Abstract
Cadmium is an environmental contaminant that enters the body through diet or cigarette smoke. It affects multiple cellular processes, including cell proliferation, differentiation, and apoptosis. Recently, cadmium has been shown to function as an endocrine disruptor, to stimulate estrogen receptor α (ERα) activity and promote uterine and mammary gland growth in mice. Although cadmium exposure has been associated with the development of breast cancer, the mechanism of action of cadmium remains unclear. To address this deficit, we examined the effects of cadmium treatment on breast cancer cells. We found that ERα is required for both cadmium-induced cell growth and modulation of gene expression. We also determined that ERα translocates to the nucleus in response to cadmium exposure. Additionally, we provide evidence that cadmium potentiates the interaction between ERα and c-Jun and enhances recruitment of this transcription factor complex to the proximal promoters of cyclin D1 and c-myc, thus increasing their expression. This study provides a mechanistic link between cadmium exposure and ERα and demonstrates that cadmium plays an important role in the promotion of breast cancer.
Cadmium promotes breast cancer cell growth and gene expression by potentiating the interaction between ERalpha and c-Jun.
Breast cancer is one of the most common malignancies that occur in women in the United States, accounting for one in four female cancers. Breast cancer develops as a result of abnormal proliferation of cells in the mammary gland. Normal growth of mammary epithelial cells is modulated by circulating levels of estrogen, a female hormone produced by the ovaries. The activity of estrogen is mediated by the estrogen receptors (ERs), ERα and ERβ. These two isoforms of ER display considerable homology in their DNA binding domains and COOH-terminal domains but are highly variable in their NH2-terminal trans-activation function-1 domains (3,4). Both ERα and ERβ bind to the active form of estrogen, 17β-estradiol (E2), with similar affinities. They function as ligand-gated transcription factors that control the expression of genes necessary for growth, development, and homeostasis. However, the two forms of ER play very different roles in breast cancer. ERα has long been known as a crucial regulator of growth and development of the mammary gland and also as a key prognostic marker for breast cancer development. Although ERβ has been reported to be present in breast cancer cells, less is known about its role in breast cancer development.
Breast cancer can be classified as either ER positive or ER negative, depending on the presence or absence of ERα. Development of ER-positive breast cancer is controlled by the activity of the ERs and the circulating levels of 17β-E2. The role of ERα in hormone-refractory breast tumorigenesis and the underlying mechanism of this role are unclear. Endocrine disruptors, including heavy metals such as cadmium (Cd), may be involved in ERα-mediated modulation of hormone-refractory breast tumorigenesis (1,2,5,6,7). Although Cd exposure has been associated with the development of breast cancer, the nature of this relationship remains unclear (1,2,5,6,7).
Cd is derived from various industrial sources, including present and former mining activities and the production of alloys, batteries, fertilizers, pigments, and combustion byproducts. Human exposure results from consumption of contaminated water and food or inhalation of cigarette smoke and metal smoldering fumes. Cd is part of the family of heavy metals that exists as a natural component of the Earth’s crust. Thus, it is present in varying concentrations in all ecosystems. In general, heavy metals cannot be degraded and often accumulate in biota, water, and soil. Because heavy metals form complexes with organic compounds, high levels of heavy metals have been shown to be dangerous to all living organisms. Heavy metals often alter the activity of biological molecules and hinder their ability to function properly by binding to nitrogen-, oxygen-, or sulfur-containing groups, ultimately interfering with cellular processes. Common toxicities affecting higher eukaryotic cells include impaired mitochondrial function, enhanced levels of free radicals, and DNA damage, all of which can result in death or malignant transformation. Exposure to elevated levels of Cd and other heavy metals has been associated with an increased incidence of breast cancer development (6,8). In particular, Cd, nickel, mercury, and selenium have been shown to have estrogenic effects and are collectively referred to as metalloestrogens.
Recent studies have suggested that metalloestrogens may function as endocrine disruptors, perturbing the normal hormonal cycle and altering the development of the mammary gland, resulting in neoplastic growth (1,2). Cd is the best characterized of the metalloestrogens. Specifically, animal studies have shown that Cd exposure can increase uterine weight and density of the mammary gland, as well as induce changes in the lining of the uterus, which are all early signs of tumorigenesis (2). Cd can mimic the action of estrogen in breast cancer cells by binding to the ER and mediating receptor activation in vitro (1,6,7). Although Cd is known to bind to ERα, its role in ERα-mediated gene expression and breast cancer cell growth remains unclear.
This study aims to further investigate the mechanism of Cd-induced breast cancer cell proliferation and gene expression and to determine the role of ERα in this process. We provide several lines of evidence that suggest that the effects of Cd on breast cancer cell growth and gene expression are dependent on ERα. Our results show that Cd promotes the nuclear localization of ERα and enhances the binding of ERα to the promoters of cyclin D1 (CycD1) and c-myc. We also demonstrate that Cd potentiates the interaction between ERα and c-Jun, a member of the activating protein (AP)-1 family of transcription factors. Taken together, our results suggest that Cd promotes the ERα/c-Jun interaction and increases recruitment of the ERα/c-Jun complex to promoter regions, thereby directly regulating the expression of Cd-inducible genes.
Results
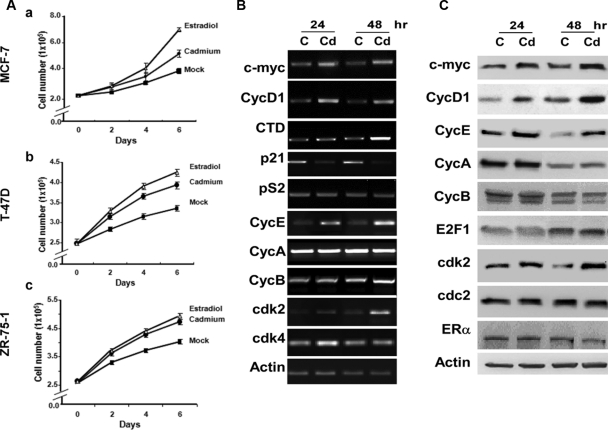
To determine whether Cd has an effect on ER-positive breast cancer cell growth, three ER-positive breast cancer cell lines, MCF-7, T-47D, and ZR-75-1, were plated in six-well plates in hormone-deprived media and treated with 10−6m cadmium chloride (CdCl2), 10−8m 17β-E2, or were mock treated with PBS (Fig. 1A). For each cell type, the optimal Cd concentration was determined using a titration assay in which cells were plated in a 96-well plate and treated with varying concentrations of CdCl2, ranging from 10−12m to 10−5m (data not shown). The results shown in Fig. 1A indicate that Cd enhanced the growth rate of all three ER-positive breast cancer cell lines (MCF-7, T-47D, and ZR-75-1), relative to the mock control. As expected, cells treated with E2 displayed a significant increase in cell growth, compared with mock-treated cells. These results were further confirmed by indirect cell proliferation assay using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) (data not shown). Although Cd affects cell proliferation to a lesser degree than E2, our results do indicate that Cd stimulates ER-positive breast cancer cell growth.
Figure 1.
