SUMMARY
The first dog-associated outbreak of rabies in swine in China (Hunan province) has been diagnosed and the related virus isolated. Sequence analysis showed that the pig isolate was a genotype 1 rabies virus with a very high nucleotide identity to local dog isolates.
On 11 February 2006, frozen brain specimens of two pigs suspected of dying from rabies were received in our laboratory at the Military Veterinary Research Institute (MVRI), Academy of Military Medical Sciences for confirmation. They originated from a rural pig farm in Yongzhou city, Hunan province. The specimens were tested by the fluorescent antibody test (FAT), mouse inoculation test (MIT) and RT–PCR. The FAT and MIT procedures were based on the WHO protocols [1] using brain smears and FITC-conjugated anti-rabies monoclonal antibody (made in our laboratory) for the FAT. Results showed that the two brain tissues were positive for rabies antigen. A litter of suckling mice inoculated intracranially with the brain tissue suspensions died of rabies 15–17 days post inoculation. Brain smear from each dead mouse was positive for rabies virus (RABV) by the FAT method. The pig rabies virus isolate was designated HuNPN01. For testing by RT–PCR, total RNA was extracted from homogenates of the two pig brain tissues by TRIzol® (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions, and reverse transcription (RT) was carried out using a mixture of 6-mer random primers, oligo-d(T)15 and M-MuLV reverse transcriptase RNase H– (New England Biolabs Inc., Ipswich, MA, USA). The RT product was used to amplify almost the full N gene of rabies virus with an expected size of 1353 bp using Ex-Taq DNA polymerase (Takara Ltd, Dalian, Liaoning, China) and two primers (forward NF: 5′-GTCGAATTCATGGATGCCGACAAG ATTGTATTCAAAG-3′ and reverse NR: 5′-GTAGGATCCTGAGTCACTCGAATATGTCTTGTTTAG-3′). The forward primer and the reverse primer match with 28 nt and 30 nt of the N gene respectively. The PCR was performed in a Peltier Thermal Cycler PTC-200 (Bio-Rad DNA Engine®, Hercules, CA, USA) under the following conditions, 94°C for 2 min, then 35 cycles of 94°C for 2 min, 60°C for 30 s, 72°C for 60 s, and finally 72°C for 10 min, followed by storage at 4°C. Both specimens demonstrated clear amplification of a 1353 bp amplicon of the RABV N gene (data not shown). The PCR amplicon of brain tissue of the isolate HuNPN01 was cloned into pMD18-T vector and subsequently sequenced by a commercial sequencing service (Takara Ltd). The sequence, including the two matching primers, has been deposited in GenBank (accession number DQ496219).
The Yongzhou farmer neither reported the outbreak to the official veterinary administration at its start, nor took any measures to halt the disease during its course. The outbreak was almost over when he sent the two heads (one dead from the disease and the other killed when moribund) to our local laboratory at the Veterinary College of Hunan Agricultural University on 9 February, 4 days before the last pig death on 13 February. After laboratory confirmation that both samples were positive for RABV, a retrospective epidemiological investigation was carried out by visiting the affected farm. The farm originally contained 56 fattening pigs weighing about 50 kg, raised in four pens linked together in a line. On 19 December 2005, the farmer's neighbour's dog was found to be acting strangely, roaming around pen 2 and leaving a bleeding bite on the snout of a pig through the pen wall bars. Twenty days later, on 8 January 2006, the injured pig in pen 2 developed a furious disease (referred to as the index case) and started to bite other pigs in the same group (n=16, the farmer could not remember clearly the location or severity of the pig bites). The index pig died 2 days after developing rabies symptoms. Although the farmer was unable to remember accurately the dates and sequence of each onset of the subsequent cases, it appears that during the next few days, two of the pigs bitten by the index pig in pen 2 developed the disease. The cases increased in the pen at the rate of 1–2 cases per day with a few furiously rabid pigs jumping the pen walls to bite pigs in the two adjoining pens. About 12 days later an onset of the disease was noticed in pens 1 and 3, indicating that the incubation period of the disease was about 10 days. The farmer reported that the disease was not present in pen 4 since no affected pig jumped into this pen. Almost all infected pigs developed furious rabies except the last one, which died suddenly on 13 February without any symptoms. The clinical manifestations included hyperexcitation, roaring and attacks on other pigs within the herd. Some affected pigs exhibited spasms in response to tactile, auditory and visual stimuli when people visited and when water was poured into their drinking tanks. All deaths occurred 2–3 days following the appearance of symptoms, with some carcasses being almost destroyed by the bites of rabid pigs. The outbreak lasted 35 days from 8 January to 13 February 2006, and resulted in 20 pig deaths including one culled when moribund. Twelve cases occurred in pen 2, and eight in the two adjoining pens. The remaining 36 pigs remained healthy and gained weight to around 70–80 kg by the end of March. During the outbreak no control measures were applied, and therefore the spread of the disease within the herds was not stopped by destruction of the first cases.
