SUMMARY
An outbreak of cryptosporidiosis associated with exposure to outdoor swimming-pool water affected an estimated 800–1000 individuals. PCR products were obtained from faecal specimens from 30 individuals who tested positive for Cryptosporidium oocysts. RFLP and sequencing analyses showed that all individuals were infected with Cryptosporidium parvum. Among the infected individuals, five had just swum in an adjacent indoor pool during the same period, and had no identified contact with individuals linked to the outdoor pool. With the use of subgenotyping based on analysis of three mini- and microsatellite loci, MS1, TP14, and GP15, we could identify two sources of exposure. One subtype was associated with the outdoor pool and another with the indoor pool. These data demonstrate that the use of mini- and microsatellite loci as markers for molecular fingerprinting of C. parvum isolates are valuable in the epidemiological investigation of outbreaks.
INTRODUCTION
Outbreaks of infectious diarrhoea associated with exposure to water from public swimming pools and recreational lakes have been frequently reported. Contamination of the water may happen through faecal accidents from swimmers, as well as free-living animals grazing in areas close to lakes. Weather conditions such as heavy rain can also affect the water quality. Certain pathogens, including Cryptosporidium spp., Giardia intestinalis, toxigenic Escherichia coli, Shigella spp. and norovirus, have been commonly involved [1–7].
Since cryptosporidia are resistant to chlorine at the levels used in water treatment, contamination of public water supplies and swimming pools constitutes a high risk for outbreaks. The low infective dose is another contributing factor that facilitates the risk for waterborne transmission [3]. Of 26 waterborne outbreaks in England and Wales reported during 1992–1995, Cryptosporidium spp. were identified as the probable cause in 14 outbreaks associated with public water supplies and swimming pools [2].
We have recently described an outbreak of cryptosporidiosis associated with an outdoor swimming pool [7]. Simultaneously with this outbreak, another five children fell ill with diarrhoea after having swum in an adjacent indoor pool used for rehabilitative therapy. All five children were positive for Cryptosporidium spp. in faecal samples. Since none of them had visited the outdoor pool, the question as to whether these individuals belonged to the same outbreak was posed. In this paper we describe the use of a microsatellite-based genotyping method to study whether there was any relationship between Cryptosporidium isolates obtained from stool samples of individuals who had swum in the two different pools.
METHODS
The outbreak
In late August 2002, an outbreak of cryptosporidiosis associated with an outdoor swimming pool occurred in a municipality close to Stockholm, Sweden. The facility comprises two pools with a total volume of 700 m3, one large (25×25 m) used for swimming and one small for mainly preschool children. During the period 20–30 August, 4585 bathers visited the facility. Epidemiological and clinical data on this outbreak have recently been described [7]. In brief, an estimated 800–1000 individuals (mainly children aged 6–12 years) were affected and the attack rate was about 50%. The median incubation period for individuals who visited the pool just once was 5 days (range 2–13 days). The frequency of secondary transmission within households was 10%.
The swimming facility was investigated and filtration systems and chlorination records for the outbreak period were examined without any adverse observations [7]. Incidences of loose faecal stool in the pool were observed and were most probably the cause of the outbreak. However, none of the samples from pool water and filters was positive for cryptosporidial oocysts or bacterial enteropathogens [7].
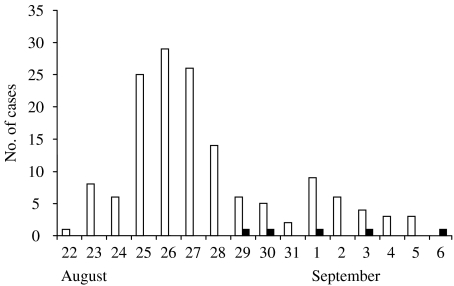
Stool specimens from 60 individuals were analysed for parasites with the use of standard techniques. Forty-two out of these 60 individuals were positive for Cryptosporidium spp., five of whom had only visited a hospital indoor pool used for rehabilitative therapy which was located about 3 km from the outdoor pool. This indoor pool was rather small (12×5 m) and was only used for outpatient rehabilitation in the daytime. In the evenings, the pool was open for a swimming school. The five children who had visited the indoor pool were aged 3–5 years, previously healthy, and had no other risk factors for cryptosporidiosis. They had no other relationships to each other than just having swum in the same pool. The dates of onset of symptoms for cases associated with the outdoor pool were from 22 August to 5 September, and for cases associated with the indoor pool from 29 August to 6 September (Fig. 1).
