SUMMARY
The purpose of this study was to document the dynamics of HIV-1 subtypes in Scotland over a 6-year period. Viral RNA from all-new diagnoses was amplified by nested PCR and sequenced in the gag and/or env regions. Subtype was assigned by phylogenetic analysis, and aligned with demographic data including likely route and geographical origin of infection. We present data on 80% of all new diagnoses in Scotland between April 2000 and April 2006. Within the background of an expanding epidemic, subtype B predominates in men who have sex with men and intravenous drug users but there is a small but consistent number of UK-acquired infections in these risk groups caused by non-B subtypes. In heterosexuals, non-B subtypes acquired abroad, especially Africa, are still the largest group but again UK-acquired numbers are rising. The social and clinical significance of the spread of non-B subtypes in different ethnic and risk groups remains to be established.
INTRODUCTION
The global HIV/AIDS epidemic is dominated by members of the HIV-1 group M (Main), groups N, O and HIV-2 representing only a minority of infections. The M group is further subdivided into nine subtypes denoted by the letters A–D, F–H, J and K, subtypes E and I have been reclassified. Subtypes A and F are further subdivided into sub-subtypes A1, A2 and F1, F2, and due to their relative similarity it has proposed that subtypes D and B could represent subdivisions of the same subtype [1]. Subtypes have traditionally had a broadly geographical and risk group distribution, suggestive of a limited number of local introductions [2]. In addition to subtypes, an increasing number of inter-subtype recombinant viruses have been described. Some are unique to individuals, but others have been shown to maintain their characteristic mosaic structure following transmission, and have been termed circulating recombinant forms (CRF). So far at least 21 have been described [3, 4].
The importance of subtype is both clinical and epidemiological. Epidemiologically, the epidemic of HIV in the Scotland (and the United Kingdom) was established in men who have sex with men (MSM) and intravenous drug users (IDU) and was almost exclusively caused by subtype B [5]. In contrast, in the majority of the world the primary route of transmission has been, and continues to be, heterosexual contact, and non-B subtypes predominate [6]. Since 2000 the most common route of infection in newly diagnosed cases in Scotland has been heterosexual contact [7].
Knowledge of the subtype of infecting virus combined with route and geographical origin of infection can therefore inform health-care services of changes in behaviour and assist in the targeting of measures to reduce transmission. Clinically, knowledge of the subtype may affect patient management at a number of levels. Observational evidence suggests that non-B subtypes are more able to transmit by the heterosexual route [8]; that the rates of clinical decline especially relative to viral load may vary with subtype [9]; that different subtypes may respond differently to drug therapy [10] and that some subtypes may be inherently more able to develop resistance to therapy [11]. Since much of this is currently unproven, it is prudent to monitor the situation such that evidence can be accrued.
In April 2000, a surveillance system for HIV subtype was established [12] the purpose of which was to document the changing patterns of HIV subtype in all new diagnoses in Scotland. Here we present the data generated over the 6-year period, April 2000–April 2006.
METHODS
Design
The design is a subtype analysis of HIV infection among newly diagnosed individuals in Scotland over a 6-year period and the alignment of the resulting data with corresponding demographic and risk information.
Patients and samples
Virus from all new HIV diagnoses, confirmed by Scotland's two specialists testing laboratories, was eligible for subtype analysis using residual sera or plasma specimens submitted for HIV diagnosis and viral load testing, respectively.
Laboratory methods
RNA was extracted from patient sera or plasma and amplified in the p17/p24 region of the gag gene by nested RT–PCR and sequenced. Sequences were aligned with a homologous region from reference strains obtained from the Los Alamos Database and subtype assigned by association on phylogenetic trees [12]. If the subtype was unequivocally B or C it was reported as such. If not, further sequencing was attempted in the v3/v4 region of the env gene to provide supporting evidence [12]. The subtype was then reported using all available data. Additional sequencing was particularly necessary for subtype A viruses since this subtype contributes to several different CRF which are difficult to distinguish using the gag region alone.