Cd-induced ER+ breast cancer cell proliferation and gene expression. Panel A, MCF-7 (a), T-47D (b), and ZR-75-1 (c) cells were plated in six-well plates and treated with 10−6m CdCl2 (•), 10−8m 17β-E2 (▵), or mock treated with PBS (▪). Cell number was monitored every 2 d by cell counting using a hemacytometer. P < 0.05 between Cd and mock-treated cells. Panel B, MCF-7 cells were hormone deprived and treated with 10−6m CdCl2 (Cd) or mock treated [control (C)] for 24 and 48 h. Total RNA was isolated, and gene expression was monitored by semiquantitative RT-PCR. Panel C, Total cell lysates were analyzed for protein expression by Western blot analysis, and data are representative of several independent experiments.
To dissect the molecular mechanism of Cd’s action on breast cancer cell growth, we examined whether Cd was able to induce ER target gene expression. MCF-7 cells were hormone deprived and treated with 10−6m CdCl2. Total RNA was isolated, and the expression levels of several ER target genes were analyzed by semiquantitative RT-PCR (Fig. 1B). Cd-treated MCF-7 cells expressed higher levels of CycD1 and c-myc as early as 24 h after exposure, whereas cathepsin D (CTD) displayed a more delayed response (48 h). These three genes have been shown to be important in the development and progression of breast cancer (11,12,13,14,15,16,17). Cd exposure had no detectable effect on the expression level of pS2, an estrogen-responsive gene that is commonly used as a prognostic marker of breast cancer (18,19). We also examined the expression of p21, a cell cycle inhibitor that has been shown to be regulated by ERα (12,20) and found that expression of p21 was decreased by Cd exposure. Consistent with its inhibitory role in the cell cycle, reduction of p21 levels may serve as an alternative mechanism by which Cd promotes cell proliferation. Alternatively, reduced p21 levels may work in conjunction with elevated levels of CycD1 and c-myc to promote cell growth.
The observation that Cd stimulated breast cancer cell proliferation led us to expand our analysis to other genes directly involved in cell cycle regulation. To this end, we included cyclins and cyclin-dependent kinases (CDKs) in our analysis. Our results show that expression of cyclin E1 (CycE), cdk2, and cdk4 increased after Cd treatment, whereas the other cyclins and CDKs showed no significant changes in their expression levels after treatment (Fig. 1B).
Western blot analysis was used to determine whether the protein levels of these genes were affected by Cd treatment (Fig. 1C). Cells treated with 10−6m CdCl2 also displayed higher CycD1 and c-myc protein levels at 24 h after treatment. However, no significant change in the ERα protein level was observed. Cd also induced the expression of CycE and cdk2 proteins. These results establish that Cd promotes MCF-7 cell proliferation through induction of ER target genes (i.e. CycD1 and c-myc) and non-ER target genes (i.e. CycE and cdk2), and that these increases are not dependent on elevated levels of ERα (Fig. 1C).
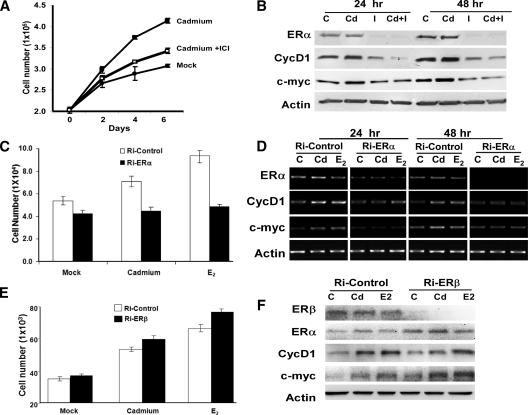
These results led us to question whether the Cd-induced effect requires ERα. To establish a link between Cd-induced cell proliferation and ERα, we treated MCF-7 cells with ICI 182,780 (ICI) to decrease endogenous levels of ERα. ICI is a pure antiestrogen that binds to the ER and promotes receptor degradation. MCF-7 cells were treated with either 10−6m CdCl2 or 10−6m CdCl2 in the presence of 10−8m ICI, or were mock treated with PBS. Cells were harvested and submitted to direct cell counting and Western blot analysis to monitor cell proliferation and ERα levels, respectively (Fig. 2, A and B). Cells treated with ICI (I or Cd+I) expressed significantly lower levels of ERα after 24 h of treatment (Fig. 2B). Reduced levels of ERα corresponded with decreased Cd-induced cell proliferation, suggesting that the effect of Cd treatment is, in part, dependent on the presence of ERα (Fig. 2, A and B). Consistent with the growth results, ICI diminished the effect of Cd on CycD1 and c-myc expression at both the protein and mRNA levels (Fig. 2B and Supplemental Fig. 1A published on The Endocrine Society’s Journals Online web site at http://endo.endojournals.org).
Figure 2.
Cd-induced cell proliferation and gene expression depends on ERα. Panel A, MCF-7 cells were hormone deprived and treated with 10−6m CdCl2 (▪), 10−6m CdCl2 in the presence of 10−8m ICI (□), or mock treated with PBS (•). Cell number was monitored every 2 d by cell counting using a hemacytometer. P < 0.05 between Cd and Cd + ICI. Panel B, MCF-7 cells were hormone deprived and treated with 10−6m CdCl2 (Cd), 10−8m ICI (I), 10−6m CdCl2 plus 10−8m ICI (Cd+I), or mock treated [control (C)] for 24 or 48 h. Protein expression was analyzed using Western blot analysis. Panel C, MCF-7 cells were plated in 24 wells and transfected with either Ri-ERα (black) or Ri-Control (white). Cells were treated with 10−6m CdCl2 or 10−7m 17β-E2 or mock treated with PBS (C), and cell growth was analyzed 4 d after treatment; each data point represents four replicate samples. D, siRNA-transfected cells were also collected 24 and 48 h after treatment for gene expression analysis using semiquantitative RT-PCR. E, MCF-7 cells were plated and transfected with either Ri-ERβ (black) or Ri-Control (white). Cells were treated with 10−6m CdCl2 or 10−7m 17β-E2 or mock treated with PBS (C). Cell growth was analyzed 4 d after treatment, and each data point represents four replicate samples. F, siRNA-transfected cells were also collected 48 h after treatment for protein analysis using Western blot analysis.
To determine whether Cd-induced breast cancer cell proliferation is dependent on ERα, we silenced ERα expression using siRNA, and cell growth was analyzed 2, 3, and 4 d after treatment (Fig. 2C and data not shown). Cells transfected with siRNA directed against ERα (Ri-ERα) did not grow in response to the Cd or estrogen (E2), whereas cells transfected with a scrambled siRNA control (Ri-control) continued to be stimulated by Cd and estrogen treament (Fig. 2C). Consistent with the growth results, Ri-ERα also diminished the effects of Cd-induced CycD1 and c-myc expression at the mRNA level (Fig. 2D).
Next, we examined the role of ERβ in Cd-induced cell growth using small interfering RNA (siRNA) to deplete endogenous ERβ expression. Cell growth was monitored 2, 3, and 4 d after treatment (Fig. 2E and data not shown). Interestingly, cells transfected with Ri-ERβ displayed an increase in cell proliferation under all three conditions (mock, Cd, and estrogen treated). Furthermore, our results also suggest that ERβ depletion has no effect on Cd or estrogen response (Fig. 2E). Western blot analysis was used to monitor the ERβ expression. Cells transfected with Ri-ERβ expressed significantly lower levels of ERβ 48 h after treatment; however, depletion of ERβ did not have any significant effects on the expression of CycD1 and c-myc at the protein level (Fig. 2F).
