SUMMARY
Hepatitis E virus (HEV) is a zoonotic pathogen of which several species of animals are considered to be reservoirs. Thirty-eight faecal samples, obtained from 22 species of animals including birds in a wildlife first-aid centre in Eastern China, were tested for HEV RNA. Our survey revealed that in total 28·9% (95% confidence interval 14·5–43·4) of the faecal samples from various mammals and birds were HEV RNA positive. Sequence and phylogenetic analyses of the 11 isolates demonstrated that all sequences clustered in genotype 4 with 96–100% identity to each other. In addition, serum samples from seven animal handlers have shown that five (71·4%) were seropositive. The findings imply that cross-species infection of HEV had probably occurred in this zoo-like location, and moreover, birds can be infected naturally with mammalian HEV.
INTRODUCTION
Hepatitis E virus (HEV), a member of the genus Hepevirus, is a non-enveloped virus with a positive-stranded RNA genome about 7·2 kb in length [1]. HEV is believed to be transmitted by the faecal–oral route, and outbreaks of hepatitis E are attributed to water contaminated with HEV. HEV and antibodies to HEV have been reportedly found in a wide variety of animals, especially swine. A hypothesis has arisen that zoonosis is involved in the transmission of HEV, especially for the cases in non-endemic areas. Recently, more direct evidences for zoonotic HEV transmission were reported [2, 3]. Previous serological studies suggest that individuals who are closely working with swine are at particularly high risk of HEV infection [4–8].
HEV isolates were divided into four distinct genotypes according to sequence and phylogenetic analyses. Genotype 1 was previously believed to be prevalent only in humans, but has reportedly been recently detected in a pig in Cambodia [9]. Genotype 2 has only been identified in humans in Mexico and Africa (Nigeria, Chad). Genotype 3 is prevalent in swine herds and humans throughout the world. Chinese genotype 4 HEV was first detected in humans in China in 1993 [10] and is mainly distributed in China, Japan, India, Indonesia, and Vietnam. It also has a wide host range, being prevalent in humans, swine, and other animals. Avian HEV was first discovered in chickens in 2001 and its nucleotide sequence is distinct from, mammalian HEV strain (sharing only about 60% homology) [11]. To date, no study has suggested infection of mammalian HEV in birds or avian HEV in mammals, and moreover, little is known about HEV transmission in zoo or zoo-like environments. The present study was carried out to investigate the cross-species infection of HEV, based on our retrospective field survey in a wildlife first-aid centre in Anhui Province, Eastern China.
MATERIALS AND METHODS
Sampling
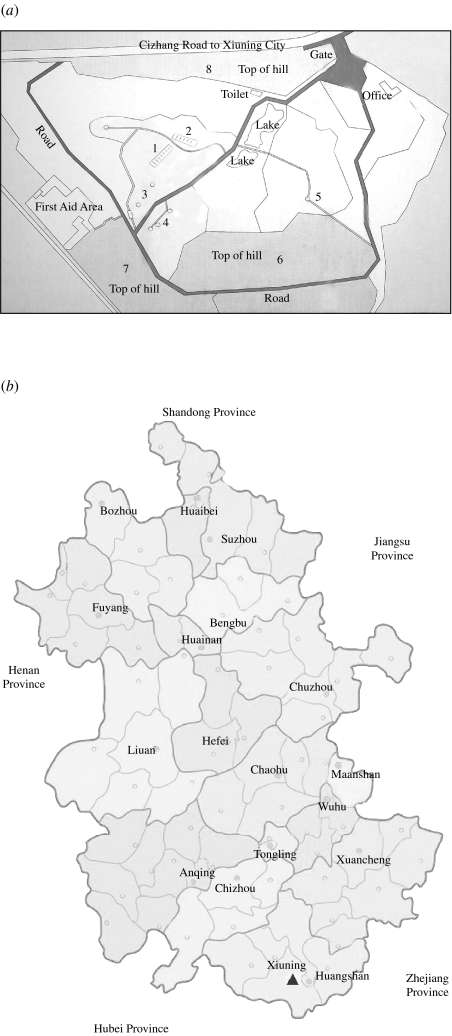
