Abstract
The ability of fungi to generate polarized cells with a variety of shapes likely reflects precise temporal and spatial control over the formation of polarity axes. The bud site selection system of Saccharomyces cerevisiae represents the best-understood example of such a morphogenetic regulatory system. However, the extent to which this system is conserved in the highly polarized filamentous fungi remains unknown. Here, we describe the functional characterization and localization of the Aspergillus nidulans homolog of the axial bud site marker Bud3. Our results show that AnBud3 is not required for polarized hyphal growth per se, but is involved in septum formation. In particular, our genetic and biochemical evidence implicates AnBud3 as a guanine nucleotide exchange factor for the GTPase Rho4. Additional results suggest that the AnBud3–Rho4 module acts downstream of the septation initiation network to mediate recruitment of the formin SepA to the site of contractile actin ring assembly. Our observations provide new insight into the signaling pathways that regulate septum formation in filamentous fungi.
THE filamentous fungi form mycelial colonies that consist of networks of branched hyphae that grow by apical extension. In the higher fungi (i.e., Ascomycota and Basidiomycota), hyphae are compartmentalized by the formation of cross-walls, or septa. It has long been suspected that the presence of septa allows filamentous fungi to partition cellular environments within a hypha to support colony homeostasis and reproductive development (Gull 1978). The process of septum formation is similar to cytokinesis of animal cells, in that it coordinated with mitosis and requires formation of a contractile actin ring (CAR) (Balasubramanian et al. 2004). By analogy to the yeasts Saccharomyces cerevisiae and Schizosaccharomyces pombe, the CAR likely provides a landmark that guides deposition of the septal wall material. However, unlike these yeasts, the septum is not subsequently degraded and cells remain attached. Furthermore, in most filamentous fungi, a small pore is retained to enable communication between adjacent hyphal compartments. Septum formation has been studied in several filamentous fungi, including Aspergillus nidulans (Harris 2001; Walther and Wendland 2003). Upon germination of asexual conidiospores in A. nidulans, the first few rounds of parasynchronous nuclear division are not accompanied by septation until cells reach an appropriate size/volume (Harris et al. 1994; Wolkow et al. 1996). Subsequently, the first septum forms near the junction of the spore and germ tube (Harris et al. 1994). Deposition of the septal wall material is tightly coupled to assembly and constriction of the CAR, which in turn requires persistent signals from mitotic nuclei (Momany and Hamer 1997). As A. nidulans hyphae continue to grow by apical extension, each parasynchronous round of mitosis in multinucleate tip cells is followed by formation of septa in the basal region of the compartment (Clutterbuck 1970). Because tip and intercalary hyphal cells are multinucleate, not all of the individual mitotic events within the tip cell are capable of triggering septation.
Genetic analyses have identified several functions required for septum formation in A. nidulans, including the septation initiation network (SIN), the septins, and a formin (Harris 2001). The SIN is a cascade of three protein kinases that is activated by a small GTPase (Krapp and Simanis 2008). In A. nidulans, the component kinases of the SIN are arranged in the pathway SepH→SepL→SidB, with SepM amd MobA serving as cofactors that regulate SepL and SidB, respectively (Kim et al. 2006, 2009). Although SIN components localize to the spindle pole bodies, this does not appear to be a prerequisite for their subsequent recruitment to the septation site (Kim et al. 2009). Functional analysis of SepH, ModA, and SidB demonstrate that the SIN is required for assembly of the CAR (Bruno et al. 2001; Kim et al. 2006). Nevertheless, the upstream activators of the SIN and its downstream effectors remain unknown. However, localization of the septin AspB and the formin SepA to the septation site have been shown to require SepH (Sharpless and Harris 2002; Westfall and Momany 2002). AspB initially appears as a single ring that does not constrict, but splits into a double ring flanking the septum (Westfall and Momany 2002). AspB is not required per se for assembly of the CAR (Westfall and Momany 2002). On the other hand, SepA is a dynamic component of the CAR that is required for its assembly (Sharpless and Harris 2002), presumably because of its ability to nucleate actin filaments.