Patient demographic and risk information
Since 1985 HIV testing laboratories throughout Scotland have reported cases of HIV seropositivity to Health Protection Scotland (HPS). Reporting is estimated to be 99% complete and the information held on each case includes gender, date of birth, soundex code of surname, first part of postal code of residence, date of earliest positive specimen and risk exposure; data are collected through the use of a national HIV test form. For cases whose risk status, as recorded on request forms by the attending clinician, is injecting drug use and/or homosexual/bisexual male, no active surveillance to verify this information is undertaken. For cases whose risk status is recorded as either child of infected mother or blood/blood product recipient, verification is conducted through liaison with the UK's Institute of Child Health (which coordinates UK-wide, clinician-based, HIV paediatric and obstetric surveillance systems) and the Scottish National Blood Transfusion Service, respectively. Where heterosexual exposure is the only risk indicated or where incomplete information is provided, contact is made with the patient's physician. For the former group, the enquiry seeks to elucidate the risk status of the case's sexual partner(s) (e.g. IDU), in which part of the world the likely exposure occurred and, if this was outside the United Kingdom, whether or not the case was an indigenous Scot who travelled abroad on vacation/business.
RESULTS
General trends
The number of HIV diagnoses has risen steadily over the period of this study from 134 in 2000/2001 to its current peak of 400 in 2005/2006 (Table 1). This increase has been in both subtype B and non-B infections, but the rise is much greater in the latter group, such that, of those that have been assigned a subtype, the percentage of non-B infections has increased from 34% (41/121) in 2000/2001 to approximately parity (120/247, 49%) in 2005/2006. Indeed at its height, in 2004/2005 non-B subtypes predominated (157/301, 52%).
Table 1.
Incidence of B and non-B HIV infections in Scotland by risk group, origin and year of diagnosis
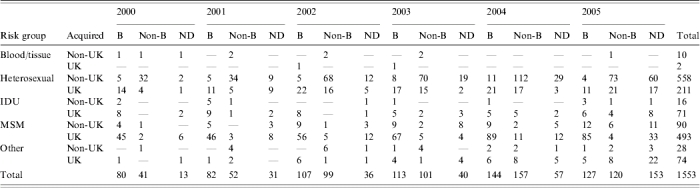
IDU, Intravenous drug users; MSM, men who have sex with men.
ND; No data (either failed to amplify or no sample was available); MSM, includes 12 cases where risk is MSM & IDU, all subtype B. Other (2 haemophiliac, 24 MTC/perinatal, 76 risk not yet established).
UK vs. non-UK acquired infections
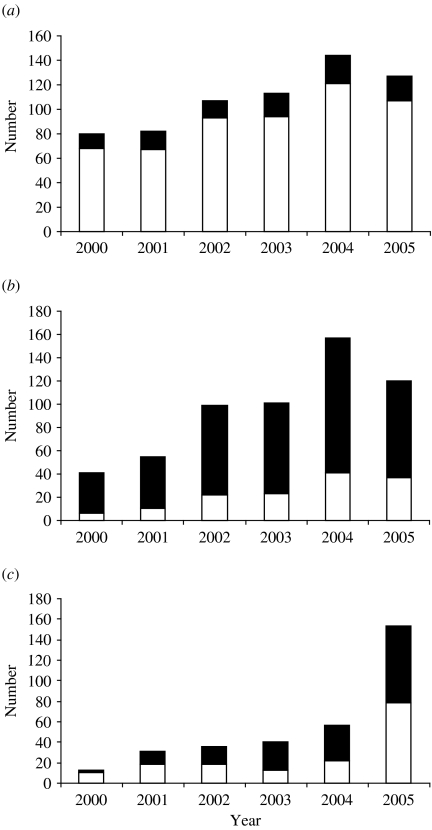
Subtype B infections were mainly acquired within the United Kingdom, the number of non-UK-acquired subtype B infections remaining fairly constant over the study period (Fig. 1a), with an annual median of 17 (range 12–23). In contrast, the majority of non-B subtype infections were acquired outside the United Kingdom (Fig. 1b). However, there was a notable rise in the number of non-B infections diagnosed over the period of the study and this was matched by a rise in those acquired within the United Kingdom. Although the percentage of non-B infections acquired in the United Kingdom was low (14·6%, 6/41) in 2000/2001, the next year it rose and thereafter remained fairly constant, with a median of 23·2% (range 22·2–30·8%).
