Abstract
Heterochromatin is notable for its capacity to propagate along a chromosome. The prevailing model for this spreading process postulates that silencing proteins are first recruited to silencer sequences and then spread from these sites independently of the silencers. However, we found that in Saccharomyces cerevisiae silencers also influence the extent of silenced chromatin domains. We compared the abilities of two different silencers, HMR-E and a telomeric repeat, to promote silencing and found that the HMR-E silencer contributed to an increased steady-state association of Sir proteins over a region of several kilobase pairs compared to the telomeric repeat, even though both silencers recruited similar levels of Sir proteins. We also discovered that, although the HMR-E silencer alone was sufficient to block transcription of the HMR locus, a secondary silencer, HMR-I, boosted the level of Sir proteins at HMR, apparently beyond the level necessary to repress transcription. Finally, we discovered that a tRNAThr gene near HMR-I helped maintain silenced chromatin and transcriptional repression under conditions of reduced deacetylase activity. This study highlights the importance of auxiliary elements, such as HMR-I and the tRNAThr gene, in enhancing the association of Sir silencing proteins with appropriate genomic locations, thereby buffering the capacity of silenced chromatin to assemble under suboptimal conditions.
SILENCED chromatin and some other specialized chromatin states have the capacity to propagate along a chromosome. This ability is important for assembling extended chromatin domains but must be controlled to prevent one chromatin domain from encroaching into another. The prevailing model of the spreading process postulates that silencing proteins are first recruited to specific sequences, termed silencers, and then spread from these sites through interactions with nucleosomes (Hoppe et al. 2002; Rusche et al. 2002; Grewal and Elgin 2007). Thus, the primary role of silencers is thought to be the nucleation of silenced chromatin, and the spreading process is thought to occur independently of the silencer. However, we recently discovered that at least one silencer in Saccharomyces cerevisiae can promote the assembly of silenced chromatin at a step after recruitment (Lynch and Rusche 2009). We have now compared the abilities of various silencers and combinations of silencers to shape the steady-state distributions of silencing proteins in the yeast S. cerevisiae and find that silencers differ in their abilities to promote the steady-state association of silencing proteins with neighboring nucleosomes.
In S. cerevisiae, domains of silenced chromatin are found at the silent mating-type, loci (HMR and HML) and at most telomeres (reviewed in Rusche et al. 2003). At the silent mating-type loci, silenced chromatin maintains haploid cell identity by preventing the expression of extra copies of the mating-type genes. This silenced chromatin also protects HMR and HML from cutting by the HO endonuclease that triggers mating-type switching (reviewed in Haber 1998). The role of silenced chromatin at subtelomeric domains is less well understood, although it is speculated to contribute to the stability of the ends of the chromosomes.
The structural components of silenced chromatin are the silent information regulator (SIR) proteins, Sir2p, Sir3p, and Sir4p, which are recruited to the chromosome by silencers. At the HMR locus, the HMR-E and HMR-I silencers flank auxiliary copies of the a mating-type genes, and at the HML locus the HML-E and HML-I silencers flank copies of the α mating-type genes. Each of these four silencers is composed of binding sites for the origin recognition complex (ORC) and for Rap1p, Abf1p, or both. In contrast, multiple Rap1p binding sites embedded within the terminal telomeric repeats facilitate the recruitment of Sir proteins to the chromosome ends. In addition, binding sites for ORC and Abf1p occur in the “core X” subtelomeric element associated with all telomeres, and these binding sites contribute to silencing at some (Fourel et al. 1999; Pryde and Louis 1999) but not all telomeres (Mondoux and Zakian 2007). Following recruitment to silencers, the Sir protein complex is thought to spread along the chromosome by repetitive cycles of histone deacetylation and binding in a process referred to as sequential deacetylation (Hoppe et al. 2002; Rusche et al. 2002). Sir2p, a histone deacetylase, generates hypoacetylated nucleosomes that are preferentially bound by Sir3p and Sir4p, which in turn recruit an additional molecule of Sir2p to the chromatin (Hoppe et al. 2002; Rusche et al. 2002). Thus, the Sir protein complex is predicted to spread away from the silencers in a linear step-wise manner.
Our discovery that the HMR-E silencer promotes the establishment of silenced chromatin over several kilobases more efficiently than does the telomeric repeat at chromosome VI-R (Lynch and Rusche 2009) suggests that spreading may not be strictly linear. We hypothesize that proteins associated with the HMR-E silencer promote a looped or compact arrangement of the chromatin fiber that brings the silencer-associated Sir complex into close proximity with multiple nucleosomes at once, such that multiple nucleosomes can be deacetylated independently of one another, enabling assembly to occur in a nonlinear fashion. Consistent with this model, chromosome conformation capture (3C) experiments have indicated that silenced chromatin adopts either a looped or compact chromatin structure at the HMR locus (Valenzuela et al. 2008; Miele et al. 2009).
To explore how silencers shape the steady-state distribution of Sir proteins at HMR, we have now examined the association of silencing proteins with HMR in the presence and absence of three known regulatory sequences—two silencers and a boundary element. The HMR-E silencer is composed of binding sites for ORC, Rap1p, and Abf1p and is required for silencing at HMR. A second silencer, HMR-I, is not required for the silencing of HMRa1, although it does contribute to the silencing of a reporter gene located in the place of HMRa1 (Rivier et al. 1999). Like HMR-E, HMR-I has binding sites for ORC and Abf1p. However, HMR-I does not have a binding site for Rap1p and cannot recruit Sir proteins to DNA on its own (Rusche et al. 2002). A third important element at HMR is a tRNAThr gene, designated tT(AGU)C in the Saccharomyces Genome Database, which is located ∼1 kb beyond the HMR-I silencer. This gene acts as a boundary to the spread of silenced chromatin (Donze et al. 1999; Donze and Kamakaka 2001; Oki and Kamakaka 2005). Both the recruitment of RNA polymerase III and the depletion of histones in the vicinity of the tRNAThr gene contribute to barrier function (Donze and Kamakaka 2001; Dhillon et al. 2009). Additionally, the tRNAThr gene is important in establishing sister chromatid cohesion at HMR (Dubey and Gartenberg 2007), and cohesins have been implicated in the regulation of silenced chromatin (Lau et al. 2002; Suter et al. 2004). Thus, the HMR-E and HMR-I silencers together with the tRNAThr boundary are thought to shape the domain of silenced chromatin at HMR.
For comparison, we examined the steady-state distribution of Sir proteins at telomere VI-R, where spreading is more likely to occur in a linear fashion. Telomere VI-R is one of the most strongly silenced telomeres (Mondoux and Zakian 2007) and lacks subtelomeric elements known to antagonize silencing, such as the X-combinatorial repeats and Y′ elements. Telomere VI-R does have a core X element, containing ORC and Abf1p binding sites, located within 350 bp of the terminal repeat. It is not known whether ORC and Abf1p contribute to the recruitment of Sir proteins to telomere VI-R, but the deletion of the core X element only reduced silencing of a URA3 reporter by twofold (Mondoux and Zakian 2007). A gene of unknown function, YFR057W, whose promoter is ∼1 kb from the terminal repeat, is silenced by the Sir proteins (Vega-Palas et al. 2000). No other functional elements are known to exist within 5 kb of the terminal repeat at telomere VI-R. Therefore, once Sir proteins have been recruited to the end of the chromosome (through Rap1p associated with the terminal repeat, and perhaps ORC and Abf1p in the adjacent core X element), the propagation of silenced chromatin along the chromosome is not expected to be influenced by either positive or negative elements.
In this study, we analyzed the contributions of each of these regulatory elements to the steady-state distribution of Sir proteins and examined how they contribute to the known biological functions of silencing at HMR. Consistent with our previous report, we discovered that the HMR-E silencer by itself promotes silenced chromatin more efficiently over a region of several kilobases compared to a telomeric repeat. Additionally, we discovered that although the HMR-E silencer alone was sufficient to maintain high levels of Sir proteins and block transcription of the HMR locus, the HMR-I silencer boosts the level of Sir proteins at HMR and modestly extends the domain of silenced chromatin. Intriguingly, we also discovered a role for the tRNAThr gene in promoting Sir protein association with the HMR locus under conditions of weakened silencing.
MATERIALS AND METHODS
Yeast strains and plasmids:
Yeast strains used in this study (Table 1) were derived from W303-1b. The following alleles were described previously: sir2Δ∷TRP1 and sir4Δ∷HIS3 (Rusche and Rine 2001), LEU2∷sir2-N345A (Imai et al. 2000; Armstrong et al. 2002), hmr-ΔE (silencer deletion 358–223, YAB71; and 546–30, YAB65) (Brand et al. 1987), ΔtRNAThr (Donze et al. 1999), and TELVI-R∷Stuffer and TELVI-R∷HMR-E (Lynch and Rusche 2009).
TABLE 1.