A total of 38 faecal samples of animals (including 30 mammals and eight birds) were obtained from a wildlife first-aid centre (Table), a zoo-like location lying in the mountain area of Eastern China, in October 2006. According to the veterinarian of the centre, these animals showed no evidence of recent illness. Surrounded by hills, the centre has an area of 150 000 m2. Figure 1 shows a map of this centre and the rearing sites for different animals (Fig. 1a) and the locality of the centre in Anhui Province (Fig. 1b). The cages and pens for the animals are located on the hillsides or the flat at the foot of the hills. In the centre, there are 31 tufted deer (decentralized rearing), 24 Sika deer (decentralized rearing), four Reeves' muntjac (decentralized rearing), one black muntjac (decentralized rearing), one David's deer (reared in pens), about 70 birds (reared in cages or decentralized rearing at lake) and 40 other animals (excluding deer, reared in cages or pens). Fresh samples were carefully collected to avoid any contamination. The deer were randomly selected and confined when they returned from the hill to the rearing site, until we obtained faecal samples. The animals which had been sampled were marked in order to avoid repeating the same sampling. The deer were reared in a decentralized manner (i.e. free to come and go as they wish), returning to the rearing site only occasionally. Thus, we performed random sampling of 50% of unmarked deer at each occasion, for a duration of 2 days in October 2006. During the 2 days of sampling, no more than 50% of the deer had visited the rearing site. Unfortunately, due mainly to time constraints, we were able to sample only 19 deer in total. With regard to other species, such as David's deer, red dog, porcupine, yak, water buffalo, clouded leopard, cassowary, parrot, white crane, green peafowl, crowned crane, silver pheasant, ostriches and red-crowned crane, only a single sample was obtained for each species because of limited population sizes. With respect to Asiatic black bears, grey wolves, rhesus macaques and stump-tailed macaques, we were able to obtain only a single sample from each, because of technical difficulties (e.g. ensuring the safety of animal handlers). Similarly, the birds reared at the lake were not sampled. All the samples were converted to 10% (w/v) suspensions in PBS (0·01 m, pH 7·2–7·4) immediately following the sampling. Finding the existence of HEV RNA in the animals' faecal samples, we contacted all seven workers including a veterinarian and six feeders in the centre and collected serum samples from them in December 2006. All human study subjects provided informed consent. These samples were shipped, frozen, to our laboratory and stored at −30°C prior to analysis.
Table.
Prevalence of hepatitis E virus in 38 animals in the wildlife first-aid centre
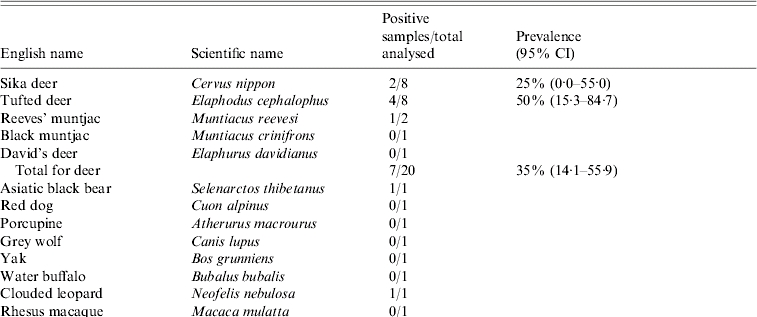
Fig. 1.