Fig. 1.
Origin of infection of (a) subtype B, (b) subtype non-B and (c) untyped HIV infections in Scotland by year of diagnosis. ■, Non-UK; □, UK.
Specimens without subtype
During the first 5 years of this study the number of samples from which no subtype data was obtained was 177 (15·3%). Most of these were samples which failed to amplify, only 45/177 (25%) specimens were not available for testing. In the final year of data presented, a far higher proportion of specimens have not yet been assigned a subtype (153/400, 38%). The majority (112/153, 73%) is due to a failure to receive a sample, a situation which will take some time to rectify. Despite this, the proportion failing to amplify in 2005/2006 was equivalent to those in previous years (41/153, 27%).
Non-B subtypes
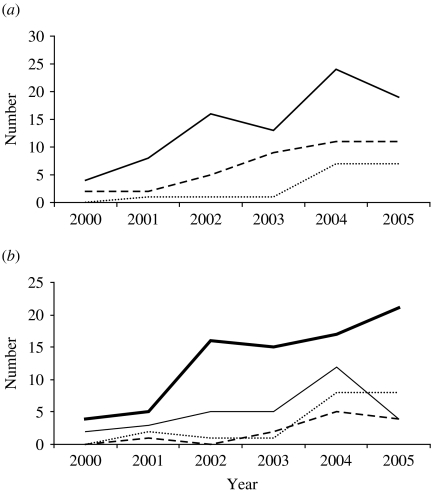
The most numerous non-B subtype was consistently subtype C (Table 2), which annually constituted approximately half of all non-B infections. The next most common non-B subtype was subtype A when combined with its various CRF. Other individual subtypes, although diverse, constituted only a minority of isolates each year. Most non-B subtype infections were acquired by heterosexual contact, both within and outside the United Kingdom (Fig. 2). The rise in the numbers of non-B infections acquired exclusively within the United Kingdom has risen steadily and in recent years, although the heterosexual contact is the most common transmission route, significant numbers were regularly found in all other risk groups.
Table 2.
Non-B subtypes of HIV in Scotland by risk group and year of diagnosis
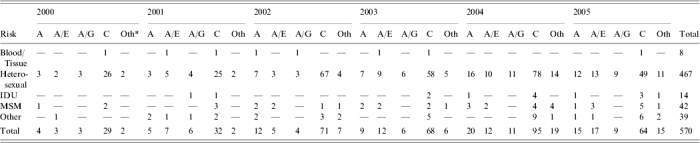
IDU, Intravenous drug users; MSM, men who have sex with men.
Other subtypes: (1) Heterosexual (19G, 11D, 2A/F, 1F, 1A/D, 1K, 1A/F, 1H, 1A/C, 1A/G/D). (2) IDU (1A/B). (3) MSM (2G, 1A/D, 1F, 1A/B, 1A/B/A, 1A/B/D). (4) Other (3G, 2A/B).
Fig. 2.
Trends in UK-acquired non-subtype B HIV infections diagnosed in Scotland, by (a) subtype and (b) risk group. (a)  , Subtype C; - - -, subtype A (includes CRF A/E and A/G); ……, other subtypes. (b)
, Subtype C; - - -, subtype A (includes CRF A/E and A/G); ……, other subtypes. (b)  , Heterosexual;
, Heterosexual;  , men who have sex with men (MSM); - - -, intravenous drug users (IDU); ……, other.
, men who have sex with men (MSM); - - -, intravenous drug users (IDU); ……, other.