Strains used in this study
| Strain | Genotype | Source |
|---|---|---|
| LRY1007 (W303) | MATα ade2-1 can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1 | R. Rothstein |
| LRY0800 | W303 MATα sir2Δ∷TRP1 LEU2∷sir2-N345A | |
| LRY0804 | W303 MATα LEU2∷sir2-N345A | |
| LRY1021 | MATahis4 | P. Schatz |
| LRY1068 | W303 MATα sir2Δ∷TRP1 | |
| LRY1815 | W303 MATα hmr-ΔI LEU2∷sir2-N345A | |
| LRY2148 | W303 MATα TELVIR∷STUFFER | |
| LRY2150 | W303 MATα TELVIR∷HMR-E | |
| LRY2302 | W303 MATα HMR-ΔtRNAThr | |
| LRY2303 | W303 MATα HMR-ΔtRNAThrLEU2∷sir2-N345A | |
| LRY2309 | W303 MATα hmr-ΔI ΔtRNAThr | |
| LRY2352 | W303 MATα hmr-ΔI ΔtRNAThrLEU2∷sir2-N345A | |
| LRY2315 | W303 MATα hmr-ΔI | |
| LRY2316 | W303 MATα hmr-ΔI LEU2∷sir2-N345A | |
| LRY2379 | W303 MATα [pJR831; PGAL1-HO; URA3]a | |
| LRY2384 | W303 MATα hmr-ΔI [pJR831; PGAL1-HO; URA3]a | |
| LRY2467 | W303 MATa [pJR831; PGAL1-HO; URA3]a | |
| LRY2482 | W303 MATα sir4Δ∷HIS3 [pJR831; PGAL1-HO; URA3]a | |
| YAB65 | W303 MATα hmr-ΔE (546–30; 516-bp deletion) | A. Brand |
| YAB71 | W303 MATα hmr-ΔE (358–223; 135-bp deletion) | A. Brand |
Brackets denote transformation with the indicated plasmid. See materials and methods for plasmid details.
To create the HMR-ΔtRNAThr allele used in this study, a previously described ΔtRNAThr deletion was amplified from ROY1681 (Donze et al. 1999) genomic DNA by PCR with Pfu Turbo DNA polymerase (Stratagene) using primer sequences 5′-gcagcttactcccaaga gtgc and 5′-gcaaggattgataatgtggtag. The PCR product was digested with XhoI and NdeI and cloned into a plasmid (pJR1270) bearing the EcoRI–HindIII fragment of HMR, lacking both the HMR-I silencer and a Ty1 LTR, in a pUC18 vector backbone to generate pLR0575. To replace the missing HMR-I silencer and Ty1 LTR with wild-type sequences, wild-type genomic DNA (LRY1007) was amplified by PCR using primer sequences 5′-gatgtgtttgtacatttggc and 5′-tcgacgtcggatttgcg. The PCR product was digested with MfeI and PstI and cloned into pLR0575 to create pLR0667.
To create the hmr-ΔI ΔtRNAThr construct, mutagenesis was performed on pLR0667 using primers 5′-ctttctactgcgataaagttattatttagattacagctagcgaaaatttgtcaacgaagttagagaaag and 5′-ctttctctaacttcgttgacaaattttcgctagctgtaatctaaataataactttatcgcagtagaaag, inserting a NheI site (boldface letters) in place of the HMR-I silencer, to generate pLR0683. The 305-bp deletion of HMR-I was filled with the same size fragment from the TRP1 open reading frame DNA by amplifying wild-type genomic DNA (LRY1007) using primer sequences 5′-cacgatgctagcactccg aaatacttggttggc and 5′-gagtcggctagccttccaacccagtcagaaatc, which contain NheI sites (boldface letters). The PCR product was digested with NheI and cloned into plasmid pLR0683 to generate pLR0690.
Finally, to create the hmr-ΔI construct, pLR0690 was digested with MfeI and PstI and the resulting fragment containing the HMR-I silencer deletion as well as the Ty1 LTR was cloned into pLR0689, which contains the EcoRI–NdeI fragment of HMR including the tRNAThr gene in a pUC18 vector backbone, to generate pLR0691.
To integrate the mutant HMR alleles into their native locus in the yeast genome, plasmids pLR0667, pLR0690, and pLR0691 were digested with EcoRI and NdeI and used to transform a yeast strain in which HMR was replaced by URA3 (LRY2177). Approximately 10 OD equivalents of transformed cells were resuspended in 50 mL of rich medium (YPD) and allowed to recover overnight at 30°. To select for integrants in which the URA3 marker was lost, 2 OD equivalents of cells were plated directly onto medium containing 5-FOA. Correct integration of HMR mutant alleles was confirmed by PCR and Southern blotting.
The plasmid pJR831 contains the HO endonuclease gene under the control of the GAL1 promoter in a YCp50 vector backbone and was a gift from Jasper Rine (University of California Berkeley).
Chromatin immunoprecipitation:
Chromatin immunoprecipitations were performed as previously described (Rusche and Rine 2001) using 10 OD equivalents of cells and 3 μl of rabbit polyclonal antiserum to recombinant LacZ-Sir2p or LacZ-Sir3p (rabbits 2931 and 2934, respectively; gifts from J. Rine, University of California Berkeley). Cells were grown in rich medium (YPD). Samples were collected in logarithmically growing cultures at an OD600 of ∼1.0 (±0.2). Cells were treated with 1% formaldehyde for 20 min to cross-link proteins to DNA, after which the cross-linking reaction was quenched by the addition of glycine to a final concentration of 0.125 m. Quantitative real-time PCR was performed as previously described (Lynch et al. 2005), except PHO5 was used as a control locus. First, the amount of product generated using each primer set was determined compared to a standard curve of input DNA. Then, relative IP values were determined by taking the ratio of the query locus to PHO5 or a silencer, as indicated. Reported values represent averages of two to six independent immunoprecipitations derived from independent cultures. Sequences of the oligonucleotides are given in Table 2.
TABLE 2.
Oligos used in this study
| Region | Sequence 1 | Sequence 2 |
|---|---|---|
| RT–PCR | ||
| NTG1 | caaggttcctcgatttagtg | gactccagatcagacaagaac |
| ACT1 | cagcgcttgcaccatccc | gagcttcatcaccaacgtaggag |
| YFR057w | caatagcctttcaaagcatac | gctttgttacgcttgcacttg |
| HMRa1 | atggaaagtaatttgactaaagtag | ccaaactcttacttgaagtggag |
| GIT1 | gttgctgacgcttcactac | gaagactgctactacagaagtc |
| Chromatin IP | ||
| PHO5 | cttgaacgatgattacgag | caagaagtcacgagcatg |
| HMR (−)1 kb | gcaatgactagagaactatcg | gatctgaaggttcagtaactc |
| HMR-E | gcaatagatcatgtactaaac | ctgcgcttattctcaaacg |
| HMR 1 kb | caatacatctccttatatcaaag | caatctcagtacctagaatg |
| HMR 2 kb | gttgatcataagtctcttc | ctatgtgtttatacaattgc |
| HMR 3 kb | ctacaatgcaaccccac | tcgacgtcggatttgcg |
| HMR 4 kb | gcgcagctatttcaattttgg | caattctaacataggatggcag |
| HMR 5 kb | cattcgacgcctactacagaac | gtaatgctggaccaggtgatatg |
| HMR 6 kb | cattgcgtccggttttgctc | gttggaaactctagtcgacac |
| TEL6R 0 kb | ctgagttcggatcactacacac | gatcattgaggatctataatcaac |
| TEL6R 1 kb | gtaggaatgcgaaaggatctgtc | gtgctaaaggaatccccagagac |
| TEL6R 2 kb | gacggaaagagggcagaaag | cagcgcacgtttgtttgatg |
| TEL6R 3 kb | gagttttgtagtagcgatccgac | gtagtgtaaccataagaaatccag |
| TEL6R 4 kb | cgtacttagagtaaccatagc | cagcaaaataaccactggtgtttaag |
| TEL∷HMR-E 0 kb | gcaatagatcatgtactaaac | gtggatgcacagttcagag |
| TEL∷HMR-E 1 kb | gaccttcataggatgtaagtag | catatcactaacttctctcagatc |
| TEL∷HMR-E 2 kb | gacggaaagagggcagaaag | cagcgcacgtttgtttgatg |
| TEL∷HMR-E 3 kb | gagttttgtagtagcgatccgac | gtagtgtaaccataagaaatccag |
| TEL∷STF 1 kb | caatagcctttcaaagcatac | gctttgttacgcttgcacttg |
| TEL∷STF 2 kb | caaattgcaggcaaataaacac | gcatgatgatccccaataac |
| TEL∷STF 3 kb | gacatgaatctcctatcgttc | gataaatggacctgtccttc |
Reverse transcriptase PCR:
RNA was isolated from logarithmically growing cells via the hot phenol method (Schmitt et al. 1990). To remove contaminating DNA, RNA was treated with rDNase I using a DNA-free kit (Ambion). RNA was converted to cDNA using M-MLV reverse transcriptase (Invitrogen) as previously described (Hickman and Rusche 2007), with the exception that 400 ng of DNase-treated RNA was used in each reaction. The resulting cDNA was quantified by real-time PCR using genomic DNA isolated from wild-type yeast (LRY1007) for the standard curve. Transcript levels of query genes were normalized to the transcript levels of the control gene NTG1. Reported values represent the averages of four independent RNA samples, each analyzed in duplicate PCR reactions.
Mating assay:
One optical density equivalent of logarithmically growing MATα haploid cells was collected by centrifugation and resuspended in 100 μl of minimal medium (YM). A 10-fold dilution series was prepared in YM, and 3 μl from each dilution was spotted onto rich medium (YPD) to verify that all samples were diluted equivalently. To assess mating proficiency, an equivalent volume of MATa tester cells (LRY1021), suspended in rich medium (YPD) at a dilution of 10 OD equivalents per mL, was added to the dilution series. A total of 3 μl of the resulting mixture was spotted onto minimal medium (YM) to select for prototrophic diploids. To assay mating in the presence of nicotinamide, MATα haploid cells were grown to late log phase in medium supplemented with nicotinamide (NAM) (Sigma) and plated with MATa tester cells on minimal medium containing NAM. Plates were imaged after 2 days at 30°.