To further assess the role of ERα in Cd-induced cell proliferation, we analyzed the effects of Cd on the growth of MDA-MB-453 breast cancer cells. MDA-MB-453 cells are ERα negative but ERβ positive. The cells were treated with varying concentrations of CdCl2 ranging from 10−12m to 10−5m, or were mock treated with PBS (Supplemental Fig. 1B). Cell proliferation was measured with the MTT assay 4 and 6 d after treatment. As expected, Cd exposure did not promote growth of MDA-MB-453 cells. Taken together, our results demonstrate that Cd-induced breast cancer cell growth is dependent on ERα, and not ERβ.
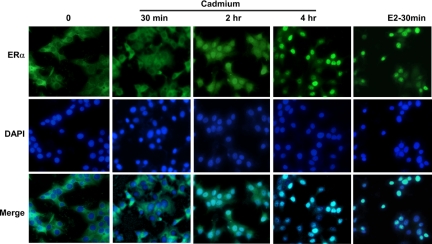
Given that Cd-induced cell proliferation and gene expression may be dependent on ERα, we postulated that Cd may stimulate ERα nuclear localization. To determine whether Cd promotes the nuclear translocation of ERα, MCF-7 cells were hormone deprived for 2 d and treated with 10−6m CdCl2 (Cd) or 10−8m E2, or were mock treated for 0.5, 2, and 4 h (Fig. 3). Cells were harvested by fixation in formaldehyde, and ERα was examined by immunofluorescence and counter-stained with 4′,6-diamidino-2-phenylindole (DAPI) to identify nuclei. ERα proteins are normally localized in both the cytoplasm and nucleus; however, when cells are hormone deprived, a majority of the ERα proteins are found in the cytoplasm (Fig. 3). As shown in Fig. 3, Cd promotes the nuclear translocation of ERα, starting at 1 h and peaking at 4 h after treatment (Fig. 3). As expected, E2 treatment also stimulates the translocation of ERα to the nucleus. However, the translocation effect was much quicker under estrogen condition, with maximum translocation observed after 30 min of treatment (Fig. 3). These data suggest that Cd acts similarly to estrogen in promoting ERα nuclear localization. Once inside the nucleus, the Cd-ERα complex may regulate the expression of Cd-induced genes (i.e. CycD1 and c-myc).
Figure 3.
Cd promotes translocation of ERα to the nucleus. MCF-7 cells were plated in six-well plates containing untreated glass cover-slips under hormone-deprived conditions. Twenty hours later, cells were treated with 10−6m CdCl2 or 10−8m 17β-E2. Cells were harvested by fixation in 10% formalin 0, 0.5, 2.0, and 4.0 h after treatment. ERα (FITC) and nuclear staining (DAPI) were analyzed by immunofluorescence.
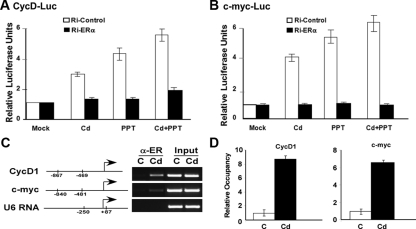
Because Cd promotes translocation of ERα to the nucleus, we questioned whether ERα is required for the expression of Cd-induced genes. To address this issue, we analyzed the trans-activation of CycD1 and c-myc promoters in the presence and absence of ERα. We performed a reporter gene assay by transfecting MCF-7 cells with a plasmid containing a luciferase gene directly linked to either the CycD1 (1.7 kb) or the c-myc (2.1 kb) promoter sequences along with siRNA directed against ERα or a scramble siRNA control. Cells transfected with an empty vector served as a negative control. After transfection, cells were treated with either Cd, propyl pyrazole triol (PPT), a highly selective ERα agonist, or Cd in the presence of PPT. Cells mock treated with PBS served as the negative control. Results in Fig. 4, A and B, demonstrate that Cd treatment modestly increases the transcriptional activity at both the CycD1 and c-myc promoters by 3- and 4-fold, respectively, in cells transfected with the Ri-Control. As expected, PPT also trans-activated the two ERα-regulated promoters (4.5-fold for CycD1 and 5.3-fold for c-myc). Interestingly, the combination of Cd and PPT only resulted in a slightly higher trans-activation, suggesting the absence of synergistic effects between the two agonists (Fig. 4, A and B). Similarly, growth and gene expression analyses of cells treated with both Cd and PPT also indicate these agonists likely have additive rather than synergistic effects (Supplemental Fig. 2). On the other hand, cells transfected Ri-ERα displayed a significant decrease in the trans-activation at both the CycD1 and c-myc promoters, suggesting that Cd induction of CycD1 and c-myc transcription is dependent on the presence of ERα. This result was also confirmed by transfecting cells with the CycD1 and c-myc luciferase reporter construct, followed by ICI treatment. Similar to Ri-ERα, cells treated with ICI expressed significantly less ERα (Fig. 2B), and the trans-activation of both CycD1 and c-myc promoters were decreased by ICI even in the presence of Cd and 17β-E2 (Supplemental Fig. 3).
Figure 4.
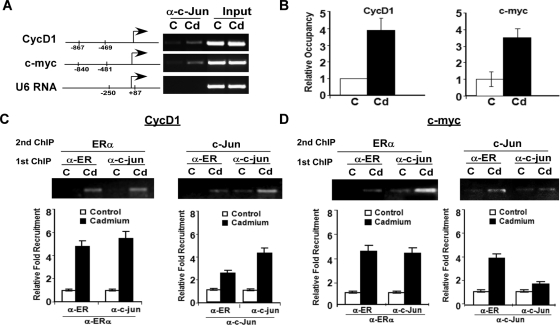
Cd activates target gene expression through ERα. MCF-7 cells were transfected with either Ri-ERα (black) or Ri-Control (white) along with (panel A) CycD1-luc or (panel B) c-myc-luc. Renilla luciferase construct was cotransfected as an internal control. After transfection, cells were treated with 10−6m CdCl2, PPT, 10−6m plus PPT, or mock treated [control (C)]. Luciferase activity was measured using a dual luciferase assay. Statistics represent triplicate samples. Panel C, MCF-7 cells were hormone deprived and treated with 10−6m CdCl2 (Cd) or mock treated with PBS (C), followed by fixation in formaldehyde. ChIP analysis was performed with α-ERα antibodies, and ERα recruitment to target gene promoters was determined by promoter-specific primers and semiquantitative RT-PCR. Primer positions are depicted on the promoter diagram for each gene. Input represents total chromatin before immunoprecipitation. Panel D, Band intensities of PCR product were quantitated using Quantity One software (Bio-Rad) and statistics represent fold changes of three independent experiments.
Because Cd increased the transcriptional activity of both CycD1 and c-myc promoters, we questioned whether Cd exposure promotes ERα recruitment to target gene promoters using a chromatin immunoprecipitation (ChIP) assay. Results in Fig. 4, C and D, show an increase in ERα occupancy at the proximal promoter regions of both CycD1 and c-myc upon Cd treatment. However, promoter sequences of genes that are not regulated by ERα, such as the U6 RNA promoter, were not detected in the ERα immunoprecipitates. These results establish a direct link between ERα and Cd-induced breast carcinogenesis.