Fig. 1.
Date of onset of gastrointestinal symptoms in 147 primary cases associated to an outdoor swimming-pool outbreak (□), and five cases associated to an indoor swimming-pool outbreak (■).
Stool specimens from 30 individuals positive for Cryptosporidium spp. were fixed with ethanol [8] and saved for molecular analysis. The remaining 12 samples were not available for further analysis. Two of the 30 saved specimens were from the individuals who had visited the indoor pool. The study was approved by the Ethics Committee of the Karolinska Institute (Solna, Sweden).
DNA purification
Parasite oocysts were disrupted with a bead beater, and total DNA was isolated from the sporozoites with the QIAmp DNA mini-kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol.
PCR–RFLP analysis and DNA sequencing
The COWP gene was amplified by PCR with the primers cry-9 (5′-GGA CTG AAA TAC AGG CAT TAT CTT G-3′) and cry-15 (5′-GTA GAT AAT GGA AGA GAT TGT G-3′) as previously described [9]. For the restriction fragment analysis, PCR products were digested with RsaI (New England Biolabs, Beverly, MA, USA) and then analysed on agarose gels.
For species verification, the18S ribosomal RNA gene was amplified as previously described [10] using the primers Cp18S-108 (5′-GTT ATA GTT TAC TTG ATA ATC TT-3′) and Cp18S-1031 (5′-TGA AGG AGT AAG GAA CAA CC-3′). The resulting amplicons were sequenced with the ABI PRISM Big Dye Terminator v3.0 (Applied Biosystems, Foster City, CA, USA). The reaction products were purified and concentrated through ethanol and sodium acetate precipitation, and the resulting pellets were resuspended in formamide before product separation was performed on an ABI PRISM 3100 Genetic Analyzer (Applied Biosystems). The DNA sequences were processed using the Vector NTI program suite 9 (Informax Inc., Oxford, UK). Edited sequences were compared to public databases (GenBank non-redundant nucleotide databases) using the BLAST family of algorithms with default parameters.
Mini- and microsatellite analysis
To further elucidate the genetic status of the strains involved in the outbreak, we tested the samples with three different micro- and minisatellite markers: MS1, TP14, and GP15. We used basically the same methods as previously described by Mallon et al. [11, 12]. The sequences of the primers used for amplification were: MS1B: 5′-AAT TAG TCG ACC TCT CAA CAG TTG G-3′, and MS1D: 5′-GGA ACA CCA TCC AAG AAC CAA AGG T-3′; TP14C: 5′-CTA ACG TTC ACA GCC AAC AGT ACC-3′, and TP14D: 5′-GTA CAG CTC CTG TTC CTG TTG-3′; GP15A: 5′-GCC GTT CCA CTC AGA GGA AC-3′, and GP15E: 5′-CAC ATT ACA AAT GAA GTG CCG C-3′. The MS1 is based on a repeat region of the hsp70 gene [13], and GP15 is based on a gene coding for a glycoprotein [14]. The numbers and sizes of the alleles amplified in each sample were determined by using one primer of each pair labelled with 6-FAM, and followed by analysis on an ABI PRISM 3100 Genetic Analyzer with the aid of the GeneScan Analysis Software (Applied Biosystems).
All molecular tests were performed on coded samples, which implies that the investigator had no knowledge of the epidemiological data of the individual cases.
RESULTS
Species identification
Based on the size of the restriction fragments, all tested isolates could be characterized as Cryptosporidium parvum. Amplification of a part of the 18S rRNA gene, followed by sequence analysis of five randomly selected samples, including one connected with the indoor pool, showed that all sequenced samples had sequences identical to the C. parvum 18S rRNA gene.