HO endonuclease cleavage assay:
For experiments testing DNA cutting by the HO endonuclease, cells were grown in selective, supplemented medium lacking uracil (CSM) (MP Biomedicals) in 2% raffinose. Cells were brought to an OD600 of ∼0.8 (±0.1), and then arrested in S phase by the addition of hydroxyurea (HU) (US Biological) directly to the medium at a final concentration of 200 mm for 4 hr. For the induction of PGAL1-HO endonuclease, galactose was added to the medium at a final concentration of 2%. Cells were collected at various times after the addition of galactose.
To detect cleavage by HO endonuclease, genomic DNA was isolated from cells by phenol extraction. Approximately 40 to 45 μg of genomic DNA was digested overnight at 37° with HindIII at a concentration of 2 units per μg DNA. DNA fragments were separated on 0.7% agarose gels, depurinated in 0.25 m HCl for 8 min, denatured in 0.5 m NaOH and 1 m NaCl for 30 min, and neutralized in 0.5 m Tris (pH 7.4) and 3 m NaCl for 30 min. DNA was transferred to Zeta Probe nylon membranes (Bio-Rad) by capillary action. For sequence specific hybridization, DNA probes were generated by PCR using total yeast genomic DNA as a template. Probes were labeled with [α-32P] dCTP using the RediPrime II DNA labeling kit (Amersham).
RESULTS
HMR-E increases the association of Sir proteins over a region of several kilobases:
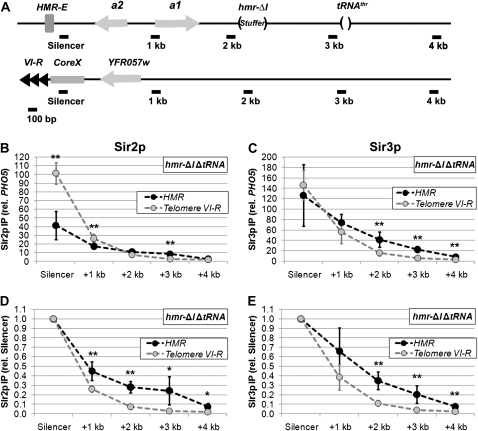
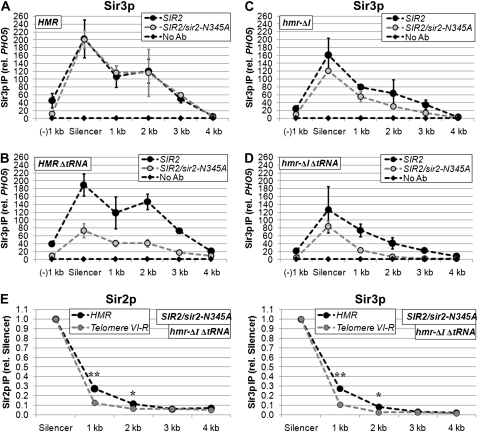
Our previous work demonstrated that, although Sir proteins are recruited to the HMR-E silencer and the terminal repeats of telomere VI-R at similar rates, they assemble more rapidly across the entire HMR locus than they do across a similarly sized domain adjacent to telomere VI-R (Lynch and Rusche 2009). Moreover, this faster rate of assembly at HMR does not depend on the secondary silencer HMR-I (Lynch and Rusche 2009). We hypothesized that this increased rate of assembly reflects the ability of the HMR-E silencer to promote the association of Sir proteins with nucleosomes over a region of a few kilobases. A prediction of this model is that at steady state, the enrichment of Sir proteins would be elevated at sites within a few kilobases of HMR-E compared to sites at similar distances from telomere VI-R. To test this prediction, we compared Sir protein levels at equivalent distances from the HMR-E silencer and telomere repeat by chromatin immunoprecipitation. To eliminate the potential effects of HMR-I and the tRNAThr gene, these elements were deleted (Figure 1A). Similar levels of Sir3p associated with the two silencer sequences (Figure 1C), yet consistently higher levels of Sir3p, were associated with sites 1, 2, or 3 kb from HMR-E compared to sites at similar distances from the telomeric repeats. In the case of Sir2p, a significantly higher enrichment was detected at the telomeric repeat compared to the HMR-E silencer (Figure 1B). Nevertheless, Sir2p levels were comparable 2 kb from the sites of recruitment, and there was actually more Sir2p 3 kb from the HMR-E silencer than 3 kb from the telomere repeat. To facilitate the comparison of the two silencers' abilities to promote the assembly of silenced chromatin, Sir protein levels were normalized to the levels at the respective silencers (Figure 1, D and E). For both Sir2p and Sir3p, the decline in Sir protein association over a given distance was less steep when silenced chromatin was initiated by HMR-E compared to telomere VI-R. Therefore, although the telomeric repeat is as effective as HMR-E at recruiting Sir proteins (if not more so), the telomeric repeat is less efficient at promoting the association of Sir proteins with neighboring nucleosomes.
Figure 1.—
HMR-E increases the enrichment of Sir proteins over a region of several kilobases. (A) Diagrams of HMR and subtelomere VI-R. Black bars indicate locations of amplicons used to quantify DNA isolated by chromatin IP. The approximate distances (in kilobase pairs) from the adjacent silencers are given. The HMR locus contains a previously described 85-bp deletion of the tRNAThr gene (Donze et al. 1999) and a 305-bp deletion of the HMR-I silencer, which is filled in with an equivalent length DNA from the TRP1 open reading frame (stuffer). In addition, a 448-bp Ty1 long-terminal repeat sequence located between the HMR-I silencer and the tRNAThr gene, which is missing in most modified HMR loci (Rusche et al. 2002), was restored. (B) Association of Sir2p with telomere VI-R and an HMR locus lacking the HMR-I silencer and tRNAThr gene. DNA coprecipitated with Sir2p from strain LRY2309 was analyzed by quantitative real-time PCR using the amplicons shown in A. Values represent the average of five independent immunoprecipitations analyzed in duplicate PCR reactions and normalized to a control locus (PHO5). An unpaired t-test was used to determine whether the enrichments were significantly different at equivalent distances from the telomere repeat and HMR-E. One asterisk indicates a *P-value < 0.05; **P-value < 0.01. (C) Association of Sir3p with telomere VI-R and an HMR locus lacking the HMR-I silencer and tRNAThr gene. DNA associated with Sir3p was isolated and analyzed in the same experiments as in B. (D and E) The relative enrichments of Sir2p and Sir3p with telomere VI-R and the modified HMR locus were normalized to the adjacent silencers rather than PHO5.
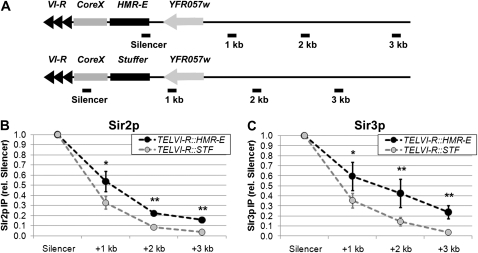
One explanation for the difference between the two silencers is genomic context. To determine whether HMR-E also promotes the association of Sir proteins with neighboring nucleosomes in the context of telomere VI-R, we examined a strain in which a 430-bp fragment containing the HMR-E silencer was integrated adjacent to telomere VI-R (Figure 2A). In this strain, higher enrichments of Sir proteins were observed over a region of several kilobases compared to a control strain, which had an equivalent-sized piece of silencing-neutral stuffer DNA (Figure 2, B and C). At this modified telomere, the terminal repeats and core X element likely cooperated with HMR-E to recruit Sir proteins to the chromosome. Nonetheless, the levels of Sir protein enrichment observed at the telomere-localized HMR-E silencer were similar to the levels at the telomeric repeat sequences in the control strain (supporting information, Figure S1), suggesting that the efficiency of Sir protein recruitment was comparable in the two strains. Therefore, the HMR-E silencer promoted the association of Sir proteins with neighboring nucleosomes over a region of several kilobases to a greater extent than did the terminal repeat at telomere VI-R.
Figure 2.—
A transposed HMR-E silencer increases the association of Sir proteins at telomere VI-R. (A) Diagrams of modified telomere VI-R loci. Either the entire HMR-E silencer (431 bp) or an equivalent-sized fragment of the TRP1 ORF was integrated into telomere VI-R. (B) Relative association of Sir2p with modified telomere VI-R loci. DNA associated with Sir2p was isolated from strains LRY2150 and LRY2148, (TELVI-R∷HMR-E and TELVI-R∷STF, respectively). Sir2p-associated DNA was quantified by real-time PCR using the indicated amplicons. Data were analyzed as in Figure 1, D and E and represent the averages of two independent immunoprecipitation experiments and at least four PCR reactions. (C) Relative association of Sir3p with modified telomere VI-R loci. Sir3p-associated DNA was isolated in the same experiments and analyzed as in B.
It is well established that silencing is less easily disrupted at HMR than at telomeres, and it has been assumed that this difference results from HMR-E having a greater ability to recruit Sir proteins. However, our results indicate that, if anything Sir proteins are recruited at higher levels immediately adjacent to telomere VI-R compared to HMR-E. Therefore, the critical difference between HMR-E and the telomere repeat must be in the ability of HMR-E to promote the association of Sir proteins with neighboring nucleosomes.