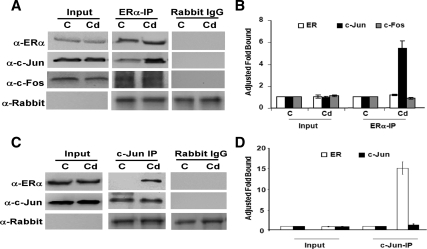
Because ERα is normally recruited to the CycD1 and c-myc promoters via the AP-1 family of transcription factors, we analyzed whether Cd treatment promotes ERα/AP-1 interaction in ERα-positive breast cancer cells by coimmunoprecipitation. MCF-7 cells were treated with 10−6m CdCl2, cell lysates were incubated with anti-ERα antibodies, and the presence of AP-1 proteins was analyzed by Western blot analysis (Fig. 5, A and B). The results in Fig. 5, A and B, show that, although c-Fos is found in complex with ERα, no significant difference was observed between Cd-treated and mock-treated cells. Strikingly, a greater fraction of c-Jun coprecipitated with ERα in the Cd-treated cells. This ERα/c-Jun interaction was confirmed by reverse coimmunoprecipitation with c-Jun. In this case, the amount of ERα found in complex with c-Jun was greater in Cd-treated cells than in the control cells (Fig. 5, C and D). Taken together, these results suggest that Cd promotes ERα/c-Jun complex formation.
Figure 5.
Cd promotes ERα/c-Jun interaction. MCF-7 cells were hormone deprived and treated with 10−6m CdCl2 (Cd) or mock treated [control (C)]. Panel A, Cells were harvested for coimmunoprecipitation 24 h after treatment. Cell lysates were immunoprecipitated with either α-ERα or normal rabbit IgG. IP complexes were separated with SDS-PAGE. Proteins interacting with ERα were analyzed by Western blot analysis of ERα, c-Jun, c-Fos, and the IgG heavy chain. The input represents 10% of total protein. Panel B, Band intensities of experiment in panel A were quantitated using Quantity One (Bio-Rad), and statistics represent three independent experiments (P < 0.05). Panel C, Cell lysates were reverse coimmunoprecipitated with either α-c-Jun or normal rabbit IgG. IP complexes were separated, and analysis of proteins interacting with c-Jun was determined by Western blot analysis of ERα, c-Jun, and IgG heavy chain. The input represents 10% of total protein. Panel D, Band intensities of experiment in panel C were quantitated using Quantity One (Bio-Rad), and statistics represent three independent experiments.
Having demonstrated that Cd can promote the ERα/c-Jun interaction and that Cd promotes ERα recruitment to the promoters of target genes, we evaluated whether c-Jun is directly involved in ERα-mediated transcription after Cd treatment. To address this question, we examined the recruitment of c-Jun to the promoter regions of CycD1 and c-myc using a second ChIP. The occupancy of c-Jun on the promoter regions of both c-myc and CycD1 is expected because both regions analyzed contain an AP-1 response element; however, the results presented in Fig. 6, A and B, show that higher levels of c-Jun were recruited to the CycD1 and c-myc promoters in Cd-treated cells in comparison with untreated cells. In contrast, c-Jun occupancy was not observed at the U6 RNA promoter. To determine whether c-Jun and ERα are part of the same transcription complex, we performed a ChIP re-ChIP assay. MCF-7 cells were treated with 10−6m Cd, and cells were harvested for ChIP analysis with the first antibody (either ERα or c-Jun). After the first ChIP assay, the DNA-protein complexes were extracted and re-ChIPed with the second antibody (either ERα or c-Jun). Our results confirm that Cd promotes the recruitment of both ERα and c-Jun to the same promoter region, suggesting that they are likely part of the same transcription complex (Fig. 6, C and D).
Figure 6.
c-Jun is recruited to target gene promoters. Panel A, MCF-7 cells were hormone deprived and treated with 10−6m CdCl2 (Cd) or mock treated [control (C)]. ChIP analysis was performed with α-c-Jun antibodies, and c-Jun recruitment to target gene promoters was determined by promoter-specific primers using semiquantitative RT-PCR. Panel B, Band intensities of PCR product were quantitated using Quantity One (Bio-Rad). Panels C and D, ChIP re-ChIP was used to determine whether ERα and c-Jun are part of the same promoter complex. ChIP assay was performed as in A with antibodies against ERα or c-Jun. Protein-DNA complexes of the first ChIP were extracted and re-ChIPed with the second antibody. Normal rabbit serum was used as a negative control. The occupancy of ERα/c-Jun complexes on target promoters was analyzed using promoter-specific primers and semiquantitative RT-PCR. Band intensities of PCR products were quantitated using Quantity One (Bio-Rad).
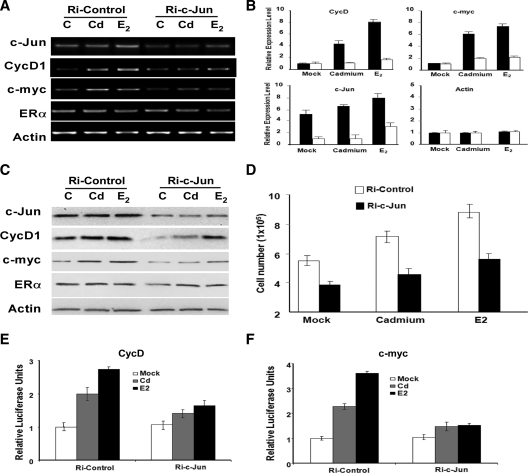
To understand the role of c-Jun in Cd-induced gene expression, we analyzed the expressions of CycD1 and c-myc after siRNA-mediated silencing of c-Jun (Fig. 7, A–C). Expression analysis at the protein and mRNA levels confirmed that the expressions of CycD1 and c-myc are decreased in cells transfected with Ri-c-Jun. To further discern the role of c-Jun in Cd-induced cell growth, we transfected MCF-7 cells with either Ri-c-Jun or a control siRNA. Cells were treated with either Cd, E2, or mock treated with PBS (C), and growth was monitored 3 d after treatment (Fig. 7D). Although the depletion of c-Jun diminishes the levels of agonist-induced cell growth and gene expression, cells continue to display some level of Cd and estrogen responsiveness. This may be attributed to the incomplete silencing of c-Jun by the siRNA. Taken together, results from this study strongly suggest Cd promotes the binding of ERα to c-Jun to regulate the expression of Cd-induced gene and promote breast cancer cell growth.
Figure 7.
Cd-induced gene expression and cell growth is dependent on c-Jun. MCF-7 cells were transfected with Ri-c-Jun (white) or Ri-Control (black). After transfection, cells were hormone deprived and treated with 10−6m CdCl2 (Cd), 10−7m 17β-E2, or mock treated [control (C)]. Gene expression was monitored by (panel A) semiquantitative and (panel B) quantitative PCR. Data are representative of fold changes in three independent experiments. Panel C, Protein expression was monitored by Western blot analysis. Panel D, MCF-7 cells were plated and transfected with either Ri-c-Jun (black) or Ri-Control (white). Cells were treated with 10−6m Cd or 10−7m 17β-E2 or mock treated (Mock). Cell growth was analyzed 4 d after treatment, and each data point represents four replicate samples. Panels E and F, The requirement of c-Jun in Cd-induced CycD1 and c-myc expression was analyzed using a Luciferase reporter assay. MCF-7 cells were transfected with CycD1-luc or c-myc-luc construct along with either Ri-c-Jun or a scrambled siRNA control. Renilla luciferase construct was cotransfected as an internal control. After transfection, cells were treated with 10−6m Cd, 10−7m 17β-E2, or mock treated (Mock). Statistics in both panels E and F represent triplicate samples.