In S. cerevisiae and S. pombe, formins such as SepA are typically activated by Rho GTPases, such as Rho1 and Cdc42 (e.g., Dong et al. 2003; Martin et al. 2007). However, neither Cdc42 nor Rac1 is required for septum formation in A. nidulans, and Cdc42 does not localize to septation sites (Virag et al. 2007). One promising candidate for a GTPase that could activate SepA is Rho4, which appears to be specific to filamentous fungi (Rasmussen and Glass 2005). In Neurospora crassa, Rho4 is a dynamic component of the CAR; its absence prevents CAR assembly, whereas constitutive activation permits spurious formation of extra CARs (Rasmussen and Glass 2005). On the basis of these results, it was suggested that Rho4 is a likely activator of formins such as SepA at septation sites.
Because SepA simultaneously localizes to hyphal tips and septation sites in A. nidulans (Sharpless and Harris 2002), we have been interested in the identification of functions that determine patterns of cell wall deposition in hyphal cells. In this context the bud site selection system of S. cerevisiae provides an important paradigm. S. cerevisiae cells display two distinct budding patterns that are controlled by mating type (Freifelder 1960; Chant 1999). Mating type a or α cells employ an axial budding pattern whereby the previous bud site serves as a template for the next bud. As a result, a chain of chitinous bud scars decorates the cell surface. In contrast, mating type a/α cells employ a bipolar budding pattern whereby buds emerge from either the distal or proximal pole of the cell (the proximal pole is defined by the presence of the birth scar, Chant and Pringle 1995). Accordingly, bud scars cluster at either pole but are not necessarily adjacent to each other. Extensive genetic analyses have provided a fairly detailed understanding of the molecular mechanisms that underlie the axial and bipolar budding patterns. For the axial pattern, the cell wall protein Axl2 serves as a landmark whose function is facilitated by its association with Axl1 and the septin-interacting proteins Bud3 and Bud4 (Chant and Herskowitz 1991; Chant et al. 1995; Chant 1999; Lord et al. 2002; Gao et al. 2007; Park and Bi 2007). For the bipolar pattern, the paralogous cell wall proteins Bud8 and Bud9, which bear no homology to Axl2, serve as distal and proximal pole markers, respectively (Chant 1999; Harkins et al. 2001; Kang et al. 2004; Park and Bi 2007). Furthermore, the membrane proteins Rax1 and Rax2 form complexes with Bud8 and Bud9, which facilitates their function (Kang et al. 2004). The positional information generated by the landmark proteins Axl2, Bud8, or Bud9 is subsequently relayed to the Ras-like Bud1/Rsr1 GTPase module via the guanine nucleotide exchange (GEF) factor Bud5 (Kang et al. 2001, 2004; Krappmann et al. 2007). This results in localized activation of the Rho-like GTPase Cdc42, which acts via multiple effectors to recruit components of the morphogenetic machinery to the specified bud site (Chant 1999; Park and Bi 2007).
Despite the importance of the bud site selection regulatory module in specifying the budding pattern of S. cerevisiae yeast cells, it remains unclear whether it is used for a similar regulatory purpose in other fungi. Ashbya gossypii is a hemiascomycete fungus closely related to S. cerevisiae that is only capable of forming hyphae (Philippsen et al. 2005). The A. gossypii Bud3 homolog, which can function in S. cerevisiae, appears to function as a landmark for septum formation and also controls the position of the contractile actin ring (Wendland 2003). In A. gossypii and Candida albicans, another hemiascomycete capable of forming true hyphae, Bud1/Rsr1 homologs appear to function at the hyphal tip to specify the direction of hyphal extension (Bauer et al. 2004; Hausauer et al. 2005). Although limited to hemiascomycetes, these studies suggest that the components of the bud site selection regulatory module may have a broader function within the fungal kingdom.