The HMR-I silencer increases the association of Sir proteins within HMR:
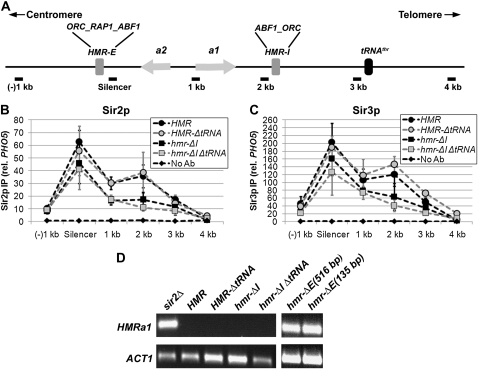
The differences in the abilities of HMR-E and the terminal repeat at telomere VI-R to promote the distribution of Sir proteins over several kilobases may reflect the different roles of these two loci in the biology of S. cerevisiae. The repression of HMRa1 is thought to be critical for an individual cell to mate. In contrast, no deleterious consequence is known to arise from the expression of YFR057w, a gene subject to Sir-mediated repression at the native telomere VI-R (Vega-Palas et al. 2000). Thus, it may be more important to have high levels of Sir proteins distributed across HMR to maintain repression. Consistent with the importance of repressing HMRa1, two additional elements, HMR-I and a tRNAThr gene, are present at HMR and act in conjunction with HMR-E. To determine how these elements contribute to the silenced domain at HMR, we compared the distributions of Sir2p and Sir3p at wild-type and modified HMR alleles in which the HMR-I silencer and tRNAThr gene were deleted individually and in combination (Figure 1A).
The HMR-I silencer contains binding sites for ORC and Abf1p, both of which interact with Sir proteins and are predicted to stabilize the association of the Sir complex with chromatin. However, HMR-I cannot recruit the Sir complex on its own and is not required for silencing of HMRa1 (Rivier et al. 1999; Rusche et al. 2002). To determine how HMR-I contributes to the distribution of Sir proteins at HMR, the relative enrichments of Sir2p and Sir3p were examined in the presence and absence of HMR-I. The loss of HMR-I resulted in a considerable reduction of Sir2p and Sir3p in the immediate vicinity of the HMR-I silencer (2 kb) and more modest decreases at the other sites (Figure 3, B and C, squares). Therefore, the HMR-I silencer increases the levels of Sir proteins at HMR.
Figure 3.—
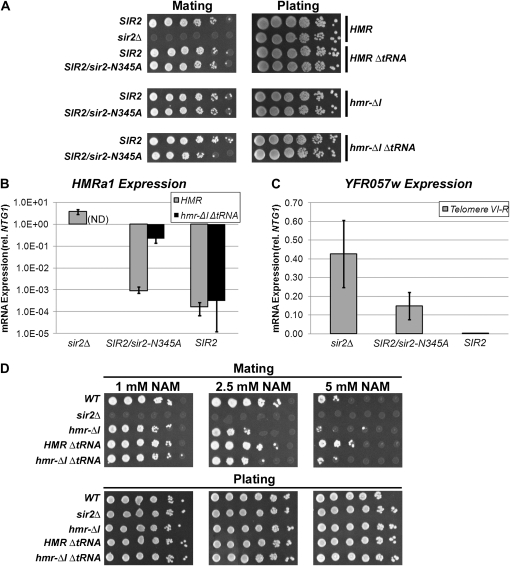
The HMR-I silencer, but not the tRNAThr gene, boosts Sir protein levels within HMR. (A) Diagram of the wild-type HMR locus. (B) Association of Sir2p with HMR in the presence and absence of regulatory elements. Sir2p-associated DNA was isolated by chromatin IP from strains with the following genotypes: wild-type HMR (LRY1007), HMR-ΔtRNAThr (LRY2302), hmr-ΔI (LRY2315), and hmr-ΔI ΔtRNAThr (LRY2309). Data were analyzed as in Figure 1B and represent the averages of at least three independent immunoprecipitation experiments, each analyzed in duplicate quantitative PCR reactions. Primer sets and relative distances were the same for each mutant, with the exception that the +4-kb location is 85 bases closer to the silencer in strains lacking the tRNAThr gene. (C) Association of Sir3p with HMR in the presence and absence of regulatory elements. Sir3p-associated DNA was isolated and analyzed as in B. (D) Transcription of HMRa1 in the presence and absence of regulatory elements. RNA was isolated from the same strains described above, a sir2Δ strain with wild-type HMR (LRY1068) and two different strains with large (YAB65, 516 bp) and small (YAB71, 135 bp) deletions of the HMR-E silencer. The mRNA transcripts were converted to cDNA and the resulting cDNA was amplified by conventional PCR using primer sequences specific to the coding regions of HMRa1 and ACT1. The PCR products were run on 1% agarose gels and visualized by ethidium bromide staining.
The tRNAThr gene acts as a boundary to the spread of silenced chromatin (Donze et al. 1999; Donze and Kamakaka 2001; Oki and Kamakaka 2005) and could shape the distribution of Sir proteins in two ways. First, the tRNAThr gene could prevent the Sir proteins from spreading into the telomere-proximal side of the locus. However, there is little spreading in this direction in its absence (Oki and Kamakaka 2005). In addition, the tRNAThr gene could maintain high levels of Sir proteins within the HMR cassette by preventing euchromatin from encroaching into the locus. For example, targeted histone acetyltransferases have been shown to modify histones across several kilobases (Vignali et al. 2000; Yu et al. 2006) and have been proposed to engage in a spreading reaction analogous to that of Sir proteins (Bulger 2005; Yu et al. 2006). To determine how the tRNAThr gene shapes the distribution of Sir proteins at HMR, the relative enrichments of Sir2p and Sir3p were examined in the absence of the tRNAThr gene. Sir2p and Sir3p levels were comparable to wild type within the HMR cassette (Figure 3, B and C, shaded circles). As expected, slightly higher levels of Sir proteins were observed at a site on the telomere-proximal side of the tRNAThr gene (4 kb from HMR-E), consistent with the reported boundary activity of the tRNAThr gene (Figure 3, B and C; also see ahead to Figure 4). These results indicate that the tRNAThr gene is not critical for maintaining the association of Sir proteins within HMR.
Figure 4.—
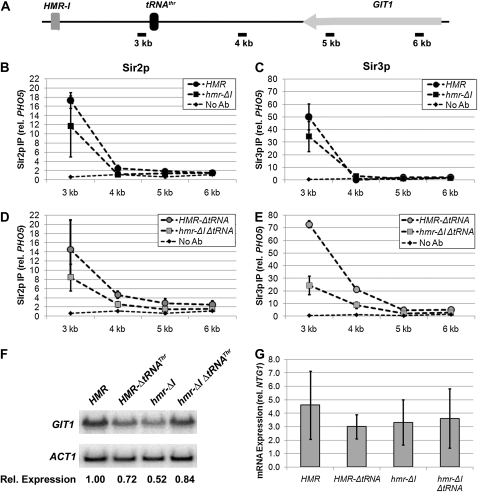
HMR-I modestly extends the range of Sir proteins at HMR. (A) Diagram of the features on the telomere-proximal side of the HMR locus. (B) Relative association of Sir2p with the telomere-proximal side of HMR in the presence and absence of HMR-I. Quantitative PCR was performed on DNA isolated in the same chromatin IP experiments as in Figure 3. Note that the scale of the y-axis differs from Figure 3 to facilitate the examination of these data. (C) Relative association of Sir3p with the telomere-proximal side of HMR, as described for B. (D and E) Relative associations of Sir2p and Sir3p with the telomere-proximal side of the HMR locus in the absence of the tRNAThr gene and in the presence or absence of the HMR-I silencer. (F) Levels of GIT1 mRNA in the presence and absence of regulatory elements, as analyzed by RNA blotting. RNA was isolated from the same strains used above with the indicated modifications at HMR, separated on an agarose gel, and transferred to a nylon membrane. The blot was probed for GIT1 and ACT1 mRNA. The values below each lane reflect the relative levels of GIT1 expression, normalized first to ACT1 and then to the value in wild-type conditions (HMR). (G) Levels of GIT1 cDNA in the presence and absence of regulatory elements, as analyzed by RT–PCR. RNA was isolated from the indicated strains and the mRNA transcripts were converted to cDNA and quantified by real-time PCR using primers specific for GIT1. Transcript levels are shown relative to the control gene (NTG1). Values represent the average of four independent RNA preparations.
It remained possible that the potential ability of the tRNAThr gene to block the encroachment of euchromatin was more important in the absence of the HMR-I silencer, which increases the association of Sir proteins. Therefore, chromatin IP was performed in strains lacking both elements. Under these conditions, there was no significant reduction in Sir protein levels compared to the single deletion of HMR-I (Figure 3, B and C, shaded squares). Therefore, even without the enhancing effect of HMR-I, the tRNAThr gene is not needed to maintain high levels of Sir protein within HMR. We conclude from these results that the HMR-I silencer, but not the tRNAThr gene, is important for maintaining high levels of Sir proteins at HMR.
The HMR-E silencer has been proposed to act in a directional manner (Zou et al. 2006a,b). Consistent with this proposal, even in the absence of both the HMR-I silencer and the tRNAThr gene, Sir proteins were distributed asymmetrically, being higher on the telomere-proximal side of HMR-E (Figure 3, B and C, compare (−)1-kb and 1-kb locations).