Discussion
Cd is an environmental contaminant that is often referred to as an endocrine disruptor due to its ability to mimic the ER and alter the expression of various estrogen target genes. Previous studies have suggested that exposure to Cd can affect mammary gland development in ovarectomized rats (2). Although there is no direct evidence that Cd can promote breast cancer in humans, it has been implicated as a cancer causing agent and has been classified by the International Agency for Research on Cancer as a potential human carcinogen. Studies have demonstrated that Cd can promote growth of MCF-7 breast cancer cells and can activate the ER in vitro (1,6,7,8,21,22). However, the mechanism of action of Cd is poorly understood. Results from this study confirm that Cd can promote cell proliferation in three human ERα-positive breast cancer cell lines, MCF-7, T-47D, and ZR-75-1 (Fig. 1A). Furthermore, our results demonstrate that Cd does not promote proliferation in ERα-negative breast cancer cells (Supplemental Fig. 1).
To fully establish a link between Cd-induced breast cancer cell growth and the ER, we used ICI and Ri-ERα to reduce the intracellular levels of ERα. We found that both ICI and Ri-ERα were able to block Cd’s action, suggesting that Cd-induced cell growth and gene expression are dependent on the ER (Fig. 2 and Supplemental Fig. 1). Consistent with previous reports, we found that multiple ER target genes (CycD1, c-myc, and CTD) are up-regulated by Cd exposure (Fig. 1, B and C) (1,6,7). Additionally, our analysis demonstrates that cells exposed to Cd also expressed higher levels of CycE, cdk2, and cdk4 and decreased levels of p21. All of these genes function as cell cycle regulators that modulate progression of cells through the G1/S transition (Fig. 1, B and C). Overexpression of any one of these cell cycle regulators results in the deregulation of growth, often leading to cancer (23,24,25,26).
Results from our reporter gene assay further support the argument that Cd stimulates the expression of CycD1 and c-myc by increasing the transcriptional activity of these target promoters. Because CycD1 and c-myc are both ER target genes, we fully expected to observe ERα occupancy at the CycD1 and c-myc promoters. Before this study, it was unclear whether Cd was responsible for stimulating the binding of ERα to these promoters or whether Cd induced nongenomic effects, leading to elevated levels of CycD1 and c-myc. Using ChIP analysis, we demonstrated that Cd enhances the recruitment of ERα to the proximal promoters of CycD1 and c-myc (Fig. 4). This finding is further supported by the observation that Cd promotes the translocation of ERα into the nucleus, consistent with ERα’s role as a transcriptional regulator (Fig. 3).
Both CycD1 and c-myc are nonclassical ER target genes, and their expression, although dependent on ERα, does not require direct binding of ERα to the DNA. Instead, ERα is recruited to their promoters via AP-1 (27,28). AP-1 is a transcription factor complex that contains proteins encoded by the proto-oncogenes c-Jun, c-Fos, and other AP-1 family members. This complex interacts with AP-1 binding sites in promoters to activate transcription of genes involved in cell growth, differentiation, and development. Previous studies have shown that c-Fos and c-Jun suppress ERα-dependent trans-activation of estrogen response element-containing promoters (29,30,31) by recruiting ERα to AP-1 promoters. ERα has been shown to cross talk with AP-1 and coregulate genes like CycD1 and c-myc (28,32). Here, we have demonstrated for the first time that Cd exposure increases the interaction between ERα and c-Jun, but not between ERα and c-Fos (Fig. 5). These data are in accordance with the recent finding that stress induced by arsenic exposure increases ERα-bound c-Jun (33). Like Cd, arsenic is a heavy metal and the ERα/c-Jun interaction has been shown to neutralize heavy metal stress to avoid apoptosis (33). In the case of Cd, this interaction stimulates the expression of genes to mediate breast cancer cell growth.
Our findings demonstrate that Cd enhances the recruitment of c-Jun to the CycD1 and c-myc promoters (Fig. 6, A and B) and that ERα is likely recruited to these promoters via its interaction with c-Jun (Fig. 6, C and D). We extended our analysis using siRNA to deplete the endogenous levels of c-Jun and further demonstrated that c-Jun is necessary for the expression of Cd-induced CycD1 and c-myc expression. This is consistent with previous studies that have reported that Cd exposure increases the binding of c-Jun to the AP-1 site, and subsequently increases the trans-activation of AP-1 promoters (34,35,36). Together with this report, the data from our study suggest that Cd is involved in facilitating the cross talk between ERα and other transcription factors, such as c-Jun, to regulate expression of Cd-induced genes. In support of our findings, a recent study has also revealed that Cd activates the Erk1/2 and Akt kinases, resulting in the phosphorylation of ERα, which has been shown to promote the ERα/c-Jun interaction (22,37,38,39). We anticipate that Cd may function to mediate protein-protein interactions between ERα and other transcription factors, providing an alternative mechanism for activating Cd-induced gene expression. This possible mechanism would also explain the failure of recent studies to demonstrate Cd’s estrogenic activity using a pure estrogen response element in a yeast estrogen screen (40,41).
In summary, the results from this study demonstrate for the first time that Cd promotes breast cancer cell growth and gene expression by potentiating the interaction between ERα and c-Jun and enhancing the binding of these transcription factors to target gene promoters. We speculate that Cd mediates ERα cross talk with other transcription factors, including Sp1, E2F, and NFκB family members, to regulate additional Cd-induced genes. It would be very interesting to investigate a possible transcription factor complex or complexes involved in regulating the expression of CycE, cdk2, and other non-ER target genes that are up-regulated in response to Cd treatment (Fig. 1, B and C). The role of ERβ in Cd-mediated growth and gene expression appears to be minimal because siRNA-mediated depletion of ERβ did not alter the Cd responsiveness of the cells (Fig. 2, E and F). The results presented in this study have established a direct link between Cd-induced ERα-regulated gene expression and breast cancer, and underscore the importance of Cd exposure in breast carcinogenesis.
Materials and Methods
Cell culture
Human MCF-7, T-47D, ZR-75-1, and MDA-MB-453 breast cancer cells were obtained from the American Type Culture Collection (Manassas, VA). All cells, except T-47D, were maintained in DMEM supplemented with 10% fetal bovine serum (FBS) (Hyclone, Logan, UT) and antibiotics. T-47D cells were maintained in RPMI 1640 supplemented with 10% FBS, sodium pyruvate, and antibiotics. All cell lines were subcultured every 3–4 d.
Cell proliferation analysis
Cells were plated at a density of 2 × 105 per well in a six-well plate and hormone deprived for 2 d. Hormone deprivation is performed by maintaining cells in media containing 10% charcoal-dextran stripped FBS (CDS). Cells were treated with 10−6m CdCl2 (Sigma, St. Louis, MO) or 10−8m 17β-E2 (Sigma), or were mock treated with 1× PBS. Cell number was determined every 2 d by cell counting in triplicate using a hemacytometer. Presented data are representative of several experiments. In the ER degradation experiments, cells were treated with 10−6m CdCl2 alone or 10−6m CdCl2 and 10−8m ICI (Sigma), or were mock treated. For the RNAi silencing, MCF- 7 cells were plated in 24-well plates and transfected with siRNA directed against ERα or c-Jun (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) using siRNA transfection reagent (Santa Cruz Biotechnology, Inc.). A scrambled siRNA sequence was used as a negative control. Twenty-four hours after transfection, cells were treated with 10−6m CdCl2, 10−7m 17β-E2 or mock treated with PBS. Cells were counted in triplicates and harvested for protein and gene expression analysis 2, 3, and 4 d after treatment.