Non-UK-acquired infections
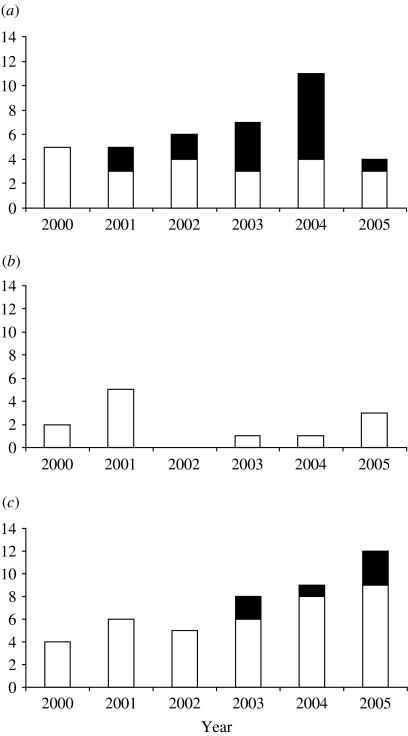
The majority of subtype B infections were acquired within the United Kingdom, but those that were not, mostly originated from the Americas or Europe. However, there were observable differences between the risk groups (Fig. 3). In 2000/2001 all non-UK- acquired subtype B infections originated in the Americas/Europe. In subsequent years increasing numbers of heterosexuals acquired their HIV from Africa/Asia, a trend which manifests itself in MSM from 2003/2004.
Fig. 3.
Origin on non-UK-acquired subtype B infections by year or diagnosis in (a) heterosexuals, (b) intravenous drug users (IDU) and (c) men who have sex with men (MSM). ■, Africa/Asia; □, Europe/Americas.
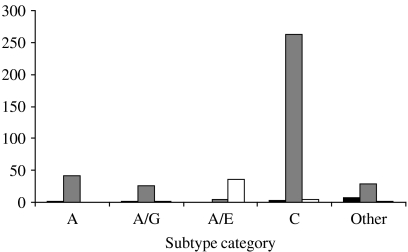
In contrast, most non-B infections acquired outside the United Kingdom originated in Africa (Fig. 4). This was true for all non-B subtypes with the exception of CRF01_AE, which dominates South-East Asia, a fact reflected by its origin in diagnoses made in Scotland.
Fig. 4.
Subtype and origin of non-B HIV subtypes acquired outside the United Kingdom. ■, Europe/Americas;  , Africa; □, Asia.
, Africa; □, Asia.
DISCUSSION
The observation that subtype B viruses are rare in the heterosexually driven epidemic in Africa, the region from which all subtypes are thought to have originated, has led to the opinion that subtype B is not well suited to this mode of transmission. Indeed, perhaps it has been out-competed in this environment by its closest relative, subtype D, which circulates in East Africa in direct competition with subtype A [13]. There are also concerns about non-B subtypes causing more aggressive disease, responding differently to anti-retroviral therapy or having lower genetic barriers to the development of drug resistance in some subtypes [14, 15]. Overall there is a fear of the unknown brought about by non-B subtypes entering a different host range via different transmission routes when experience at both an individual and population level is limited. The sensible approach therefore seems to monitor the situation until sufficient evidence accrues to make evidence-based decisions.
In this paper we have presented HIV subtype, origin of infection and risk-group data on 85% (976/1153) of all new diagnoses of HIV over a 5-year period for the whole of Scotland. The final year remains less complete although where data is available, previous trends appear to continue. Where subtype is missing, three quarters of this was due to an inability to amplify by PCR. Where data was available for such specimens, they invariably proved to have either low or undetectable viral loads. We are therefore confident that failure due to sequence variation was not a significant contributor to missing data.
In 2000 the ratio of B to non-B infections was about 2:1. Two years later this had decreased to approximately parity and has remained steady at this level. However this is against a background of a documented recent rise in the incidence of infection in MSM, a risk group dominated by subtype B viruses [7].