(a) Map of Wannan wildlife first-aid centre. The numbers in the map indicate the rearing sites for different animals. 1, Rearing cages or pens for Asiatic black bear, red dog, porcupine, grey wolf, clouded leopard, rhesus macaque and stump-tailed macaque. 2, Rearing cages for red-crowned crane, silver pheasant, crowned crane, white crane and cassowary. 3, Rearing pens for tufted deer, Reeves' muntjac, yak, water buffalo, and black muntjac. 4, Rearing pens for green peafowl and ostriches. 5, Rearing pens for Sika deer and David's deer. 6, 7, 8, Decentralized rearing area for deer. (b) Map of Anhui Province. The solid triangle shows the locality of Wannan wildlife first-aid centre.
Nucleic acid extraction and designing of polymerase chain reaction (PCR) primers
Faecal sample suspensions were clarified by centrifugation at 5000 g for 45 min, and 100-μl aliquots of the clarified material was used for viral RNA extraction. Total RNA was extracted from 100-μl aliquots of the clarified faecal suspension or human serum by using Trizol reagent (Invitrogen, Carlsbad, CA, USA) in accordance with the manufacturer's protocol. The viral RNA was finally dissolved in 20 μl RNase-free water. The primers used for mammalian HEV and avian HEV sequence amplification in this study were as previously described [12, 13]. For mammalian HEV, the primers were HEV1 [forward primer: 5′-AATTATGCC(T)CAGTAC(T)CGG(A)GTTG-3′] and HEV2 [reverse primer: 5′-CCCTTA(G)TCC(T)TGCTGA(C)GCATTCTC-3′] for the first round of PCR and HEV3 [forward primer: 5′-GTT(A)ATGCTT(C)TGCATA(T)CATGGCT-3′] and HEV4 [reverse primer: 5′-AGCCGACGAAATCAATTCTGTC-3′] for the second round. These two sets of primers were designed to produce a 348-nt segment of open reading frame (ORF) 2, nt 5996–6343 relative to swine HEV, and were capable of detecting all four mammalian HEV genotypes. For avian HEV, the primers were AHEV1 [forward primer: 5′- TCGCCT(C)GGTAAT(C)ACA(T)AATGC-3′] and AHEV2 [reverse primer: 5′- GCGTTC(G)CCG(C)ACAGGT(C)CGGCC-3′] for the first round of PCR and AHEV3 [forward primer: 5′-ACA(T)AATGCT(C)AGGGTCACCCG-3′] and AHEV4 [reverse primer: 5′-ATGTACTGA(G)CCA(G)CTG(C)GCCGC-3′] for the second round. These two sets of primers were designed to amplify a 242-nt segment of ORF2 relative to avian HEV. Letters within parentheses indicate degenerate bases.
Reverse transcriptase (RT)–PCR and development of a nested PCR
RT–PCR was performed by using M-MuLV RT (Fermentas, Hanover, MD, USA). Briefly, 5 μl RNA solution was analysed by RT–PCR, plus 4 μl 5×RT buffer, 2 μl of 10 mm (each) dNTP, 20 pmol reverse primer (primer HEV4 or AHEV4), 20 U RNase inhibitor, and 200 U RT in a total volume of 20 μl. After incubation for 60 min at 42°C, the mixture was incubated for 10 min at 70°C to denature the products and then chilled on ice. Five μl of the resulting cDNA was amplified by the universal RT–PCR assay using PerfectShot Taq (Loading dye Mix) DNA polymerase (Takara, Tokyo, Japan). The PCR parameters for the first-round PCR included a denaturation step at 95°C for 9 min, followed by 39 cycles of denaturation for 1 min at 94°C, annealing for 1 min at 42°C, extension for 2 min at 72°C, and a final incubation at 72°C for 7 min. The parameters for the second-round PCR were similar.
Nucleotide sequencing
The nested PCR products were analysed in a 1·5% agarose gel. The expected DNA band specific for the HEV was excised from the gel, purified with the AxyPrep DNA gel extraction kit (Axygen, Union City, CA, USA) and cloned into pMD T-Vector (Takara). Both strands of the inserted DNA amplicons were sequenced in a DNA analyser (Applied Biosystems 3730 DNA analyser; Invitrogen).