For the indirect cell proliferation assay, 2000 cells were plated per well in a 96-well plate in hormone-deprived media. Twenty-four hours later, cells were treated with 10−6m CdCl2 or 10−8m 17β-E2, or were mock treated. Proliferation was monitored every 2 d by incubating cells with 0.5 mg/ml MTT (Sigma) for 1.5 h. MTT crystals were dissolved in dimethylsulfoxide (Sigma) and read at 595 nm in a plate reader (Bio-Rad, Hercules, CA).
Western blot analysis
MCF-7 cells were plated and treated as described above and harvested 24 and 48 h after treatment. Cells were lysed in 1% sodium dodecyl sulfate-HEPES buffer, and the total protein concentration was determined using the Bio-Rad protein assay kit (Bio-Rad). Total protein was separated via SDS-PAGE and transferred to a poly(vinylidene difluoride) membrane. Protein expression was monitored using protein-specific antibodies: α-ER (BD, Franklin Lakes, NJ), α-ERβ, α-CycE, α-cdk2, α-p21, α-E2F1, α-c-Jun (Santa Cruz Biotechnology, Inc.), α-CycD1 (Transduction Laboratories, Lexington, KY), and α-actin (Sigma).
Coimmunoprecipitation
MCF-7 cells were plated and treated as previously described. They were harvested 48 h after Cd treatment. Cells were lysed in a 0.5% Igepal CA-630 lysis buffer and sonicated for 30 sec. Cell lysates were immunoprecipitated with α-ERα (1:1 ratio of ERα antibodies: HC-20 and MC-20; Santa Cruz Biotechnology, Inc.), α-c-Jun (Santa Cruz Biotechnology, Inc.), or normal rabbit IgG for 1.5 h and then incubated with protein A for 1 h. Protein complexes were washed three times with PBS and then separated by SDS-PAGE followed by Western blot analysis with α-ERα (Neomarkers, Fremont, CA), α-c-Jun (Santa Cruz Biotechnology, Inc.), α-c-Fos (Santa Cruz Biotechnology, Inc.), and α-rabbit IgG (Bio-Rad) antibodies.
RT-PCR analysis
MCF-7 cells were plated and treated as previously described. Cells were harvested for mRNA analysis 24 and 48 h after treatment. Total RNA was isolated using the Trizol reagent according to the manufacturer’s protocol. Three micrograms of total RNA, Moloney murine leukemia virus RT (Invitrogen, Carlsbad, CA), and oligo-dT18 primers were used in the RT reaction. Gene expression was monitored using 3 μl of cDNA and gene-specific primers by 28 cycles of semiquantitative PCR (Bio-Rad). PCR products were analyzed by agarose gel electrophoresis, and bands were visualized with a ChemiDoc Imager (Bio-Rad). The primer sequences used for RT-PCR are listed below:
CycD1F, TCCTGTGCTGCGAAGTGGAAAC
CycD1R, AAATCGTGCGGGGTCATTGC
CycEF, ATACAGACCCACAGAGACAG
CycER, TGCCATCCACAGAAATACTT
cdk2F, TTTGCTGAGATGGTGACTCGC
cdk2R, CACTGGAGGAGGGGTGAGATTAG
E2F1F, CGCATCTATGACATCACCAACG
E2F1R, GAAAGTTCTCCGAAGAGTCCACG
c-mycF, TGACACTGTCCAACTTGACCCTCTT
c-mycR, TCGCAAGACTCCAGCGCCTTCTC
CTDF, GGTGGAATACTTTGCCTGCCTTC
CTDR, CCGAGACGCTCCCAGACATC
p21F, GAACTTCGACTTTGTCACCGAG
p21R, CTTCCTCTTGGAGAAGATCAGC
pS2F, TTGGAGCAGAGAGGAGGCAATG
pS2R, TTAGGATAGAAGCACCAGGGGAC
cyclin AF, CCCCCAGAAGTAGCAGAGTTTGTG
cyclin AR, GCTTTGTCCCGTGACTGTGTAGAG
cdk4F, AAGAGTGTGAGAGTCCCCAATGG
cdk4R, GATTTTGCCCAACTGGTCGG
actinF, GAGAAAATCTGGCACCACACC
actinR, ATACCCCTCGTAGATGGGCAC
For quantitative RT-PCR analysis of gene expression, gene sequences were amplified in the presence of SYBR Green fluorophore (Superarray, Frederick, MD) and detected using the iCycler real-time PCR equipment (Bio-Rad). Fluorescent values after each elongation step were collected along with a melting curve analysis at the end of the PCR. Fold difference (N) was calculated using the equation: n = E(CT-treated − CT control) between treated cells and mock-treated (control) cells. E is defined as the PCR efficiency of the specific primer pair used and was determined by the standard curve [E =10(1/slope of standard curve)]. CT is the threshold cycle, which is defined as the minimum cycle number at which the fluorescence of the PCR product is first detected.
Reporter gene assay
The reporter gene assay with promoter-luc was performed by transfecting MCF-7 cells with the firefly luciferase reporter plasmid, pBV-myc promoter (2.1 kb) or pA3-CycD1 promoter (1.7 kb), and with the pRL-SV40 Renilla luciferase plasmid (Promega) using Lipofectamine (Invitrogen) according to the manufacturer’s protocol. Five hours after transfection, the medium was changed to hormone-deprived media containing 10−6m CdCl2, 10−7m 17β-E2, or 1× PBS. Cells were harvested at 48 h after transfection and analyzed using a dual luciferase assay kit (Promega). All reporter gene assays were performed in triplicate, with the entire experiment repeated at least three times. pBV-myc (2.1 kb) was obtained from Addgene and the pA3-CycD1 promoter (1.7 kb) was a kind gift of Dr. Richard Pestell (9).
For the RNAi silencing experiments, MCF-7 cells were seeded in 24-well plates and transfected firefly luciferase reporter plasmid, pBV-myc promoter (2.1 kb) or pA3-CycD1 promoter (1.7 kb), and with the pRL-SV40 Renilla luciferase plasmid (Promega, Madison, WI) in the presence of either ERα, c-Jun, ERβ, or control siRNA using siRNA transfection reagent (Santa Cruz Biotechnology, Inc.). Five hours after transfection, the medium was changed to hormone deprived, and 24 h later, cells were treated with 10−6m CdCl2, 10−7m 17α-E2, or mock treated with PBS. Cells were harvested at 48 h after treatment and analyzed using a dual luciferase assay kit (Promega).
