 Apr 25;259(8):4822–4826. [PubMed] [Google Scholar]
- Freedman L. P., Luisi B. F., Korszun Z. R., Basavappa R., Sigler P. B., Yamamoto K. R. The function and structure of the metal coordination sites within the glucocorticoid receptor DNA binding domain. Nature. 1988 Aug 11;334(6182):543–546. doi: 10.1038/334543a0. [DOI] [PubMed] [Google Scholar]
- Fried M., Crothers D. M. Equilibria and kinetics of lac repressor-operator interactions by polyacrylamide gel electrophoresis. Nucleic Acids Res. 1981 Dec 11;9(23):6505–6525. doi: 10.1093/nar/9.23.6505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giedroc D. P., Coleman J. E. Structural and functional differences between the two intrinsic zinc ions of Escherichia coli RNA polymerase. Biochemistry. 1986 Aug 26;25(17):4969–4978. doi: 10.1021/bi00365a037. [DOI] [PubMed] [Google Scholar]
- Giedroc D. P., Johnson B. A., Armitage I. M., Coleman J. E. NMR spectroscopy of 113Cd(II)-substituted gene 32 protein. Biochemistry. 1989 Mar 21;28(6):2410–2418. doi: 10.1021/bi00432a011. [DOI] [PubMed] [Google Scholar]
- Giedroc D. P., Keating K. M., Williams K. R., Coleman J. E. The function of zinc in gene 32 protein from T4. Biochemistry. 1987 Aug 25;26(17):5251–5259. doi: 10.1021/bi00391a007. [DOI] [PubMed] [Google Scholar]
- Giniger E., Varnum S. M., Ptashne M. Specific DNA binding of GAL4, a positive regulatory protein of yeast. Cell. 1985 Apr;40(4):767–774. doi: 10.1016/0092-8674(85)90336-8. [DOI] [PubMed] [Google Scholar]
- Greenfield N., Fasman G. D. Computed circular dichroism spectra for the evaluation of protein conformation. Biochemistry. 1969 Oct;8(10):4108–4116. doi: 10.1021/bi00838a031. [DOI] [PubMed] [Google Scholar]
- Johnston M., Dover J. Mutations that inactivate a yeast transcriptional regulatory protein cluster in an evolutionarily conserved DNA binding domain. Proc Natl Acad Sci U S A. 1987 Apr;84(8):2401–2405. doi: 10.1073/pnas.84.8.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston M. Genetic evidence that zinc is an essential co-factor in the DNA binding domain of GAL4 protein. Nature. 1987 Jul 23;328(6128):353–355. doi: 10.1038/328353a0. [DOI] [PubMed] [Google Scholar]
- Keating K. M., Ghosaini L. R., Giedroc D. P., Williams K. R., Coleman J. E., Sturtevant J. M. Thermal denaturation of T4 gene 32 protein: effects of zinc removal and substitution. Biochemistry. 1988 Jul 12;27(14):5240–5245. doi: 10.1021/bi00414a044. [DOI] [PubMed] [Google Scholar]
- Keegan L., Gill G., Ptashne M. Separation of DNA binding from the transcription-activating function of a eukaryotic regulatory protein. Science. 1986 Feb 14;231(4739):699–704. doi: 10.1126/science.3080805. [DOI] [PubMed] [Google Scholar]
- Laughon A., Gesteland R. F. Primary structure of the Saccharomyces cerevisiae GAL4 gene. Mol Cell Biol. 1984 Feb;4(2):260–267. doi: 10.1128/mcb.4.2.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y. S., Carey M. F., Ptashne M., Green M. R. GAL4 derivatives function alone and synergistically with mammalian activators in vitro. Cell. 1988 Aug 26;54(5):659–664. doi: 10.1016/s0092-8674(88)80010-2. [DOI] [PubMed] [Google Scholar]
- Maret W., Andersson I., Dietrich H., Schneider-Bernlöhr H., Einarsson R., Zeppezauer M. Site-specific substituted cobalt(II) horse liver alcohol dehydrogenases. Preparation and characterization in solution, crystalline and immobilized state. Eur J Biochem. 1979 Aug 1;98(2):501–512. doi: 10.1111/j.1432-1033.1979.tb13211.x. [DOI] [PubMed] [Google Scholar]
- Miller J., McLachlan A. D., Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. EMBO J. 1985 Jun;4(6):1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severne Y., Wieland S., Schaffner W., Rusconi S. Metal binding 'finger' structures in the glucocorticoid receptor defined by site-directed mutagenesis. EMBO J. 1988 Aug;7(8):2503–2508. doi: 10.1002/j.1460-2075.1988.tb03097.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Studier F. W., Moffatt B. A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986 May 5;189(1):113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]