Here, we investigate the possibility that homologs of the bud site selection proteins may provide positional information that marks the hyphal tip and/or septation sites in A. nidulans. We characterize an apparent homolog of Bud3 and show that it is required for assembly of the CAR at septation sites. Our results provide new insight into the regulation of septum formation by suggesting that AnBud3 functions downstream of the SIN as a GEF for Rho4.
MATERIALS AND METHODS
Strains, media, growth conditions, and staining:
Aspergillus nidulans strains used in this study are listed in Table 1. Minimal + vitamins (MNV) media were made according to Kafer (1977). MNV-glycerol and MNV-threonine fructose media were made as described in Pearson et al. (2004). Malt extract agar (MAG) and yeast extract glucose + vitamins (YGV) media were made as described previously (Harris et al. 1994). The reagent 5-fluoroorotic acid (5-FOA; US Biological, Swampscott, MA) was added to media at a concentration of 1 mg/ml after autoclaving. For septation and hyphal growth studies, conidia from appropriate stains were grown at 28° for 12 hr on cover slips. Hyphae attached to the cover slip were fixed using a modified standard protocol (Harris et al. 1994) [fixing solution contained 3.7% formaldehyde, 25 mm EGTA, 50 mm piperazine-N,N-bis (2 ethanesulfonic acid) (PIPES), and 0.5% dimethyl sulfoxide] for 20 min and then stained with staining solution containing both 273 nm fluorescent brightener 28 (Sigma-Aldrich, St. Louis, MI) and 160 nm Hoechst 33258 (Molecular Probes, Eugene, OR).
TABLE 1.
Strains used in this study
| Strain | Genotype | Source or reference |
|---|---|---|
| A28 | pabaA6 biA1 | FGSC (accession no. A28) |
| GR5 | pyrG89 wA3 pyroA4 | FGSC (accession no. A773) |
| TNO2A3 | pyrG89; argB2; pyroA4 nkuA∷argB | |
| AHS2 | pyrG89; argB2; Δbud3∷pry-4 pyroA4 nkuA∷argB | This study |
| AHS3 | pyrG89; argB2; Δbud3∷pyroA pyroA4 nkuA∷argB | This study |
| AHS5 | pyrG89; argB2; Δrho4∷pyroA pyroA4 nkuA∷argB | This study |
| AHS7 | pyrG89; argB2; Δmsb1∷pyroA pyroA4 nkuA∷argB | This study |
| AHS25 | pyrG89; argB2; Δrho4∷pyroA; Δbud3∷pyrG; pyroA4 nkuA∷argB | This study |
| AHS30 | sepA∷gfp∷pyr-4; pyrG89 pabaA1; Δbud3∷pyroA; yA2 | This study |
| AHS252 | yA2; argB2; pyroA4 | This study |
| AAV123.1 | pyrG89 sepA∷gfp∷pyr-4; argB2; pyroA4 ΔnkuA∷argB | Virag et al. (2007) |
| ASH630 | sepA1; pyrG89; wA3 | Lab stock |
| ACP115 | tpmA∷GFP∷pyr-4; pyrG89; wA3 | Pearson et al. (2004) |
| AKS70 | sepA∷gfp∷pyr-4; pyrG89 pabaA1; yA2 | Sharpless and Harris (2002) |
| AHS41 | pyrG89; argB2; gfp∷bud3∷pyr4; pyroA4; nkuA∷argB | This study |
| AHS43 | pyrG89;argB2;alcA∷gfp∷rho4; pyr-4; nkuA∷argB | This study |
| AHS51 | pyrG89; argB2;gfp∷bud3∷pyr4; pyroA4; sepA1 nkuA∷argB | This study |
| AHS53 | sepA1; tpmA∷GFP∷pyr4;pyrG89 | This study |
| AHS3C2 | Δbud3 suppressor | This study |
| AJM34 | sepH 1; pabaA6; lysB5; chaA1 | |
| AHS61 | pyrG89; sepH 1 | This study |
| AHS62 | pyrG89; gfp∷bud3∷pyr4; sepH 1 | This study |
Construction of gene replacement strains:
The bud3, rho4, and msb1 genes from strains AHS3, AHS4, and AHS7, respectively, were replaced with the pyroAA.f. marker from A. fumigatus. All gene replacements were generated using the gene targeting system developed by Nayak et al. (2006) and the gene replacement generation strategy developed by Yang et al. (2004). Oligonucleotides used in this study are listed in Supporting information, Table S1. The pyroAA.f. DNA marker fragment was PCR amplified from plasmid pTN1 (Nayak et al. 2006). DNA fragments upstream and downstream of bud3 and rho4 were amplified from the wild-type strain FGSC28 (available through the Fungal Genetics Stock Center, Kansas City, MO). High-fidelity and long template PCR systems (Roche Diagnostics, Indianapolis, IN) were used for amplifications of individual and fusion fragments, respectively, using a Px2 Hybaid or an Eppendorf Mastercycler gradient thermal cycler. The amplification conditions were according to the manufacturer's recommendations. PCR products were gel purified using the QIAquick gel extraction kit (QIAGEN, Valencia, CA). The gene replacement constructs were transformed into strain TNO2A3 and plated on supplemented minimal medium with 0.6 m KCl. Transformations were performed according to the protocol described by Osmani et al. (2006). Transformation candidates were tested for homologous integration of the gene replacement construct and the absence of the wild-type gene by diagnostic PCR as described by Yang et al. (2004). The same strategy was used to replace bud3 with the pyr-4 nutritional marker from N. crassa. The pyr-4 DNA marker fragment was amplified from plasmid pRG3. The resulting constructs were transformed into TNO2A3. The bud3 gene replacement construct with pyroAA.f marker was transformed to strain AAV123 to generate strain AHS30.
Genetic interaction experiments:
The cdc42 (ANID_07487.1), racA (ANID_04743.1), and rho4 (ANID_02687.1) gene sequences, including upstream (∼500 bp) and downstream regions (∼300 bp), were retrieved from the A. nidulans genome at the Broad Institute (http://www.fgsc.net/aspergenome.htm). These sequences were amplified (primers described in Table S1) and cloned into the pCR2.1–TOPO vector (Invitrogen, Carlsbad, CA) to generate plasmids pHS11, pHS12, and pHS13, respectively. For overexpression experiments, strain AHS3 was cotransformed with pRG3–AMA1 and each of the plasmids pHS11, pHS12, and pHS13.
Construction of GFP fusions to AnBud3 and Rho4:
To localize AnBud3, we fused GFP to the N terminus using the five-piece fusion PCR approach recently described by Taheri-Talesh et al. (2008). In addition to the retention of native promoter sequences, final constructs also contained a short linker of five glycines and alanines inserted between the GFP and AnBud3 coding sequences. In brief, the following five fragments were amplified (primers described in Table S1): (1) a 1.3-kb sequence upstream of bud3, (2) the GFP coding sequence (minus the stop codon) derived from plasmid pMCB17apx, (3) the bud3 gene plus 400 bp of downstream sequence, (4) the N. crassa pyr-4 selectable marker, also derived from pMCB17apx, and (5) a 1.3-kb sequence extending from 400 to 1700 bp downstream of bud3. Fragments 1, 3, and 5 were amplified by specific primers with 30-bp tails that were reverse complements of the adjacent fragments. Finally, the forward primer used to amplify fragment 1 and the reverse primer used to amplify fragment 5 were used to fuse the entire five-fragment gene replacement construct. The high-fidelity and long template PCR systems (Roche Diagnostics) were employed to amplify individual and fusion fragments, respectively, on a Px2 Hybaid or an Eppendorf Mastercycler gradient thermal cycler. PCR products were gel purified using the QIAquick gel extraction kit (QIAGEN). The resulting gfp∷bud3∷pyr-4 cassette was used to replace wild-type bud3 in strain TNO2A3 using the approach described by Nayak et al. (2006).