In 2000 63% (84/134) of new HIV infections were acquired in the United Kingdom but this declined steadily over the following 3 years to 51% by 2003/2004. The majority of infections acquired abroad have been assigned to the heterosexual route of infection, reflecting the position of this transmission route worldwide. Indeed the majority of heterosexually acquired infections originate outside the United Kingdom. Between 2000 and 2002 heterosexual transmission constituted 44% of all new infections, but between 2002 and 2005 this figure had risen to >50% (53%, 52% and 55% respectively).
The most common non-B subtype identified was subtype C. This reflects its position as the most common subtype worldwide, dominating southern Africa and the Indian subcontinent [15]. The next most numerous subtype was A and CRF containing sections of this subtype, i.e. CRF01_A/E, and CRF02_A/G. For both convenience and technical reasons, within this paper, these subtypes are often taken together. HIV subtyping is not an exact science and it is getting increasingly difficult to assign a virus with absolute confidence. The majority of subtype assignation is based on a few hundred bases of a single or perhaps two genes. Although we have employed what is regarded as the gold standard of subtyping, namely phylogenetic comparison, the sum of the sequence may hide sections of another subtype within its length [16]. In other cases a gag gene could be confidently assigned to subtype A, but sequence from the env gene was equally confidently assigned to subtype G. Thus, the virus was assigned A/G, irrespective of whether or not its mosaic pattern resembled CRF02_AG. If however, we were unable to generate sequence data from the env gene, it remained designated subtype A. Thus combining numbers of A's with A/*, is technically sensible, but from an epidemiological view it is confusing since pure subtype A is associated with East Africa, A/G with West Africa and A/E with South-East Asia [15].
We are fortunate in Scotland in knowing, in most cases, the likely country of origin of infection and this matched the sequence data in most cases. However, the converse, predicting the country of origin from limited sequence data can be misleading. In Scotland most of the non-UK-, non-B-acquired infections originated from Sub-Saharan Africa, irrespective of subtype. Subtype CRF01_AE was the only significant deviation from this rule, originating predominantly from Asia.
CRF are named not only because they are composed of different subtypes but also they have spread between a number of individuals and maintained their distinctive mosaic structure. With the generation of more sequence data it is becoming increasingly clear that there are other recombinant forms of HIV which may transmit as a stable form or which are unique to that individual [16]. Thus, for example, the A/G, non-CRF02_A/G described above, may have been generated within that individual or may have been acquired as a recombinant. The mixing of more individuals from different countries, harbouring a variety of HIV subtypes, provides the opportunity for the generation of more novel recombinant viruses. Thus, whilst viruses infecting individuals in the majority of the world will reflect the local homogenous risk group strain, a minority in cosmopolitan cities have the potential for developing unique viruses of unknown pathogenesis. Unfortunately it is the latter with which we in the United Kingdom are likely to have to come to terms with.
In this study we identified a wide range of HIV subtypes other than A, B, C and known CRF, indicating that our methodology is robust in being able to detect sequence diversity. Although each exotic subtype only constituted a few sporadic isolates on a population level, individually they may provide valuable information with regards pathogenesis in the individual and spread within a community.
We have documented the HIV subtype and origin of infection of about 80% of all new diagnoses in the whole of Scotland over a 6-year period. There is considerable viral diversity, subtype B representing only half of all isolates. Whilst this subtype still predominates in MSM and IDU there is a small but significant number of UK-acquired infections in these risk groups caused by non-B subtypes. In heterosexuals, non-B subtypes acquired abroad are still the largest group, but again UK-acquired numbers are rising. Subtyping is not always a straightforward or simple process and we would predict that the situation is likely to become more rather than less complicated in multicultural Britain. Therefore, to enable an assessment of the impact of subtype diversity on a clinical and epidemiological level continued monitoring is essential.
ACKNOWLEDGEMENTS
This work was funded by the National Services Division of the Scottish Executive.