The elevated levels of Sir proteins due to the HMR-I silencer are not required for silencing HMRa1:
The decrease in the association of Sir proteins observed in the absence of the HMR-I silencer suggests that transcriptional silencing may be compromised in these strains. However, quantitative mating assays, which indirectly reflect transcription of the HMR locus, revealed no obvious silencing defect in the absence of HMR-I (Rivier et al. 1999; see ahead to Figure 7A). To detect potential rare transcripts from HMR, we performed reverse transcriptase PCR on RNA isolated from strains with and without the HMR-I silencer. Controls revealed that HMRa1 was transcribed as expected in the absence of silencing in a sir2Δ strain but was undetectable by conventional or real-time PCR in the presence of SIR2 (Figure 3D). In the absence of the HMR-I silencer, no HMRa1 cDNA was detected either by conventional PCR (Figure 3D) or real-time PCR. Two control genes, ACT1 and NTG1, could be amplified (Figure 3D), indicating that cDNA synthesis was successful. Thus, HMRa1 remained silenced in the absence of HMR-I despite the reduced association of Sir proteins with the promoter. Similarly, no HMRa1 cDNA was observed in strains lacking the tRNAThr gene alone or in combination with the HMR-I silencer (Figure 3D). In contrast, HMRa1 mRNA was detected in the absence of the HMR-E silencer (Figure 3D and Brand et al. 1987). Therefore, HMR-E, but not HMR-I or the tRNAThr gene, is necessary to silence HMRa1 and maintain haploid cell identity, which is considered to be the critical function of silenced chromatin at the mating type cassettes. Additionally, these data, along with the chromatin IP results, reveal the surprising fact that more Sir proteins get recruited to HMR than are required for silencing.
Figure 7.—
Both the HMR-I silencer and the tRNAThr gene are required to maintain complete silencing of HMRa1 when deacetylation is reduced. (A) Mating ability was assessed by exposing 10-fold serial dilutions of MATα haploids to MATa tester haploids (LRY1021). The resulting diploids were selected on minimal medium. The same strains described in Figure 6 were used, as well as a MATα sir2Δ strain (LRY1068). (B) Levels of HMRa1 in a strain containing wild-type HMR and sir2Δ (LRY1068), and strains containing either wild-type HMR or hmr-ΔI ΔtRNAThr in the presence of both Sir2p and Sir2-N345Ap (LRY0804 and LRY2352) or only Sir2p (LRY1007 and LRY2309). HMRa1 levels were not assayed in a strain containing hmr-ΔI ΔtRNAThr and sir2Δ (ND). RNA was isolated from the indicated strains and the mRNA transcripts were converted to cDNA and quantified by real-time PCR using primers specific for the HMRa1. Transcript levels are shown relative to the control gene (NTG1). Values represent the average of four independent RNA preparations and are plotted on a logarithmic scale. (C) Relative levels of YFR057w mRNAs were quantified in strains with sir2Δ (LRY1068), SIR2 and sir2-N345A (LRY0804), and wild-type SIR2 (LRY1007). Data from four independent RNA isolations are plotted as in B. (D) Mating ability was assayed upon exposure to the given concentrations of nicotinamide (NAM) in the same strains as in A.
HMR-I modestly extends the range of Sir proteins on the telomere-proximal side of HMR:
The observation that the HMR-I silencer increases the association of Sir proteins in its vicinity (Figure 3, B and C) raises the possibility that the HMR-I silencer enables the Sir proteins to propagate significantly farther along the chromosome. In this case, a boundary element may be important to block the extension of silenced chromatin. To determine whether the HMR-I silencer promotes the assembly of Sir proteins on its telomere-proximal side, we measured the levels of Sir proteins on the telomere-proximal side of HMR in the presence and absence of the silencer. When the HMR-I silencer was present, a modest enrichment of Sir proteins was observed at the 3-kb site (Figure 4, B and C; the scale of the y-axis is different than in Figure 3), suggesting that the HMR-I silencer has some ability to extend the range of silenced chromatin. A slight enrichment of Sir proteins was also observed at the 4-kb site beyond the boundary, in the presence but not the absence of HMR-I (Figure 4, B and C).
To observe the potential extension of Sir chromatin over a greater distance, the chromatin IP was repeated in the absence of the tRNAThr gene. Consistent with previous observations (Oki and Kamakaka 2005; Dhillon et al. 2009), in the absence of the tRNAThr gene, Sir2p and Sir3p levels were slightly elevated at the 4-kb site (Figure 4, compare D and E to B and C). However, Sir proteins were not as elevated in the absence of the HMR-I silencer, again indicating that the HMR-I silencer promotes a modest extension of the Sir proteins on its telomere-proximal side (Figure 4, D and E).
In the strains lacking the tRNAThr gene but retaining HMR-I, Sir protein levels were also slightly elevated over background at the 5- and 6-kb locations, which reside in the open reading frame of the GIT1 gene (Figure 4A, see also Oki and Kamakaka 2005; Dhillon et al. 2009). This observation suggests that the expression of the GIT1 gene, which is reported to be repressed in the absence of the tRNAThr gene (Donze and Kamakaka 2001; Oki and Kamakaka 2005), may be less affected in the absence of the HMR-I silencer. To test this possibility, we examined the level of GIT1 mRNA in the presence and absence of HMR-I by RNA blotting and quantitative RT–PCR. Slight fluctuations in GIT1 expression were observed in each of the mutants tested (Figure 4, F and G). However, GIT1 levels in these strains were all within twofold of wild-type levels, and no statistically significant differences were observed. Therefore, we conclude that the occasional spread of Sir proteins into the GIT1 open reading frame, as occurs in the absence of the tRNAThr gene, has little impact on the expression of GIT1.
HMR is resistant to HO endonuclease in the absence of HMR-I:
HMR-I clearly enhances the association of Sir proteins with HMR but is not required for the transcriptional repression of the HMRa1 gene. In addition to silencing of mating-type genes, which is critical to maintaining haploid cell identity, a second function of silenced chromatin at the HM loci is to protect DNA from being cut by the HO endonuclease during mating-type switching (reviewed in Haber 1998). S. cerevisiae cells change their mating type by site-directed recombination. The expression of the HO endonuclease during G1 of the cell cycle results in the formation of a double strand break at MAT, which is subsequently repaired via recombination with the HM locus of opposite mating type (reviewed in Haber 1998). Recombination is facilitated by conserved sequences found at all three locations (MAT, HML, and HMR) that include the recognition site for the HO endonuclease (Figure 5A). It is thought that silenced chromatin protects the HM loci from being cut by the HO endonuclease, ensuring that recombination only occurs at MAT (Klar et al. 1981; Strathern et al. 1982; Loo and Rine 1994).
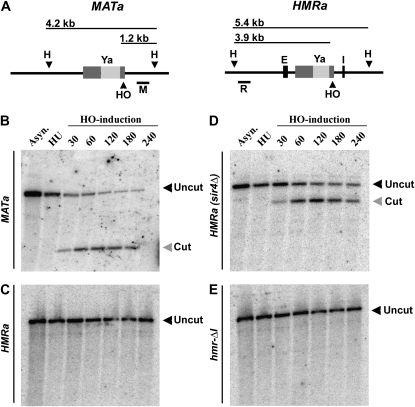
Figure 5.—
HMR is resistant to HO-endonuclease digestion in the absence of HMR-I. (A) Diagram of the expected DNA fragment sizes generated by HO endonuclease (HO) and HindIII (H) digestion at MATa and HMRa. Bars below the schematics represent the locations of DNA probes used for hybridizations to MAT (M) and HMR (R). Light shaded bands indicate a-gene specific sequence. Dark shaded bars represent sequences found at MAT, HMR, and HML, which include the recognition sequence for the HO endonuclease. (B) Time course of cutting by HO at the MATa locus. Samples were collected from a wild-type MATa strain (LRY2467) in asynchronously growing conditions (Asyn.), after S-phase arrest in HU, and at various times following induction of HO. Genomic DNA was isolated, digested with HindIII, and analyzed by Southern blotting. (C–E) Time courses of cutting by HO at the HMRa locus in strains of (C) wild-type HMR (LRY2379), (D) sir4Δ (LRY2482), and (E) hmr-ΔI (LRY2384).
The HO recognition site at HMR is <190 bases from the HMR-I silencer (Nickoloff et al. 1986). To address whether the increase in Sir protein association mediated by the HMR-I silencer helps protect HMR from HO-endonuclease digestion, we examined the ability of HO to cleave this site in a strain lacking HMR-I. We used a previously described assay in which cut intermediates are stabilized by arresting the cells with hydroxyurea (HU) (Connolly et al. 1988). Under these conditions, repair of double-strand breaks by homologous recombination is inhibited by the DNA replication checkpoint (Alabert et al. 2009). After arrest in HU, HO endonuclease was induced by the addition of galactose to the medium. Samples were collected at different times following induction, and genomic DNA was isolated, digested with HindIII (H), and analyzed by Southern blotting using probes specific to MAT (M) or HMR (R) (Figure 5A). As a control, we examined the ability of HO to cut at MATa and wild-type HMRa. Cleavage of the MATa locus was detected within 30 min of induction of HO (Figure 5B). In contrast, no cutting was detected at wild-type HMRa, even after 4 hr of HO induction (Figure 5C). In the absence of Sir proteins, HMRa was cut with similar kinetics to those observed at the unprotected MATa locus, as expected (Figure 5D). However, cleavage was not observed in the absence of HMR-I (Figure 5E). Therefore, the HMR-I-mediated increase in Sir protein levels at HMR was not necessary to protect the adjacent HO recognition sequence from being cut.