Quality control
Standard precautions were used for all procedures to reduce the possibility of sample contamination by amplified DNA molecules. Negative and positive controls were added from RT to nucleotide sequencing. All manipulations of the HEV gene were carried out in a laboratory where research on HEV had not been performed previously.
IgG and IgM anti-HEV assay
Serum samples were tested for IgG and IgM anti-HEV by using commercial ELISAs (Wan Tai Pharmaceutical Co., Beijing, China). Serum samples were diluted 1:10, and tested according to the manufacturer's instructions. A positive reaction was indicated when the signal:cut-off exceeded 1·5.
Phylogenetic analysis
The 299-nt consensus sequences were derived after primer sequences were removed from the 348-nt PCR product and were aligned using Clustal X1.8. A phylogenetic tree was constructed using the neighbour-joining method and evaluated using the interior branch test method with Mega2.0 software. Prototype HEV strains used as references in the analysis and their GenBank accession numbers are as follows: Burma 1, M80581; Burma 2, D10330; India 1, U22532; India 2, AF124407; India 3, AF459438; Mexico 1, M74506; US 1, AF060668; swUS 1, AY575857; Japan 1, AB082545; Japan 2, AB074915; and swJapan 1, AB097811, China, AJ272108; Japan wild boar, DQ079628; Japan wild deer, AB189071; Western China, XinJiang, human, NC_001434; North East China, Changchun, swine, EF077630; Changchun, human, DQ445498; East China, Shanghai, AB197674; West North China, swine, AY596311; South China, Liuzhou, human, AF103940; East China, Hefei, human, AF13491; North China, Beijing, swine, AJ428853; Beijing, human 1, AJ344190; Beijing, human 2, AJ344187; Avian, M74506.
RESULTS
Prevalence of HEV
The Table shows the prevalence of HEV RNA in animal samples. Virus detection was carried out by using RT–PCR amplification of a 348-nt segment of the ORF2 sequence and a 242-nt segment of the ORF2 sequence of avian HEV. In total, 11 out of a total of 38 samples [28·9%, 95% confidence intervals (CI) 14·5–43·4] were HEV RNA positive, using mammalian HEV primers (i.e. HEV genotypes 1–4). The RNA-positive results were seen in seven deer (35%, 95% CI 14·1–55·9) including Sika deer, Reeves' muntjac and Tufted deer which are uniquely distributed in China, two birds (25%, 95% CI 0·0–55·0) including crowned crane and silver pheasant, one Asiatic black bear, and one clouded leopard. None of the samples revealed HEV RNA-positive results for avian HEV.
With regard to seven human serum samples, HEV RNA was not detected. Nevertheless, testing anti-HEV antibodies (i.e. IgM and IgG), we found that five (71·4%, 95% CI 38·0–100·0) were positive for either IgM or IgG, suggesting that the positive subjects experienced HEV infection either recently or in the past; four were reactive for IgG, two were reactive for IgM, and one was reactive for both antibodies.
Phylogenetic analysis of HEV isolates
The PCR-amplified products of 11 isolates were sequenced. The resulting sequences were submitted to GenBank and their accession numbers are EF417580–EF417590. Sequence analyses showed that they shared 96–100% identity with each other and the results identified nine distinct nucleotide sequences. Phylogenetic analyses were conducted using the sequence alignments of these isolates and the references. Results showed that all the 11 isolates belonged to genotype 4, phylogenetically closely related to the strain (DQ079628) isolated from the Japanese wild boar (Fig. 2) and shared 90–92% identity with it. Geographically, Hefei is only about 100 km from the centre. However, the strain isolated (AF134916) from Hefei's patients was not phylogenetically closer to the 11 isolates than the others in the genotype 4 cluster and only shared 86–87% identity with them.
Fig. 2.
Phylogenetic tree constructed by alignment of the 299-nt nucleotide sequence of ORF2 from isolates in this study and 26 references of other animal and human HEV isolates, using the neighbour-joining method and evaluated using the interior branch test method with Mega2.0 software. Percent bootstrap support is indicated at each node. An avian HEV strain is included as outgroup.