Although documented in textbooks [2], rabies in swine is uncommon and has been poorly described [3–9]. It accounts for only 0·1–1·1% of the incidence of animal rabies [4, 6, 8]. The outbreak described here, causing the death of 20 out of 56 pigs is the first case report of pig rabies in China, featuring the acute and furious forms with an incubation time of 20 days for the index pig and about 10 days for contact animals. The incubation period observed in the present study is consistent with that observed by Luangtongkum et al., who reported that the incubation period of three lethal pig rabies infections caused by dog bites is 8–16 days [5]. After the appearance of symptoms, all affected pigs died within 1–3 days and showed furious clinical symptoms similar to those described previously [4–6, 8, 9], although an outbreak of this size has not been reported so far. This demonstrates that rabies can be transmitted naturally among pigs, and that swine are highly susceptible to rabies, with the disease readily spreading in pig herds if culling is not carried out at an early stage.
Rabies is an almost invariably fatal zoonotic viral disease that can affect all warm-blooded animals worldwide. Infection following dog bites accounts for China having the second highest number of human rabies cases in the world [10]. Hunan is one of the most severely affected provinces in the country, with about 400 human rabies deaths annually [11, 12]. Yongzhou is a district comprising eight counties and one city of that name. Due to the high number of free-roaming dogs, which are raised for security purposes and rarely vaccinated, this district has experienced the most severe rabies endemics in Hunan province. From 2000–2003 the annual incidence of human rabies cases in Yongzhou district was 47, 81, 75 and 108 respectively, the highest in the province and accounting for 25% of the total provincial cases [12]. In 2005 and 2006, the corresponding numbers were 36 and 64 respectively (unpublished data, Hunan Provincial Center of Disease Control). There is, however, a lack of information about the incidence of animal rabies in the district. The rabies-affected pig farm is located in a village in the southwest suburbs of Yongzhou city, where no human rabies has been reported in recent years.
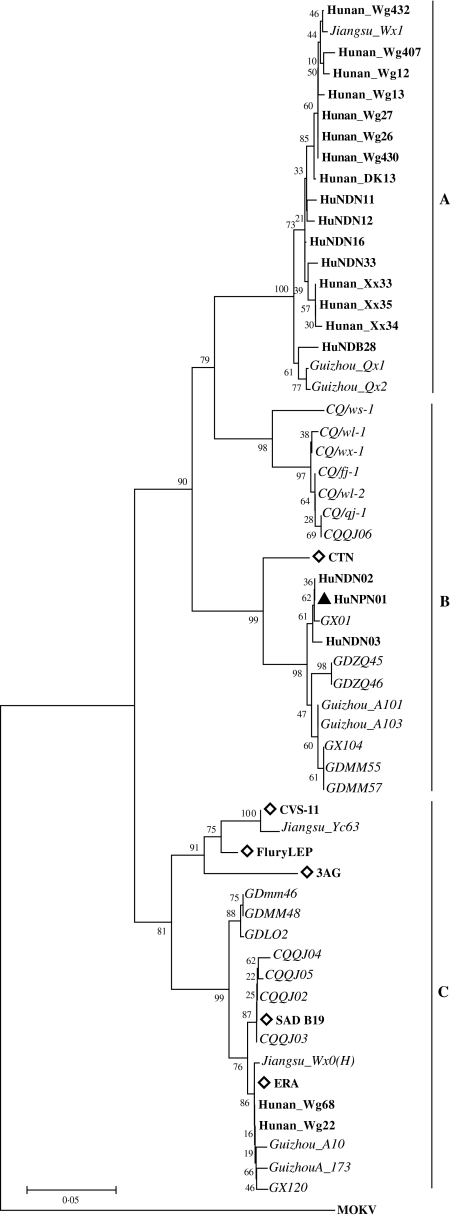
The most recent study has shown that the viruses causing rabies in China are genotype I strains consisting of three genetic subgroups [13]. The dog responsible for the pig outbreak escaped and was found dead near the house of its owner the next day after biting the index pig. Unfortunately it was buried without being reported to a veterinary diagnostic agency. In order to investigate the genetic relationship of the pig isolate with dog isolates circulating in China, an N gene-based phylogenetic analysis focusing on genotype I rabies viruses was conducted using the sequences currently available from GenBank, which were obtained from rabid dogs identified in Hunan, Guangxi, Guangdong, Guizhou, and Jiangsu provinces, and Chongqing city (see Table). The analysis showed that the Chinese rabies viruses can be classified into three major subgroups, A, B and C, and in Hunan province isolates of all three subgroups were prevalent. The pig virus HuNPN01 was segregated into subgroup B, showing the highest genetic identity with some isolates recently obtained from dogs in Hunan (HuN) and Guanxi (GX), Guizhou (GZ) and Guangdong (GD) provinces (see Fig.). Further genetic comparison using DNAstar software (DNASTAR Inc., Madison, WI, USA) showed that the partial N gene of HuNPN01 was 99·7% (370/371) identical to HuNDN02 isolated in 2005 from a rabid dog in Hengshan city near Yongzhou. Sequence comparison with four Chinese vaccine strains, CTN and 3AG for human (killed vaccine), ERA and Flury LEP for animal (live vaccine) showed that the full N gene sequence of HuNPN01, excluding the portions matching with primers, had 94·4% (1223/1295 nt), 86·3% (1118/1295 nt), 87·2% (1129 /1295 nt) and 87·3% (1131/1295 nt) identity respectively, indicating that the pig isolate did not originate from the widespread use of live vaccine in dogs.