The HMR-I silencer and tRNAThr gene cooperate to maintain Sir proteins at HMR when deacetylation is reduced:
The HMR-I silencer and tRNAThr gene are not required to maintain transcriptional repression of HMRa1, but their presence at the HMR locus suggests they have a biological function. One situation in which the increased association of Sir proteins at HMR-I might be important is when the Sir2p deacetylase has reduced activity. The deacetylase activity of Sir2p requires NAD+ (Imai et al. 2000; Landry et al. 2000; Smith et al. 2000) and is inhibited by nicotinamide (Bitterman et al. 2002). Sir2p activity may be reduced by fluctuations in the intracellular levels of these metabolites. To mimic reduced deacetylase activity, we coexpressed enzymatically inactive and wild-type alleles of SIR2, which should result in enzymatic inactivity for approximately half of Sir2p molecules. We previously found that the HMR locus is unaffected by this condition, but that silenced chromatin at telomere VI-R is disrupted (Lynch and Rusche 2009), as is repression of a reporter gene at telomere VII-L (Armstrong et al. 2002). This phenotype is not simply due to doubling the amount of Sir2p because two copies of the wild-type gene do not affect silencing at telomere VII-L (Armstrong et al. 2002).
To determine whether HMR-I or the tRNAThr gene contributed to the resistance of HMR to reduced deacetylase activity, Sir protein levels at HMR were assessed by chromatin IP in the presence of only wild-type SIR2 or both SIR2 and sir2-N345A (Figure 6 for Sir3p and Figure S2 for Sir2p). Remarkably, expression of sir2-N345A resulted in a profound decrease in Sir protein enrichment in the absence of the tRNAThr gene (Figure 6B). In contrast, a relatively modest, albeit reproducible, decrease in Sir protein association was observed in the absence of HMR-I (Figure 6C). In the absence of both the HMR-I silencer and the boundary, a more severe decrease in Sir3p distribution was observed than in either single mutant, with total Sir3p levels approaching background within 2–3 kb of the HMR-E silencer. As previously reported (Lynch and Rusche 2009), no loss of Sir3p was observed when the sir2-N345A mutant was expressed in a wild-type HMR strain. Therefore, the tRNAThr gene and the HMR-I silencer together collaborate with the HMR-E silencer to prevent the loss of Sir proteins when deacetylation is reduced.
Figure 6.—
The HMR-I silencer and tRNAThr gene help maintain silenced chromatin at HMR when deacetylase activity is reduced. (A) Sir3p association with HMR in the presence of either wild-type Sir2p alone (LRY1007) or Sir2p and catalytically inactive Sir2-N345Ap (LRY0804). Values in A–D represent the averages of at least three independent experiments analyzed as in Figure 1B. (B) Sir3p association with HMR-ΔtRNAThr in the presence of Sir2p alone (LRY2302) or Sir2p and Sir2-N345Ap (LRY2303). (C) Sir3p association with HMR-ΔI in the presence of Sir2p alone (LRY2315) or Sir2p and Sir2-N345Ap (LRY2316). (D) Sir3p association with HMR-ΔI ΔtRNAThr in the presence of Sir2p alone (LRY2309) or Sir2p and Sir2-N345Ap (LRY2352). (E) The relative enrichments of Sir2p and Sir3p with telomere VI-R and the modified HMR locus in the presence of Sir2-N345Ap (LRY2352). Data were analyzed as in Figure 1, D and E.
To determine whether HMR-E on its own retained an ability to promote the assembly of silenced chromatin over a distance under conditions of reduced deacetylase activity, we compared the distributions of Sir proteins adjacent to HMR-E or the telomeric repeat in the hmr-ΔI ΔtRNAThr strain. Indeed, although the association of Sir2p and Sir3p was reduced at both telomere VI-R and HMR in the presence of Sir2-N345Ap (Figure 6E), the level of Sir proteins 1 or 2 kb from the silencer represented a greater fraction of the level observed at the silencer at HMR compared to the telomere, suggesting that the HMR-E silencer provides some additional resistance to compromised deacetylation.
The HMR-I silencer and tRNAThr gene cooperate to maintain transcriptional silencing when deacetylation is reduced: To determine whether the reduction in Sir proteins associated with hmr-ΔI ΔtRNAThr under conditions of reduced deacetylase activity (Figure 6D) impacts silencing of HMRa1, transcription was assessed in two ways. First, a mating assay was conducted. Only when HMRa1 is silenced will MATα strains mate. MATα haploids containing different HMR alleles in combination with different SIR2 alleles were mixed with haploids of the opposite mating type and plated on medium selective for diploids. Neither the deletion of the HMR-I silencer nor the tRNAThr gene alone resulted in a detectable defect in mating (Figure 7A). However, in the absence of both HMR-I and the tRNAThr gene, an ∼10-fold defect in mating was observed in the presence of Sir2-N345Ap (Figure 7A, bottom row). To detect HMRa1 transcripts, cDNA was prepared from each of the strains and quantified by real-time PCR. Consistent with the mating assay, HMRa1 transcripts were close to the limit of detection in strains with the wild-type HMR or hmr-ΔtRNAThr alleles (Figure 7B and data not shown). In the absence of HMR-I, a very slight derepression of HMRa1 occurred in the presence of Sir2-N345Ap, although the levels of HMRa1 were ∼1% of those in a sir2Δ strain (data not shown). However, in the absence of both HMR-I and tRNAThr, the coexpression of Sir2p and Sir2-N345Ap resulted in derepression of HMRa1 levels at ∼7% of a sir2Δ strain (Figure 7B). Therefore, the HMR-I silencer and tRNAThr gene together maintain repression of HMRa1 when deacetylation is compromised.
To determine whether the HMR-I silencer and tRNAThr gene help maintain silencing of HMRa1 in environmental conditions that reduce deacetylation, mating was assessed in the presence of nicotinamide, which inhibits Sir2p (Bitterman et al. 2002). Indeed, in the presence of 5 mm nicotinamide, mating was significantly reduced in each of the strains tested (Figure 7D). However, in the absence of HMR-I, the defect in mating was 10- to 100-fold more severe compared to wild-type HMR. In contrast, in the absence of the tRNAThr gene, mating occurred at levels similar to those observed in strains containing wild-type HMR. Additionally, no further mating defect was observed in the absence of both HMR-I and the tRNAThr gene than in the absence of HMR-I alone. Therefore, the HMR-I silencer helps maintain efficient silencing of HMR in conditions of reduced Sir2p activity.
Silencing at telomeres is known to be inhibited by the coexpression of Sir2-N345Ap and Sir2p (Armstrong et al. 2002; Lynch and Rusche 2009). To assess the ability of telomere VI-R to maintain transcriptional silencing in the presence of Sir2-N345Ap, we measured the level of YFR057w mRNA in the presence and absence of Sir2-N345Ap (Figure7C). In the presence of Sir2-N345Ap, YFR057w expression was partially derepressed to ∼35% of the level in a sir2 mutant (Figure 7C). This level of induction is greater than was observed for HMRa1 in an hmr-ΔI ΔtRNAThr strain (7% of the level in a sir2 mutant; Figure 7B), consistent with silencing being more easily disrupted at telomere VI-R than HMR, although there are several caveats to this interpretation, given that different genes with different promoters are being compared.
DISCUSSION
HMR-E increases the association of Sir proteins over several kilobases:
Historically, it has been thought that silencers act by recruiting silencing proteins to a particular site in the genome and that the spreading of silencing proteins along the chromosome occurs independently of the silencer. However, we found that the HMR-E silencer acts by a process distinct from recruitment to enable the rapid establishment of silenced chromatin over several kilobases (Lynch and Rusche 2009). These initial studies focused on the establishment of silencing following the induction of high levels of Sir3p, and it was important to investigate how HMR-E and other silencers shape the steady-state distribution of Sir proteins expressed at endogenous levels. We now report that HMR-E maintains Sir proteins over several kilobases at higher levels relative to the silencer than does the telomeric repeat at chromosome VI-R. This increased level of Sir proteins is observed both when HMR-E is in its native location at HMR (Figure 1) and when it is translocated to telomere VI-R (Figure 2). Moreover, the enhanced association of Sir proteins is observed in the absence of the auxiliary silencer HMR-I (Figures 1 and 2), indicating that HMR-E achieves this increase on its own. In addition, the association of Sir proteins is enhanced on one side of HMR-E compared to the other (Figure 3). Therefore, in addition to accelerating the rate of Sir protein assembly (Lynch and Rusche 2009), the HMR-E silencer increases the steady-state association of Sir proteins over several kilobases in a directional manner.
The mechanism by which HMR-E enhances the association of Sir proteins remains to be determined. One possibility is that proteins associated with HMR-E favor the formation of a higher-order arrangement of the chromatin fiber, such as a looped or compact structure, and that this arrangement enables assembly to occur in a nonlinear, and hence more efficient, fashion. We observed a gradual decrease in Sir protein association as a function of distance from the HMR-E silencer (Figures 1, 2, and 3), a result inconsistent with the HMR-E silencer facilitating the formation of a single, defined higher-order structure. Instead, transient interactions between silencer-associated proteins and nearby nucleosomes may result in the formation of a set of related structures that enhance the assembly of silenced chromatin in a distance-dependent fashion. Moreover, the asymmetrical distribution of Sir proteins around HMR-E (Figure 3; hmr-ΔI ΔtRNA strain) could reflect a tendency of proteins at the HMR-E silencer to interact more frequently with sequences on the Abf1-binding side of the silencer, preferentially generating higher-order chromatin structures within the HMR cassette.