DISCUSSION
Accumulated evidence indicates that HEV infection is a zoonosis which involves various animal reservoirs [2–6]. Among the four distinct genotypes, genotype 4 has been dominant in China [14]. The infection is known to be enzootic in swine, where the isolates were shown to be closely associated with those from humans [5, 15, 16]. Recently, genotype 4 HEV was reported to be widely distributed in humans and swine in Eastern China [6]. In addition to the previous study, the present study showed that 35% of deer were infected with HEV, a sufficiently high figure to attract attention. Indeed, the deer have been suggested to play an important role in HEV transmission [17, 18], which is consistent with our results. It is suspected that poor rural sanitary conditions and the decentralized rearing manner of deer in the zoo-like location may have contributed to the high prevalence. We have also shown that four other species were infected with HEV, and moreover, the sequences of the 11 isolates all belonged to genotype 4, sharing 96–100% identity. For those with HEV RNA-positive results, we successfully obtained isolates from both mammals (i.e. Asiatic black bear and clouded leopard) and birds (i.e. crowned crane and silver pheasant). This showed that cross-species infection had probably occurred in the first-aid centre.
Avian HEV was first discovered in chickens in 2001 and its genetic information is rather distinct from the isolates from mammals and probably represents a fifth genotype [11]. Avian HEV has proved to be the primary causative agent of hepatitis-splenomegaly (HS) syndrome [19], an emerging chicken disease in North America. Although avian HEV shares only 50–60% nucleotide sequence identity with mammalian HEV based on full genome sequence, it shares common antigenic epitopes on the capsid protein, similar genomic organization and conserved functional motifs in ORF1 with mammalian HEV [20, 21]. Avian HEV is enzootic in chicken flocks in the United States and has the ability to cross the species barrier and infect turkeys [11, 13, 22], however, no data showed avian or mammalian HEV transmission between mammals and birds. In the present study, the two HEV isolates from birds did not react to the primer of AHEV but did react to that of mammalian HEV, and their partial sequence of ORF2 shared about 98% identity with the other nine isolates from mammals (genotype 4) in this study. Thus, this study implies that the virus may have originated from mammals and suggests that natural infection of mammalian HEV in birds perhaps exists by means of mammal-to-avian transmission. The environment of wild birds is much larger than that of mammals and wild birds can move freely from one place to another involving great distances. To the best of our knowledge, this study is the first to suggest natural infection of mammalian HEV in birds. Further studies are needed to clarify if wild birds play a role in the spreading the mammalian HEV, because the natural infection in birds could potentially result in a serious hazard warranting public health attention.
Although our survey location was in Eastern China, an HEV enzootic region, none of the animals in the present study were previously reported as HEV positive. The enzootic state implies that the crude proportion of positive samples (i.e. 28·9%) was rather high for such an area. Considering that the virus can be detected for as long as 1 month following infection in other species [23–25], we suspect that a large epizootic of HEV had probably occurred in the zoo at the time of, or shortly before, our sampling.
It should be noted that our survey results may not have sufficiently captured the whole picture of HEV infection in the zoo, because of limited sample sizes and our non-random sampling. Considering the most likely modes of HEV transmission (i.e. faecal–oral route), different living conditions and behaviours of each animal species could have also influenced our results. Thus, it should be noted that our prevalence estimates are greatly biased, probably involving sampling errors. However, this does not influence our finding suggesting that the cross-species infection had occurred and that birds were infected with mammalian HEV.
Based on seroprevalence surveys using anti-HEV antibodies, previous studies suggest that some of the people who are closely working with swine are at high risk of HEV infection [4–8]. In the present study, 71·4% (5/7) of sera from the workers in the centre were positive for anti-HEV IgG or IgM. Even though small sample size does not permit an explicit comparison, the proportion seropositive was much higher than that of the general population in Eastern China (i.e. 17·2% [26]). The high proportion could be attributed to the frequency and length of exposure (i.e. animal handlers have taken care of animals for more than 5 years). The finding supports previous studies, implying that it is vitally important to establish a way of protecting animal handlers from exposure.