ChIP
MCF-7 cells were plated in 10-cm plates and hormone deprived for 1.5 d. Cells were treated with 10−6m CdCl2 for 30 h. After treatment, the cells were fixed with formaldehyde, scraped, and collected by centrifugation. ChIP was performed as previously described (10). Briefly, equal numbers of cells were lysed and sonicated in lysis buffer to break the chromatin down to 1- to 2-kb fragments. The chromatin mixture was immunoprecipitated with antibodies specific for the proteins of interest [1:1 mixture of α-ER antibodies: HC-20 and MC-20 or α-c-Jun (Santa Cruz Biotechnology, Inc.)]. Antibody complexes were purified using either protein A or G beads. The complexes were reverse-cross-linked to release the chromatin fragments, and the DNA was purified using QiaQuick columns (QIAGEN, Valencia, CA). Occupancy at a specific promoter was detected by PCR using the promoter-specific primers listed below. PCR products ranging 300–500 bp were separated by agarose gel electrophoresis and visualized using the ChemiDoc system (Bio-Rad). The sequences of the ChIP primers are listed below:
CycD1-ChIPF, CATTCAGAGGTGTGTTTCTCCC
CycD1-ChIPR, CTCAGCGACTGCATCTTCTTTC
c-myc-ChIPF, GACACATCTCAGGGCTAAACAG
c-myc-ChIPR, GAGAGTGGAGGAAAGAAGGGTA
U6RNA-ChIPF, GAGGGCCTATTTCCCATGATTC
U6RNA-ChIPR, GAATTTGCGTGTCATCCTTGC
ChIP re-ChIP
For the re-ChIP assay, protein A-antibody complexes resulting from the first ChIP were extracted twice by incubation with 25 μl of 10 mm dithiothreitol for 20 min each at 37 C. The supernatants were pooled and diluted 10 times with re-ChIP buffer (20 mm Tris·HCl, pH 8.1; 2 mm EDTA; 150 mm NaCl; and 0.1% Triton X-100). The chromatin mixture was re-ChIPed with the second antibody: α-ER (1:1 mixture of α-ER antibodies: HC-20:MC-20), α-c-Jun, or normal rabbit IgG, (Santa Cruz Biotechnology, Inc.). Antibody complexes were purified using protein A beads. The complexes were reverse-cross-linked to release the chromatin fragments by incubation at 65 C for 4–6 h, and the DNA was purified and analyzed as previously described.
Supplementary Material
Acknowledgments
We thank Dr. Richard Pestell for his generous gift of the pA3-CycD1 promoter plasmid; Dr. Sheila Johnson-Brousseau, Dr. Mary B. Sevigny, and Dr. Mietek Kolipinski for their insightful comments and critical reading of the manuscript; and Phan Huy Vu and Angelo Wong for their technical support.
Footnotes
This work was supported by grants from the Wendy Will Case Cancer Fund and the National Cancer Institute Grants CA121983-01A1 and CA121983-01A1S1.
Disclosure Summary: The authors have nothing to disclose.
First Published Online March 10, 2010
Abbreviations: AP, Activating protein; Cd, cadmium; CdCl2, Cd chloride; CDK, cyclin-dependent kinase; ChIP, chromatin immunoprecipitation; CTD, cathepsin D; CycD1, cyclin D1; CycE, cyclin E1; E2, estradiol; ER, estrogen receptor; FBS, fetal bovine serum; ICI, ICI 182,780; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide; siRNA, small interfering RNA; PPT, propyl pyrazole triol; Ri, SiRNAi.
References
- Stoica A, Katzenellenbogen BS, Martin MB 2000 Activation of estrogen receptor-α by the heavy metal cadmium. Mol Endocrinol 14:545–553 [DOI] [PubMed] [Google Scholar]
- Johnson MD, Kenney N, Stoica A, Hilakivi-Clarke L, Singh B, Chepko G, Clarke R, Sholler PF, Lirio AA, Foss C, Reiter R, Trock B, Paik S, Martin MB 2003 Cadmium mimics the in vivo effects of estrogen in the uterus and mammary gland. Nat Med 9:1081–1084 [DOI] [PubMed] [Google Scholar]
- Mosselman S, Polman J, Dijkema R 1996 ERβ: identification and characterization of a novel human estrogen receptor. FEBS Lett 392:49–53 [DOI] [PubMed] [Google Scholar]
- Paech K, Webb P, Kuiper GG, Nilsson S, Gustafsson J, Kushner PJ, Scanlan TS 1997 Differential ligand activation of estrogen receptors ERα and ERβ at AP1 sites. Science 277:1508–1510 [DOI] [PubMed] [Google Scholar]
- Antila E, Mussalo-Rauhamaa H, Kantola M, Atroshi F, Westermarck T 1996 Association of cadmium with human breast cancer. Sci Total Environ 186:251–256 [DOI] [PubMed] [Google Scholar]
- Martin MB, Reiter R, Pham T, Avellanet YR, Camara J, Lahm M, Pentecost E, Pratap K, Gilmore BA, Divekar S, Dagata RS, Bull JL, Stoica A 2003 Estrogen-like activity of metals in MCF-7 breast cancer cells. Endocrinology 144:2425–2436 [DOI] [PubMed] [Google Scholar]
- Garcia-Morales P, Saceda M, Kenney N, Kim N, Salomon DS, Gottardis MM, Solomon HB, Sholler PF, Jordan VC, Martin MB 1994 Effect of cadmium on estrogen receptor levels and estrogen-induced responses in human breast cancer cells. J Biol Chem 269:16896–16901 [PubMed] [Google Scholar]
- Choe SY, Kim SJ, Kim HG, Lee JH, Choi Y, Lee H, Kim Y 2003 Evaluation of estrogenicity of major heavy metals. Sci Total Environ 312:15–21 [DOI] [PubMed] [Google Scholar]
- Albanese C, Johnson J, Watanabe G, Eklund N, Vu D, Arnold A, Pestell RG 1995 Transforming p21ras mutants and c-Ets-2 activate the cyclin D1 promoter through distinguishable regions. J Biol Chem 270:23589–23597 [DOI] [PubMed] [Google Scholar]
- Louie MC, Zou JX, Rabinovich A, Chen HW 2004 ACTR/AIB1 functions as an E2F1 coactivator to promote breast cancer cell proliferation and antiestrogen resistance. Mol Cell Biol 24:5157–5171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joe AK, Memeo L, McKoy J, Mansukhani M, Liu H, Avila-Bront A, Romero J, Li H, Troxel A, Hibshoosh H 2005 Cyclin D1 overexpression is associated with estrogen receptor expression in Caucasian but not African-American breast cancer. Anticancer Res 25:273–281 [PubMed] [Google Scholar]
- Margueron R, Licznar A, Lazennec G, Vignon F, Cavaillès V 2003 Oestrogen receptor α increases p21(WAF1/CIP1) gene expression and the antiproliferative activity of histone deacetylase inhibitors in human breast cancer cells. J Endocrinol 179:41–53 [DOI] [PubMed] [Google Scholar]
- Barnes DM, Gillett CE 1998 Cyclin D1 in breast cancer. Breast Cancer Res Treat 52:1–15 [DOI] [PubMed] [Google Scholar]
- Liaudet-Coopman E, Beaujouin M, Derocq D, Garcia M, Glondu-Lassis M, Laurent-Matha V, Prébois C, Rochefort H, Vignon F 2006 Cathepsin D: newly discovered functions of a long-standing aspartic protease in cancer and apoptosis. Cancer Lett 237:167–179 [DOI] [PubMed] [Google Scholar]