Table.
The 57 strains or isolates of rabies viruses used in the study except for pig isolate HuNPN01
Fig.
The 371 bp sequence of the N gene 5′-end was used for phylogenetic analysis within genotype I, which was conducted using MEGA version 3.1 (http://www.megasoftware.net/) (using the neighbour-joining algorithm) and PHYLIP version 3.63 (Seattle, WA, USA) (using the maximum parsimony method). The tree was estimated statistically by 1000 replicates of the bootstrap value and visualized by the TREEVIEW program (http://taxonomy.zoology.gla.ac.uk/rod/treeview.html). Genotype 3 MOKV was used as the outgroup and all sequence information is shown in the Table. The three major subgroups, A, B and C, correspond with published nomenclature [13]. Bold type, isolates of Hunan Province; italic type, isolates of other provinces; ▲, pig isolate HuNPN01; ◊, vaccine and challenge strains.
Our data strongly support the conclusion that the outbreak in swine was caused by the rabid dog. Moreover, the history of exposure to a dog bite, the rabies-like clinical signs, and the positive FAT, MIT and RT–PCR results indicate that rabies was only disease entity involved in the outbreak.
ACKNOWLEDGEMENTS
This work was funded by National Natural Science Foundation of China (Project no. 30600445), National 863 Programme (2006AA02Z456) and the Department of Science and Technology of Guangdong Province (Key project no. 2004A20403001).
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Meslin FX, Kaplan MM, Koprowski H. Laboratory Techniques in Rabies. 4th edn. Geneva: World Health Organization; 1996. pp. 80–93. , pp. [Google Scholar]
- 2.Morehouse LG, Straw BE, D'Allaire S, Mengeling WL, Taylor DJ. Disease of Swine. 8th edn. Ames: Iowa State Press; 1999. Rabies; pp. 247–253. , pp. [Google Scholar]
- 3.Baer GM, Olson HR. Recovery of pigs from rabies. Journal of the American Veterinary Medical Association. 1972;160:1127–1128. [PubMed] [Google Scholar]
- 4.Dhillon SS, Dhingra PN. A note on rabies in swine. Veterinary Medicine, Small Animal Clinician. 1973;68:1044. [PubMed] [Google Scholar]
- 5.Luangtongkum S et al. Rabies in swine: natural infection in three cases. Thailand Journal of Veterinary Medicine. 1986;16:159–164. [Google Scholar]
- 6.Merriman GM. Rabies in Tennessee swine. Journal of the American Veterinary Medical Association. 1966;148:809–811. [PubMed] [Google Scholar]
- 7.Mitmoonpitak C et al. Post-exposure rabies treatment in pigs. Vaccine. 2002;20:2019–2021. doi: 10.1016/s0264-410x(02)00055-5. [DOI] [PubMed] [Google Scholar]
- 8.Morehouse LG, Kintner LD, Nelson SL. Rabies in swine. Journal of the American Veterinary Medical Association. 1968;153:57–64. [PubMed] [Google Scholar]
- 9.Yates WDG, Rehmtulla AJ, McIntosh DW. Porcine rabies in Western Canada. Canadian Veterinary Journal. 1983;24:162–163. [PMC free article] [PubMed] [Google Scholar]
- 10.Tang X et al. Pivotal role of dogs in rabies transmission, China. Emerging Infectious Diseases. 2005;11:1970–1972. doi: 10.3201/eid1112.050271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu F et al. The Human rabies epidemic in Hunan province between 1999–2003. Chinese Journal of Epidemiology. 2004;25:1088. [Google Scholar]
- 12.Liu F et al. The analysis of 1282 cases of human rabies in Hunan province between 1999–2003. Practical Preventive Medicine. 2005;12:852–854. [Google Scholar]
- 13.Zhang YZ et al. Molecular characterization of rabies virus isolates in China during 2004. Virus Research. 2006;121:179–188. doi: 10.1016/j.virusres.2006.05.010. [DOI] [PubMed] [Google Scholar]
- 14.Conzelmann KK et al. Molecular cloning and complete nucleotide sequence of the attenuated rabies virus SAD B19. Virology. 1990;175:485–499. doi: 10.1016/0042-6822(90)90433-r. [DOI] [PubMed] [Google Scholar]
- 15.Le Mercier P, Jacob Y, Tordo N. The complete Mokola virus genome sequence: structure of the RNA-dependent RNA polymerase. Journal of General Virology. 1997;78:1571–1576. doi: 10.1099/0022-1317-78-7-1571. [DOI] [PubMed] [Google Scholar]
- 16.Xiong ZL et al. Detection of dog rabies viruses in Chongqing district from December of 2004 to April of 2005. Chinese Journal of Zoonoses. 2006;22:85–88. [Google Scholar]