SUMMARY OF KEY FINDINGS
In conclusion, the present study suggests cross-species infection of HEV in a zoo-like location in Eastern China involving various mammalian and avian species. In particular, birds were demonstrated to be naturally infected with mammalian HEV. A majority of animal handlers were infected either recently or in the past, implying a need to protect them from the exposures.
ACKNOWLEDGEMENTS
We thank Dr Aibin Liang and Dr Congbin Yao for their help in the test for anti-HEV IgM, and Professor Richard Davis for linguistic revision of the manuscript. This work was supported by the Key Project of Shanghai Science and Technology Committee of China under grant no. 06391912.
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Reyes GR et al. Isolation of a cDNA from the virus responsible for enterically transmitted non-A, non-B hepatitis. Science. 1990;247:1335–1339. doi: 10.1126/science.2107574. [DOI] [PubMed] [Google Scholar]
- 2.Li TC et al. Hepatitis E virus transmission from wild boar meat. Emerging Infectious Diseases. 2005;11:1958–1960. doi: 10.3201/eid1112.051041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tei S et al. Zoonotic transmission of hepatitis E virus from deer to human beings. Lancet. 2003;362:371–373. doi: 10.1016/S0140-6736(03)14025-1. [DOI] [PubMed] [Google Scholar]
- 4.Meng XJ et al. Prevalence of antibodies to hepatitis E virus in veterinarians working with swine and in normal blood donors in the United States and other countries. Journal of Clinical Microbiology. 2002;40:117–122. doi: 10.1128/JCM.40.1.117-122.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drobeniuc J et al. Hepatitis E virus antibody prevalence among persons who work with swine. Journal of Infectious Diseases. 2001;184:1594–1597. doi: 10.1086/324566. [DOI] [PubMed] [Google Scholar]
- 6.Zheng Y et al. Swine as a principal reservoir of hepatitis E virus that infects humans in eastern China. Journal of Infectious Diseases. 2006;193:1643–1649. doi: 10.1086/504293. [DOI] [PubMed] [Google Scholar]
- 7.Meng XJ et al. Prevalence of antibodies to the hepatitis E virus in pigs from countries where hepatitis E is common or is rare in the human population. Journal of Medical Virology. 1999;59:297–302. [PubMed] [Google Scholar]
- 8.Withers MR et al. Antibody levels to hepatitis E virus in North Carolina swine workers, non-swine workers, swine, and murids. American Journal of Tropical Medicine and Hygiene. 2002;66:384–388. doi: 10.4269/ajtmh.2002.66.384. [DOI] [PubMed] [Google Scholar]
- 9.Caron M et al. Identification of genotype 1 hepatitis E virus in samples from swine in Cambodia. Journal of Clinical Microbiology. 2006;44:3440–3442. doi: 10.1128/JCM.00939-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang R et al. Existing variations on the gene structure of hepatitis E virus strains from some regions of China. Journal of Medical Virology. 1995;47:303–308. doi: 10.1002/jmv.1890470403. [DOI] [PubMed] [Google Scholar]
- 11.Huang FF et al. Heterogeneity and seroprevalence of a newly identified avian hepatitis E virus from chickens in the United States. Journal of Clinical Microbiology. 2002;40:4197–4202. doi: 10.1128/JCM.40.11.4197-4202.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooper K et al. Identification of genotype 3 hepatitis E virus (HEV) in serum and faecal samples from pigs in Thailand and Mexico, where genotype 1 and 2 HEV strains are prevalent in the respective human populations. Journal of Clinical Microbiology. 2005;43:1684–1688. doi: 10.1128/JCM.43.4.1684-1688.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sun ZF et al. Genetic identification of avian hepatitis E virus (HEV) from healthy chicken flocks and characterization of the capsid gene of 14 avian HEV isolates from chickens with hepatitis-splenomegaly syndrome in different geographical regions of the United States. Journal of General Virology. 2004;85:693–700. doi: 10.1099/vir.0.19582-0. [DOI] [PubMed] [Google Scholar]