Molecular subtyping
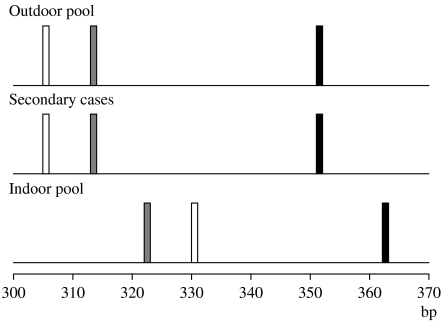
Analysis of MS1, corresponding to a repeat region of the hsp70 gene [13], was successful for all samples. In the MS1 analysis, 28 of the samples had a 351 bp allele whereas two samples had a 362 bp allele. Further analysis with the microsatellite GP15, corresponding to a region of a highly polymorphic glycoprotein antigen described by Strong et al. [14], showed that the latter two samples had a 330 bp allele whereas seven of the samples had a 305 bp allele. For the remaining 21 samples we could not get any positive results, due low sensitivity of the PCR. Attempts to use other types of buffer systems in the PCR, as suggested by Mallon et al. [11, 12], were unsuccessful. The final analysis used the TP14 marker, and once again the two divergent samples had a unique allele (322 bp) compared to the remaining 22 samples (313 bp) that gave positive results.
Decoding of the samples showed that the two unique samples originated from the individuals who had just visited the indoor pool (Fig. 2). The rest of the samples originated from patients who were linked to the outdoor swimming pool, either directly or as secondary cases (Fig. 2).
Fig. 2.
Multilocus genotyping profiles for samples from patients that had used the outdoor swimming pool, secondary cases linked to the outdoor swimming pool, and patients who had used the indoor swimming pool. Three loci were analysed, TP14 ( ), GP15 (□) and MS1 (■). The relative sizes of the different genotypes are shown.
), GP15 (□) and MS1 (■). The relative sizes of the different genotypes are shown.
DISCUSSION
Many molecular methods have been used for genotyping or fingerprinting strains of different microorganisms for epidemiological purposes. Such methods are of great value in contact tracing as well as outbreak investigations, as a complement to descriptive and analytical epidemiology. The use of molecular techniques is, however, highly dependent on sufficient discriminatory power, typability and reproducibility. A relationship between isolates from samples of two individuals does not, therefore, necessarily mean a connection if there is no epidemiological relationship. However, if two isolates are of different genotypes, it is unlikely that they are related to the same source.
The use of molecular tools to study Cryptosporidium spp. has not only reshaped the taxonomy considerably, but it has also increased our understanding about the epidemiology of the disease [15, 16]. Currently, at least seven species (C. hominis, C. parvum, C. meleagridis, C. felis, C. canis, C. suis and C. muris) are associated with human disease. In addition, two Cryptosporidium spp. genotypes, monkey and cervine, have been found in human samples. C. hominis and C. parvum dominate the number of reported cases of human cryptosporidiosis [16]. Interestingly, increasing numbers of cases of cryptosporidiosis are attributed to C. meleagridis, a species originally described in turkeys.
In the present study, we performed the first genotyping of a Cryptosporidium pool-associated outbreak in Sweden. All isolates where typing could be done contained C. parvum, suggesting that animal contamination of the water was possible, although a human source was more likely. This is in keeping with previous observations, where it has been shown that many C. parvum infections originate from humans [15]. However, population genetics studies using mini- and microsatellite markers also suggest that there might be a strong bias for substructuring between various host species [11, 12]. This implies that there might be human specific multi-locus genotypes (MLG) of C. parvum and other MLG that are found in more than one species.
Molecular typing is frequently used for species identification of Cryptosporidium in the Nordic countries as illustrated by two recent outbreaks: one in Norway and one in Denmark [17, 18]. In the Norwegian outbreak C. parvum was identified as the cause of disease, both in calves as well as humans who had been in contact with the animals [17]. In the Danish outbreak a large number of people were infected by C. hominis, as identified by PCR [18]. The source of infection was probably raw carrots, kept in water, which were served in a company canteen [18]. No further subgenotyping was performed on the samples from any of these two outbreaks. On the other hand, depending on the species, additional genotyping might be more or less complex [19]. Hunter et al. suggest that even three loci microsatellite analyses are not discriminatory enough for C. hominis. In contrast, they conclude that the use of three loci have a very high discriminatory power for C. parvum, due to higher strain diversity [19]. This latter observation is also in keeping with our results.