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Kuiken C. HIV Sequence Compendium 2002. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, LA-UR no. 03-3564; 2002. [Google Scholar]
- 2.Walker PR et al. Comparative population dynamics of HIV-1 subtypes B and C: subtype-specific differences in patterns of epidemic growth. Infection, Genetics and Evolution. 2005;5:199–208. doi: 10.1016/j.meegid.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 3.Leitner T, Leitner T, Foley B, Hahn B, Marx P, McCutchan F, Mellors J, Wolinsky S, Korber B HIV Sequence Compendium 2005. Theoretical Biology and Biophysics Group, Los Alamos National Laboratory, LA-UR no. 06-0680; 2005. HIV-1 subtype and circulating recombinant form (CRF) reference sequences; pp. 41–48. , pp. [Google Scholar]
- 4.Geretti AM. HIV-1 subtypes: epidemiology and significance for HIV management. Current Opinion in Infectious Diseases. 2006;19:1–7. doi: 10.1097/01.qco.0000200293.45532.68. [DOI] [PubMed] [Google Scholar]
- 5.Hue S et al. Genetic analysis reveals the complex structure of HIV-1 transmission within defined risk groups. Proceedings of the National Academy of Sciences USA. 2005;102:4425–4429. doi: 10.1073/pnas.0407534102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.UNAIDS http://www.unaids.org. http://www.unaids.org . Joint United Nations Programme on HIV/AIDS ( ). Accessed December 2006. [PubMed]
- 7.Health Protection Agency http://www.hpa.org.uk/infections/topics_az/hiv_and_sti/hiv/epidemiology/introduction.htm. http://www.hpa.org.uk/infections/topics_az/hiv_and_sti/hiv/epidemiology/introduction.htm . New diagnosis slide set ( ). Accessed December 2006.
- 8.Mastro TD et al. Why do subtypes segregate among persons with different risk behaviours in South Africa and Thailand? AIDS. 1997;11:113–116. doi: 10.1097/00002030-199701000-00017. [DOI] [PubMed] [Google Scholar]
- 9.Senkaali D et al. The relationship between HIV-1 disease progression and v3 serotype in a rural Ugandan cohort. AIDS Research and Human Retroviruses. 2004;20:932–937. doi: 10.1089/aid.2004.20.932. [DOI] [PubMed] [Google Scholar]
- 10.Spira S et al. Impact of clade diversity on HIV-1 virulence, antiretroviral drug sensitivity and drug resistance. Journal of Antimicrobial Chemotherapy. 2003;51:229–240. doi: 10.1093/jac/dkg079. [DOI] [PubMed] [Google Scholar]
- 11.Brenner B et al. A V106M mutation in HIV-1 clade C viruses exposed to efavirenz confers cross resistance to non-nucleoside reverse transcriptase inhibitors. AIDS. 2003;17:F1–5. doi: 10.1097/00002030-200301030-00001. [DOI] [PubMed] [Google Scholar]
- 12.Yirrell DL et al. HIV-1 Subtype in Scotland: the establishment of a national surveillance system. Epidemiology and Infection. 2004;132:693–698. doi: 10.1017/s095026880400233x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yirrell DL et al. HIV-1 subtype dynamics over a 10 year period in a rural Ugandan cohort. International Journal of STD AIDS. 2004;15:103–106. doi: 10.1258/095646204322764299. [DOI] [PubMed] [Google Scholar]
- 14.Peeters M, Toure-Kane C, Nkengasong JN. Genetic diversity of HIV in Africa: impact on diagnosis, treatment, vaccine development and trials. AIDS. 2003;17:2547–2560. doi: 10.1097/01.aids.0000096895.73209.89. [DOI] [PubMed] [Google Scholar]
- 15.Wainberg MA. HIV-1 subtype distribution and the problem of drug resistance. AIDS. 2004;18:S63–S68. doi: 10.1097/00002030-200406003-00012. [DOI] [PubMed] [Google Scholar]
- 16.Yirrell DL et al. Inter and intra-genic inter-subtype HIV-1 recombination in rural and semi-urban Uganda. AIDS. 2003;16:279–286. doi: 10.1097/00002030-200201250-00018. [DOI] [PubMed] [Google Scholar]