- Berchem G, Glondu M, Gleizes M, Brouillet JP, Vignon F, Garcia M, Liaudet-Coopman E 2002 Cathepsin-D affects multiple tumor progression steps in vivo: proliferation, angiogenesis and apoptosis. Oncogene 21:5951–5955 [DOI] [PubMed] [Google Scholar]
- Mimori K, Mori M, Shiraishi T, Tanaka S, Haraguchi M, Ueo H, Shirasaka C, Akiyoshi T 1998 Expression of ornithine decarboxylase mRNA and c-myc mRNA in breast tumours. Int J Oncol 12:597–601 [DOI] [PubMed] [Google Scholar]
- Guérin M, Barrois M, Terrier MJ, Spielmann M, Riou G 1988 Overexpression of either c-myc or c-erbB-2/neu proto-oncogenes in human breast carcinomas: correlation with poor prognosis. Oncogene Res 3:21–31 [PubMed] [Google Scholar]
- Horiguchi J, Iino Y, Takei H 1996 Expression of pS2 estrogen-inducible protein in primary breast cancer. Oncology 53:12–15 [DOI] [PubMed] [Google Scholar]
- Davidson NE, Bronzert DA, Chambon P, Gelmann EP, Lippman ME 1986 Use of two MCF-7 cell variants to evaluate the growth regulatory potential of estrogen-induced products. Cancer Res 46:1904–1908 [PubMed] [Google Scholar]
- Varshochi R, Halim F, Sunters A, Alao JP, Madureira PA, Hart SM, Ali S, Vigushin DM, Coombes RC, Lam EW 2005 ICI182,780 induces p21Waf1 gene transcription through releasing histone deacetylase 1 and estrogen receptor α from Sp1 sites to induce cell cycle arrest in MCF-7 breast cancer cell line. J Biol Chem 280:3185–3196 [DOI] [PubMed] [Google Scholar]
- Liu Z, Yu X, Shaikh ZA 2008 Rapid activation of ERK1/2 and AKT in human breast cancer cells by cadmium. Toxicol Appl Pharmacol 228:286–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brama M, Gnessi L, Basciani S, Cerulli N, Politi L, Spera G, Mariani S, Cherubini S, d'Abusco AS, Scandurra R, Migliaccio S 2007 Cadmium induces mitogenic signaling in breast cancer cell by an ERα-dependent mechanism. Mol Cell Endocrinol 264:102–108 [DOI] [PubMed] [Google Scholar]
- Malumbres M, Barbacid M 2009 Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer 9:153–166 [DOI] [PubMed] [Google Scholar]
- Berglund P, Landberg G 2006 Cyclin e overexpression reduces infiltrative growth in breast cancer: yet another link between proliferation control and tumor invasion. Cell Cycle 5:606–609 [DOI] [PubMed] [Google Scholar]
- Sutherland RL, Musgrove EA 2004 Cyclins and breast cancer. J Mammary Gland Biol Neoplasia 9:95–104 [DOI] [PubMed] [Google Scholar]
- Musgrove EA 2006 Cyclins: roles in mitogenic signaling and oncogenic transformation. Growth Factors 24:13–19 [DOI] [PubMed] [Google Scholar]
- Sabbah M, Courilleau D, Mester J, Redeuilh G 1999 Estrogen induction of the cyclin D1 promoter: involvement of a cAMP response-like element. Proc Natl Acad Sci USA 96:11217–11222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeNardo DG, Cuba VL, Kim H, Wu K, Lee AV, Brown PH 2007 Estrogen receptor DNA binding is not required for estrogen-induced breast cell growth. Mol Cell Endocrinol 277:13–25 [DOI] [PubMed] [Google Scholar]
- Shemshedini L, Knauthe R, Sassone-Corsi P, Pornon A, Gronemeyer H 1991 Cell-specific inhibitory and stimulatory effects of Fos and Jun on transcription activation by nuclear receptors. EMBO J 10:3839–3849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzukerman M, Zhang XK, Pfahl M 1991 Inhibition of estrogen receptor activity by the tumor promoter 12-O-tetradeconylphorbol-13-acetate: a molecular analysis. Mol Endocrinol 5:1983–1992 [DOI] [PubMed] [Google Scholar]
- Doucas V, Spyrou G, Yaniv M 1991 Unregulated expression of c-Jun or c-Fos proteins but not Jun D inhibits oestrogen receptor activity in human breast cancer derived cells. EMBO J 10:2237–2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safe S, Kim K, Kim K 2008 Non-classical genomic estrogen receptor (ER)/specificity protein and ER/activating protein-1 signaling pathways. J Mol Endocrinol 41:263–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi X, Borowicz S, Pramanik R, Schultz RM, Han J, Chen G 2004 Estrogen receptor inhibits c-Jun-dependent stress-induced cell death by binding and modifying c-Jun activity in human breast cancer cells. J Biol Chem 279:6769–6777 [DOI] [PubMed] [Google Scholar]
- Souza V, Escobar Md Mdel C, Gómez-Quiroz L, Bucio L, Hernández E, Cossio EC, Gutiérrez-Ruiz MC 2004 Acute cadmium exposure enhances AP-1 DNA binding and induces cytokines expression and heat shock protein 70 in HepG2 cells. Toxicology 197:213–228 [DOI] [PubMed] [Google Scholar]
- Hsiao CJ, Stapleton SR 2004 Characterization of Cd-induced molecular events prior to cellular damage in primary rat hepatocytes in culture: activation of the stress activated signal protein JNK and transcription factor AP-1. J Biochem Mol Toxicol 18:133–142 [DOI] [PubMed] [Google Scholar]
- Huang C, Zhang Q, Li J, Shi X, Castranova V, Ju G, Costa M, Dong Z 2001 Involvement of Erks activation in cadmium- induced AP-1 transactivation in vitro and in vivo. Mol Cell Biochem 222:141–147 [DOI] [PubMed] [Google Scholar]
- Baron S, Escande A, Albérola G, Bystricky K, Balaguer P, Richard-Foy H 2007 Estrogen receptor α and the activating protein-1 complex cooperate during insulin-like growth factor-I-induced transcriptional activation of the pS2/TFF1 gene. J Biol Chem 282:11732–11741 [DOI] [PubMed] [Google Scholar]
- Petz LN, Ziegler YS, Schultz JR, Nardulli AM 2004 Fos and Jun inhibit estrogen-induced transcription of the human progesterone receptor gene through an activator protein-1 site. Mol Endocrinol 18:521–532 [DOI] [PubMed] [Google Scholar]
- Johnston SR, Head J, Pancholi S, Detre S, Martin LA, Smith IE, Dowsett M 2003 Integration of signal transduction inhibitors with endocrine therapy: an approach to overcoming hormone resistance in breast cancer. Clin Cancer Res 9:524S–532S [PubMed] [Google Scholar]
- Silva E, Lopez-Espinosa MJ, Molina-Molina JM, Fernández M, Olea N, Kortenkamp A 2006 Lack of activity of cadmium in in vitro estrogenicity assays. Toxicol Appl Pharmacol 216:20–28 [DOI] [PubMed] [Google Scholar]
- Denier X, Couteau J, Baudrimont M, Hill EM, Rotchell J, Minier C 2008 In vitro study of the effects of cadmium on the activation of the estrogen response element using the YES screen. Mar Environ Res 66:108–110 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.