A troublesome observation in our outbreak report was the fact that five individuals who had not swum in the outdoor pool, but in an adjacent indoor pool, got cryptosporidiosis during the same period. These five people had not, as far as is known, had any contact with individuals involved in the outbreak. Therefore, whether there was any connection between these two episodes became a matter of discussion. With the use of subgenotyping based on microsatellite methods, a possible association between the two outbreaks could, however, be ruled out. Samples originating from the patients that had been to the indoor pool corresponded to MLG4, previously described by Mallon et al. [11]. This MLG belongs to subgroup B, which encompasses not only human isolates but also bovine and ovine isolates [12]. However, to our knowledge MLG4 isolates have not been recovered from any other source than humans up to now. Given the circumstances under which the individuals seem to have been infected, it is unlikely that the contamination of the therapy pool was of animal origin. The samples from visitors to the large public outdoor swimming pool were not of any MLG previously described [11, 12].
Epidemiological investigation and sampling from infected individuals strongly suggests that the swimming pools were indeed the sources of these outbreaks, although no oocysts were found in water samples from the pools [7]. Such outbreaks have been frequently reported and unlike many other microorganisms Cryptosporidium oocysts are highly resistant to chlorine which combined with a low infectious dose make them ideal for waterborne transmission [2–7]. The reason why two separate swimming-pool outbreaks occurred so close to each other both in space and time is difficult to explain. The late summer was very hot in Sweden for this year and the pools were crowded with people, which might have increased the risk of faecal contamination. The maintenance of the pools seemed adequate regarding filtration systems and chlorination and microbiological examination of the water showed no contamination [7]. However, incidences of loose stool were observed at several occasions at least in the outdoor swimming pool and the recommendations for the management of faecal accidents [20] had not been followed. Both the outdoor and indoor pools were closed immediately when the outbreaks were detected. The free chlorine concentration was raised to 3 mg/l (pH 7·7–7·4) and the filters were back-flushed once a day for 7 days. Before the pools were reopened, the free chlorine concentration was raised to 15 mg/l for 16 h [7]. The importance of following guidelines for management of faecal accidents was strictly emphasized and information of correct hygiene precautions for individuals visiting public swimming pools was instituted. The fact that the indoor pool was used for outpatient rehabilitation during daytime and children's swimming school in the evenings may raise concern and further underscores the importance of hygienic precautions, e.g. never swimming until full recovery from a diarrhoeal illness, implementation of adequate toilet, hand-washing and nappy-changing routines, and warnings against swallowing pool water.
One limitation in our study is that faecal Cryptosporidium isolates from only two of the five children who had swum in the indoor pool were available for molecular subtyping. However, the fact that both isolates were of the same unique subtype, differing from the subtype associated with the outdoor swimming pool, strongly suggests that there were in fact two sources of exposure.
In conclusion, our report clearly shows that the use of mini- and microsatellites as markers for molecular fingerprinting of C. parvum isolates can be of great value in epidemiological investigations of outbreaks due to Cryptosporidium, and can be used to separate outbreaks that otherwise would be difficult, if not impossible, to distinguish.
ACKNOWLEDGEMENTS
We thank Katarina Näslund and Lillemor Carlsson for valuable technical assistance. This work is part of Med-Vet-Net (FOOD-CT-2004-506122), a Network of Excellence for the Integrated Research on the Prevention and Control of Zoonoses, funded within the 6th European Union Framework Programme.
DECLARATION OF INTEREST
None.