HMR-I impacts Sir protein levels at HMR but does not affect transcriptional silencing:
The Abf1p and ORC-binding sequences of HMR-I are conserved in a related species of yeast, S. paradoxus, although the surrounding sequences are highly diverged (Teytelman et al. 2008). Therefore, HMR-I probably has a biological function that positively impacts the fitness of a yeast cell. In keeping with this hypothesis, we found that the HMR-I silencer does elevate the levels of Sir proteins within the HMR cassette (Figure 3) and helps maintain the Sir proteins at HMR when deacetylase activity is reduced (Figure 6C). However, despite the reduced association of Sir proteins in the absence of HMR-I, transcriptional silencing remained effective (Figure 3D; Brand et al. 1987). Similarly, we found that HMR remained protected from cleavage by the HO endonuclease in the absence of HMR-I (Figure 5E). Thus, it remains unclear how the increased association of Sir proteins with HMR due to HMR-I contributes to the biological function of this silencer. One possibility is that HMR-I protects the locus against loss of silencing under suboptimal conditions, such as a reduction in deacetylase activity, as discussed below.
It is striking that the HMR cassette appears to recruit more Sir proteins than are necessary to maintain silencing. In fact, transcriptional repression remains strong when the association of Sir proteins with the a1 promoter is reduced to one-quarter of the maximum observed at the HMR-E silencer (Figure 6). Although it is not clear whether every cell is affected similarly by the loss of HMR-I given that the enrichments of Sir proteins observed by chromatin IP represent an average over the population, a fair number of cells must be depleted of Sir proteins at the promoter. Therefore, Sir proteins may not need to be present continuously throughout HMR to block transcription and thus may not act by hindering access of RNA polymerase II to the promoter, as was originally proposed. Instead, Sir proteins may generate modifications of histones that persist even when Sir proteins dissociate from the promoter. Alternatively, Sir proteins associated with the HMR-E silencer may act from a distance to prevent RNA polymerase II from initiating transcription. A final possibility is that Sir proteins, which have the capacity to multimerize, are normally present in “super-stoichiometric” amounts and thus, even with the decrease observed in the absence of HMR-I, every promoter remains associated with Sir proteins.
The tRNAThr gene plays both positive and negative roles in the regulation of silencing:
The tRNAThr gene at HMR is notable as one of the few characterized boundary elements in S. cerevisiae. tRNA genes have also been shown to separate heterochromatin from other chromatin domains at centromeres and mating-type loci in Schizosaccharomyces pombe (Noma et al. 2006; Scott et al. 2006, 2007) and to block upstream activator sequences from acting on promoters in S. cerevisiae (Simms et al. 2008), indicating that these genes may have conserved functions in partitioning domains of chromatin. However, our analysis reveals that, although the tRNAThr gene does block the spread of Sir proteins, it probably is not the major mechanism controlling the extent of silenced chromatin. In the absence of the tRNAThr gene, Sir2p and Sir3p were only marginally enriched on the telomere-proximal side of HMR (Figure 4). Therefore, it appears that silenced chromatin naturally decays over a distance of 1 or 2 kb even without encountering a specific boundary element. The tRNAThr gene thus serves as a backup mechanism to check the propagation of silenced chromatin when it extends beyond its usual limit.
We also found that the tRNAThr gene helped maintain Sir proteins within the HMR cassette when Sir2p activity was compromised (Figure 6, B and D), although it had no effect on the distributions of Sir2p or Sir3p in the presence of wild-type SIR2 (Figure 3, B and C). At least two mechanisms could account for this role of the tRNAThr gene in promoting the association of Sir proteins with HMR. One possibility is that the tRNAThr gene blocks the spread of euchromatin, and in particular acetyltransferases, into the silenced locus. It has been suggested that acetyltransferases participate in a spreading reaction similar to that of Sir proteins (Bulger 2005). For example, acetyltransferases targeted to a particular sequence can acetylate histones over several kilobases (Vignali et al. 2000; Yu et al. 2006), and this long-range acetylation is disrupted by nucleosome excluding sequences (Yu et al. 2006). Thus, the tRNAThr gene may block the spread of euchromatin into HMR. This effect may be particularly pronounced in the absence of HMR-I, which helps to maintain Sir proteins at HMR (Figure 6D).
A second possibility is that the ability of the tRNAThr gene to recruit cohesins may help maintain silenced chromatin when deacetylation is reduced. The tRNAThr gene adjacent to HMR promotes the association of cohesin proteins with the silenced HMR locus (Dubey and Gartenberg 2007). Although the loss of cohesins in the absence of the tRNAThr gene has no impact on silencing of HMRa1 (Chang et al. 2005), it remains possible that it reduces the ability of Sir proteins to remain associated with HMR under conditions of reduced deacetylation.
A final possibility we do not favor is that the increased spreading of a finite pool of Sir proteins reduces the availability of these proteins to associate with normally silenced domains. This model has been suggested in other contexts (Kimura et al. 2002; Suka et al. 2002; van Leeuwen et al. 2002), but seems less likely in the case of the hmr ΔI ΔtRNA strain, given that Sir proteins do not associate significantly with sequences beyond the position of the deleted tRNA gene (Figure 6D).
The biological functions of HMR-I and the tRNAThr gene:
The contributions of HMR-I and the tRNAThr gene to silencing are only observed in the context of reduced Sir2p activity. Under normal laboratory conditions, HMR-E alone is strong enough to silence HMRa1, maintain haploid cell identity, and protect against HO endonuclease cleavage. Why then, is it necessary to have an auxiliary silencer to boost Sir protein association and a boundary to block the subsequent spreading? Our experiments with reduced Sir2p deacetylase activity offer some clues. In contrast to laboratory growth conditions, in nature yeast are subjected to variations in available nutrients. The direct link between Sir2p activity and NAD+ metabolism suggests that under some conditions deacetylation may be compromised and consequently yeast may have evolved insulating mechanisms for maintaining silenced chromatin at HMR in such circumstances. Indeed, in oxidative stress conditions, silencing at HMR is improved upon overexpression of Sir2p (Oberdoerffer et al. 2008). Similarly, the natural boost in Sir protein levels conveyed by HMR-I (and in some cases the tRNAThr gene) may have a similar effect. Furthermore, the apparent overabundance of Sir proteins at HMR may mitigate the loss of Sir protein enrichment at silenced loci that occurs during aging (Lin et al. 2009), thus delaying the onset of sterility in older cells.
Perspective:
These studies extend our previous work by demonstrating that, in addition to accelerating the rate of assembly of silenced chromatin, the HMR-E silencer increases the steady-state level of Sir proteins within several kilobase pairs of the silencer. Moreover, this work reinforces our previous conclusion that in the absence of such a silencer the ability of Sir proteins to spread is limited (Lynch and Rusche 2009; Rusche and Lynch 2009). For example, even when the tRNAThr boundary element was deleted, robust levels of Sir proteins were detected only within a few kilobase pairs of the HMR-E and HMR-I silencers (Figures 3 and 4). This limited capacity to spread probably mitigates the potentially toxic effects of fortuitous assembly and spreading of Sir proteins at inappropriate genomic locations. At the same time, these limitations increase the importance of silencers, such as HMR-E and HMR-I, in stabilizing the associations of Sir proteins in appropriate locations, particularly when deacetylation is compromised. Thus, these elements probably serve to buffer the capacity of silenced chromatin to assemble under suboptimal conditions.
Acknowledgments
We thank Jasper Rine for providing the PGAL1-HO (pJR831) plasmid and the α-Sir2p and α-Sir3p antibodies used in this study and three anonymous reviewers for helpful suggestions. This research was supported by a grant from the National Institutes of Health (GM073991).
Supporting information is available on line at http://www.genetics.org/cgi/content/full/genetics.109.113100/DC1.