- 14.Wang Y. Epidemiology, molecular biology and zoonosis of genotype IV hepatitis E in China [in Chinese] Chinese Journal of Epidemiology. 2003;24:618–622. [Google Scholar]
- 15.Lu L, Li C, Hagedorn CH. Phylogenetic analysis of global hepatitis E virus sequences: genetic diversity, subtypes and zoonosis. Reviews in Medical Virology. 2006;16:5–36. doi: 10.1002/rmv.482. [DOI] [PubMed] [Google Scholar]
- 16.Satou K, Nishiura H. Transmission dynamics of hepatitis E among swine: potential impact upon human infection. BMC Veterinary Research. 2007;3:9. doi: 10.1186/1746-6148-3-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu C et al. Using improved technology for filter paper-based blood collection to survey wild Sika deer for antibodies to hepatitis E virus. Journal of Virological Methods. 2007;142:143–150. doi: 10.1016/j.jviromet.2007.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsuura Y et al. Prevalence of antibody to hepatitis E virus among wild sika deer, Cervus nippon, in Japan. Archives of Virology. 2007;152:1375–1381. doi: 10.1007/s00705-007-0965-6. [DOI] [PubMed] [Google Scholar]
- 19.Billam P et al. Systematic pathogenesis and replication of avian hepatitis E virus in specific-pathogen-free adult chickens. Journal of Virology. 2005;79:3429–3437. doi: 10.1128/JVI.79.6.3429-3437.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haqshenas G et al. The putative capsid protein of the newly identified avian hepatitis E virus shares antigenic epitopes with that of swine and human hepatitis E viruses and chicken big liver and spleen disease virus. Journal of General Virology. 2002;83:2201–2209. doi: 10.1099/0022-1317-83-9-2201. [DOI] [PubMed] [Google Scholar]
- 21.Huang FF et al. Determination and analysis of the complete genomic sequence of avian hepatitis E virus (avian HEV) and attempts to infect rhesus monkeys with avian HEV. Journal of General Virology. 2004;85:1609–1618. doi: 10.1099/vir.0.79841-0. [DOI] [PubMed] [Google Scholar]
- 22.Sun ZF et al. Generation and infectivity titration of an infectious stock of avian hepatitis E virus (HEV) in chickens and cross-species infection of turkeys with avian HEV. Journal of Clinical Microbiology. 2004;42:2658–2662. doi: 10.1128/JCM.42.6.2658-2662.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanaka H et al. Molecular investigation of hepatitis E virus infection in domestic and miniature pigs used for medical experiments. Xenotransplantation. 2004;11:503–510. doi: 10.1111/j.1399-3089.2004.00170.x. [DOI] [PubMed] [Google Scholar]
- 24.Cooper K et al. Identification of genotype 3 hepatitis E virus (HEV) in serum and fecal samples from pigs in Thailand and Mexico, where genotype 1 and 2 HEV strains are prevalent in the respective human populations. Journal of Clinical Microbiology. 2005;43:1684–1688. doi: 10.1128/JCM.43.4.1684-1688.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Halbur PG et al. Comparative pathogenesis of infection of pigs with hepatitis E viruses recovered from a pig and a human. Journal of Clinical Microbiology. 2001;39:918–923. doi: 10.1128/JCM.39.3.918-923.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dong C et al. Identification of genetic diversity of hepatitis E virus (HEV) and determination of the seroprevalence of HEV in eastern China. Archives of Virology. 2007;152:39–746. doi: 10.1007/s00705-006-0882-0. [DOI] [PubMed] [Google Scholar]