REFERENCES
- 1.Yoder JS et al. Surveillance for waterborne-disease outbreaks associated with recreational water – United States, 2001–2002. Morbidity and Mortality Weekly Report. Surveillance Summaries. 2004;50:1–22. [PubMed] [Google Scholar]
- 2.Furtado C et al. Outbreaks of waterborne infectious intestinal disease in England and Wales, 1992–1995. Epidemiology and Infection. 1998;121:109–119. doi: 10.1017/s0950268898001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen X-M et al. Cryptosporidiosis. New England Journal of Medicine. 2002;346:1723–1731. doi: 10.1056/NEJMra013170. [DOI] [PubMed] [Google Scholar]
- 4.Puech MC et al. A statewide outbreak of cryptosporidiosis in New South Wales associated with swimming at public pools. Epidemiology and Infection. 2001;126:389–396. doi: 10.1017/s0950268801005544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lemmon JM, McAnulty JM, Bawden-Smith J. Outbreak of cryptosporidiosis linked to an indoor swimming pool. Medical Journal of Australia. 1996;165:613–616. doi: 10.5694/j.1326-5377.1996.tb138664.x. [DOI] [PubMed] [Google Scholar]
- 6.Kramer MH et al. First reported outbreak in the United States of Cryptosporidiosis associated with a recreational lake. Clinical Infectious Diseases. 1998;26:27–33. doi: 10.1086/516271. [DOI] [PubMed] [Google Scholar]
- 7.Insulander M et al. An outbreak of cryptosporidiosis associated with exposure to swimming pool water. Scandinavian Journal of Infectious Diseases. 2005;37:354–360. doi: 10.1080/00365540410021072. [DOI] [PubMed] [Google Scholar]
- 8.Jongwutiwes S et al. Simple method for long-term copro-preservation of Cryptosporidium oocysts for morphometric and molecular analysis. Tropical Medicine & International Health. 2002;7:257–264. doi: 10.1046/j.1365-3156.2002.00854.x. [DOI] [PubMed] [Google Scholar]
- 9.Spano F et al. PCR-RFLP analysis of the Cryptosporidium oocyst wall protein (COWP) gene discriminates between C. wrairi and C. parvum, and between C. parvum isolates of human and animal origin. FEMS Microbiology Letters. 1997;150:209–217. doi: 10.1016/s0378-1097(97)00115-8. [DOI] [PubMed] [Google Scholar]
- 10.Enemark HL et al. Molecular characterization of Danish Cryptosporidium parvum isolates. Parasitology. 2002;125:331–341. doi: 10.1017/s0031182002002226. [DOI] [PubMed] [Google Scholar]
- 11.Mallon M et al. Population structures and the role of genetic exchange in the zoonotic pathogen Cryptosporidium parvum. Journal of Molecular Evolution. 2003;56:407–417. doi: 10.1007/s00239-002-2412-3. [DOI] [PubMed] [Google Scholar]
- 12.Mallon ME et al. Multilocus genotyping of Cryptosporidium parvum Type 2: population genetics and sub-structuring. Infection, Genetics and Evolution. 2003;3:207–218. doi: 10.1016/s1567-1348(03)00089-3. [DOI] [PubMed] [Google Scholar]
- 13.Khramtsov NV et al. Cloning and analysis of a Cryptosporidium parvum gene encoding a protein with homology to cytoplasmic form Hsp70. Journal of Eukaryotic Microbiology. 1995;42:416–422. doi: 10.1111/j.1550-7408.1995.tb01605.x. [DOI] [PubMed] [Google Scholar]
- 14.Strong WB, Gut J, Nelson RG. Cloning and sequence analysis of a highly polymorphic Cryptosporidium parvum gene encoding a 60-kilodalton glycoprotein and characterization of its 15- and 45-kilodalton zoite surface antigen products. Infection and Immunity. 2000;68:4117–4134. doi: 10.1128/iai.68.7.4117-4134.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiao L et al. Cryptosporidium taxonomy: recent advances and implications for public health. Clinical Microbiology Reviews. 2004;17:72–97. doi: 10.1128/CMR.17.1.72-97.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caccio SM et al. Unravelling Cryptosporidium and Giardia epidemiology. Trends in Parasitology. 2005;21:430–437. doi: 10.1016/j.pt.2005.06.013. [DOI] [PubMed] [Google Scholar]
- 17.Robertson L et al. A small outbreak of human cryptosporidiosis associated with calves at a dairy farm in Norway. Scandinavian Journal of Infectious Diseases. 2006;38:810–813. doi: 10.1080/00365540600606457. [DOI] [PubMed] [Google Scholar]
- 18.Ethelberg S et al. Cryptosporidiosis outbreak associated with eating in a canteen, Denmark, August 2005. Eurosurveillance. 2005;10:E051027.4. doi: 10.2807/esw.10.43.02822-en. [DOI] [PubMed] [Google Scholar]
- 19.Hunter PR et al. Subtypes of Cryptosporidium parvum in humans and disease risk. Emerging Infectious Diseases. 2007;16:82–88. doi: 10.3201/eid1301.060481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anon. Responding to faecal accidents in disinfected swimming venues. Morbidity and Mortality Weekly Report. 2001;50:416–417. [PubMed] [Google Scholar]