References
- Alabert, C., J. N. Bianco and P. Pasero, 2009. Differential regulation of homologous recombination at DNA breaks and replication forks by the Mrc1 branch of the S-phase checkpoint. EMBO J. 28 1131–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong, C. M., M. Kaeberlein, S. I. Imai and L. Guarente, 2002. Mutations in Saccharomyces cerevisiae gene SIR2 can have differential effects on in vivo silencing phenotypes and in vitro histone deacetylation activity. Mol. Biol. Cell 13 1427–1438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitterman, K. J., R. M. Anderson, H. Y. Cohen, M. Latorre-Esteves and D. A. Sinclair, 2002. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J. Biol. Chem. 277 45099–45107. [DOI] [PubMed] [Google Scholar]
- Brand, A. H., G. Micklem and K. Nasmyth, 1987. A yeast silencer contains sequences that can promote autonomous plasmid replication and transcriptional activation. Cell 51 709–719. [DOI] [PubMed] [Google Scholar]
- Bulger, M., 2005. Hyperacetylated chromatin domains: lessons from heterochromatin. J. Biol. Chem. 280 21689–21692. [DOI] [PubMed] [Google Scholar]
- Chang, C. R., C. S. Wu, Y. Hom and M. R. Gartenberg, 2005. Targeting of cohesin by transcriptionally silent chromatin. Genes Dev. 19 3031–3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly, B., C. I. White and J. E. Haber, 1988. Physical monitoring of mating type switching in Saccharomyces cerevisiae. Mol. Cell. Biol. 8 2342–2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon, N., J. Raab, J. Guzzo, S. J. Szyjka, S. Gangadharan et al., 2009. DNA polymerase epsilon, acetylases and remodellers cooperate to form a specialized chromatin structure at a tRNA insulator. EMBO J. 28 2583–2600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donze, D., and R. T. Kamakaka, 2001. RNA polymerase III and RNA polymerase II promoter complexes are heterochromatin barriers in Saccharomyces cerevisiae. EMBO J. 20 520–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donze, D., C. R. Adams, J. Rine and R. T. Kamakaka, 1999. The boundaries of the silenced HMR domain in Saccharomyces cerevisiae. Genes Dev. 13 698–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubey, R. N., and M. R. Gartenberg, 2007. A tDNA establishes cohesion of a neighboring silent chromatin domain. Genes Dev. 21 2150–2160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourel, G., E. Revardel, C. E. Koering and E. Gilson, 1999. Cohabitation of insulators and silencing elements in yeast subtelomeric regions. EMBO J. 18 2522–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grewal, S. I., and S. C. Elgin, 2007. Transcription and RNA interference in the formation of heterochromatin. Nature 447 399–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haber, J. E., 1998. Mating-type gene switching in Saccharomyces cerevisiae. Annu. Rev. Genet. 32 561–599. [DOI] [PubMed] [Google Scholar]
- Hickman, M. A., and L. N. Rusche, 2007. Substitution as a mechanism for genetic robustness: the duplicated deacetylases Hst1p and Sir2p in Saccharomyces cerevisiae. PLoS Genet. 3 e126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoppe, G. J., J. C. Tanny, A. D. Rudner, S. A. Gerber, S. Danaie et al., 2002. Steps in assembly of silent chromatin in yeast: Sir3-independent binding of a Sir2/Sir4 complex to silencers and role for Sir2-dependent deacetylation. Mol. Cell. Biol. 22 4167–4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai, S., C. M. Armstrong, M. Kaeberlein and L. Guarente, 2000. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403 795–800. [DOI] [PubMed] [Google Scholar]
- Kimura, A., T. Umehara and M. Horikoshi, 2002. Chromosomal gradient of histone acetylation established by Sas2p and Sir2p functions as a shield against gene silencing. Nat. Genet. 32 370–377. [DOI] [PubMed] [Google Scholar]
- Klar, A. J., J. N. Strathern and J. B. Hicks, 1981. A position-effect control for gene transposition: state of expression of yeast mating-type genes affects their ability to switch. Cell 25 517–524. [DOI] [PubMed] [Google Scholar]
- Landry, J., J. T. Slama and R. Sternglanz, 2000. Role of NAD(+) in the deacetylase activity of the SIR2-like proteins. Biochem. Biophys. Res. Commun. 278 685–690. [DOI] [PubMed] [Google Scholar]
- Lau, A., H. Blitzblau and S. P. Bell, 2002. Cell-cycle control of the establishment of mating-type silencing in S. cerevisiae. Genes Dev. 16 2935–2945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, Y. Y., J. Y. Lu, J. Zhang, W. Walter, W. Dang et al., 2009. Protein acetylation microarray reveals that NuA4 controls key metabolic target regulating gluconeogenesis. Cell 136 1073–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo, S., and J. Rine, 1994. Silencers and domains of generalized repression. Science 264 1768–1771. [DOI] [PubMed] [Google Scholar]
- Lynch, P. J., H. B. Fraser, E. Sevastopoulos, J. Rine and L. N. Rusche, 2005. Sum1p, the origin recognition complex, and the spreading of a promoter-specific repressor in Saccharomyces cerevisiae. Mol. Cell. Biol. 25 5920–5932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch, P. J., and L. N. Rusche, 2009. A silencer promotes the assembly of silenced chromatin independently of recruitment. Mol. Cell. Biol. 29 43–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miele, A., K. Bystricky and J. Dekker, 2009. Yeast silent mating type loci form heterochromatic clusters through silencer protein-dependent long-range interactions. PLoS Genet. 5 e1000478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondoux, M. A., and V. A. Zakian, 2007. Subtelomeric elements influence but do not determine silencing levels at Saccharomyces cerevisiae telomeres. Genetics 177 2541–2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickoloff, J. A., E. Y. Chen and F. Heffron, 1986. A 24-base-pair DNA sequence from the MAT locus stimulates intergenic recombination in yeast. Proc. Natl. Acad. Sci. USA 83 7831–7835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noma, K., H. P. Cam, R. J. Maraia and S. I. Grewal, 2006. A role for TFIIIC transcription factor complex in genome organization. Cell 125 859–872. [DOI] [PubMed] [Google Scholar]
- Oberdoerffer, P., S. Michan, M. McVay, R. Mostoslavsky, J. Vann et al., 2008. SIRT1 redistribution on chromatin promotes genomic stability but alters gene expression during aging. Cell 135 907–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oki, M., and R. T. Kamakaka, 2005. Barrier function at HMR. Mol. Cell 19 707–716. [DOI] [PubMed] [Google Scholar]
- Pryde, F. E., and E. J. Louis, 1999. Limitations of silencing at native yeast telomeres. EMBO J. 18 2538–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivier, D. H., J. L. Ekena and J. Rine, 1999. HMR-I is an origin of replication and a silencer in Saccharomyces cerevisiae. Genetics 151 521–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche, L. N., and J. Rine, 2001. Conversion of a gene-specific repressor to a regional silencer. Genes Dev. 15 955–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche, L. N., A. L. Kirchmaier and J. Rine, 2002. Ordered nucleation and spreading of silenced chromatin in Saccharomyces cerevisiae. Mol. Biol. Cell 13 2207–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche, L. N., A. L. Kirchmaier and J. Rine, 2003. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu. Rev. Biochem. 72 481–516. [DOI] [PubMed] [Google Scholar]
- Rusche, L. N., and P. J. Lynch, 2009. Assembling heterochromatin in the appropriate places: A boost is needed. J. Cell. Physiol. 219 525–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt, M. E., T. A. Brown and B. L. Trumpower, 1990. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 18 3091–3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, K. C., S. L. Merrett and H. F. Willard, 2006. A heterochromatin barrier partitions the fission yeast centromere into discrete chromatin domains. Curr. Biol. 16 119–129. [DOI] [PubMed] [Google Scholar]
- Scott, K. C., C. V. White and H. F. Willard, 2007. An RNA polymerase III-dependent heterochromatin barrier at fission yeast centromere 1. PLoS ONE 2 e1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simms, T. A., S. L. Dugas, J. C. Gremillion, M. E. Ibos, M. N. Dandurand et al., 2008. TFIIIC binding sites function as both heterochromatin barriers and chromatin insulators in Saccharomyces cerevisiae. Eukaryot. Cell 7 2078–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, J. S., C. B. Brachmann, I. Celic, M. A. Kenna, S. Muhammad et al., 2000. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc. Natl. Acad. Sci. USA 97 6658–6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strathern, J. N., A. J. Klar, J. B. Hicks, J. A. Abraham, J. M. Ivy et al., 1982. Homothallic switching of yeast mating type cassettes is initiated by a double-stranded cut in the MAT locus. Cell 31 183–192. [DOI] [PubMed] [Google Scholar]
- Suka, N., K. Luo and M. Grunstein, 2002. Sir2p and Sas2p opposingly regulate acetylation of yeast histone H4 lysine16 and spreading of heterochromatin. Nat. Genet. 32 378–383. [DOI] [PubMed] [Google Scholar]
- Suter, B., A. Tong, M. Chang, L. Yu, G. W. Brown et al., 2004. The origin recognition complex links replication, sister chromatid cohesion and transcriptional silencing in Saccharomyces cerevisiae. Genetics 167 579–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teytelman, L., M. B. Eisen and J. Rine, 2008. Silent but not static: accelerated base-pair substitution in silenced chromatin of budding yeasts. PLoS Genet. 4 e1000247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela, L., N. Dhillon, R. N. Dubey, M. R. Gartenberg and R. T. Kamakaka, 2008. Long-range communication between the silencers of HMR. Mol. Cell. Biol. 28 1924–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen, F., P. R. Gafken and D. E. Gottschling, 2002. Dot1p modulates silencing in yeast by methylation of the nucleosome core. Cell 109 745–756. [DOI] [PubMed] [Google Scholar]
- Vega-Palas, M. A., E. Martin-Figueroa and F. J. Florencio, 2000. Telomeric silencing of a natural subtelomeric gene. Mol. Gen. Genet. 263 287–291. [DOI] [PubMed] [Google Scholar]
- Vignali, M., D. J. Steger, K. E. Neely and J. L. Workman, 2000. Distribution of acetylated histones resulting from Gal4–VP16 recruitment of SAGA and NuA4 complexes. EMBO J. 19 2629–2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, Q., J. Sandmeier, H. Xu, Y. Zou and X. Bi, 2006. Mechanism of the long range anti-silencing function of targeted histone acetyltransferases in yeast. J. Biol. Chem. 281 3980–3988. [DOI] [PubMed] [Google Scholar]
- Zou, Y., Q. Yu and X. Bi, 2006. a Asymmetric positioning of nucleosomes and directional establishment of transcriptionally silent chromatin by Saccharomyces cerevisiae silencers. Mol. Cell. Biol. 26 7806–7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou, Y., Q. Yu, Y. H. Chiu and X. Bi, 2006. b Position effect on the directionality of silencer function in Saccharomyces cerevisiae. Genetics 174 203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]