The plasmid pHS31, containing alcA(p)∷gfp∷rho4, was constructed in two steps. An N-terminal sequence from rho4 that corresponds to amino acids 1–261 was amplified from wild-type strain A28. Cloning sites for AscI and PacI were incorporated onto the ends of the amplified fragment. The PCR product was gel purified and cloned into pCR2.1–TOPO to generate pHS30. The resulting plasmid was digested with AscI and PacI (New England Biolabs), and the liberated rho4 fragment ligated into pMCB17apx (Efimov 2003) to generate pHS31. Thereby, the N terminus of rho4 was fused to GFP, which in turn is expressed under the control of alcA(p). Upon transformation into strain TNO2A3, homologous integration of this construct generates a single full-length copy of rho4 regulated by alcA(p), plus a truncated version controlled by native promoter sequences.
AnBud3 guanine nucleotide exchange assays:
MBP-tagged AnBud3 and Rho4 constructs were cloned by RT–PCR using the primers (sequences are provided in Table S1) DJ_An0113_NcoI_f and DJ_An0113_NotI_TGA_rev for bud3, and DJ_An2687rho4_NcoI_f andDJ_HindIII_An2687rho4_r for rho4, along with cDNA prepared from vegetative hyphae. Total RNA was obtained by TRIzol extraction (Invitrogen) and cDNA prepared using RevertAid M-MuLV Reverse Transcriptase (Fermentas). cDNA was subcloned into pJet1.2/blunt vector (Fermentas). AnBud3 and Rho4 constructs were digested with either NcoI and NotI (bud3) or NcoI and HindIII (rho4) and inserted into a modified pMalc2x vector (Vogt and Seiler 2008), which was digested accordingly to generate plasmids pMal_AnBUD3 and pMal_AnRHO4. MBP–AnBud3 and MBP–Rho4 fusion proteins were expressed and purified as previously described (Vogt and Seiler 2008).
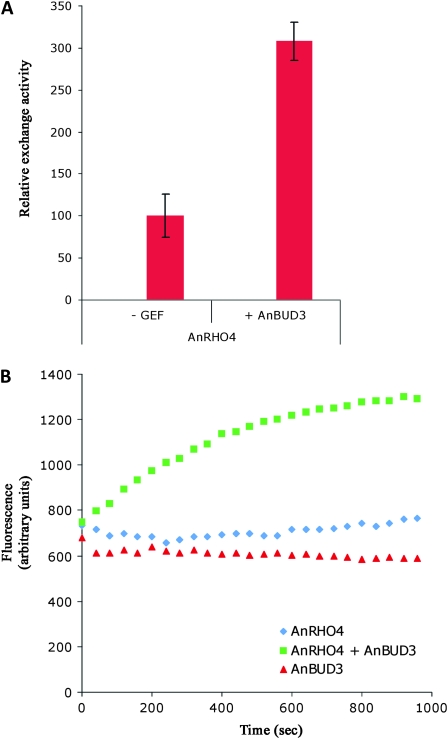
Guanine nucleotide exchange assays were performed by fluorometric determination of mant-GDP (a fluorescently labeled GTP analog) incorporation as described (Abe et al. 2000) using a Tecan Infinite 200 spectrophotometer at 21°. The reaction was started by adding 0.1 μm mant–GDP and 1 μm MBP–Bud3 to 1 μm Rho4 in 30 mm Tris, pH 7.5, 5 mm MgCl2, 10 mm NaH2PO4/K2HPO4, 3 mm DTT, which was preequilibrated for 5 min at 21°. Fluorescence intensity (λexc = 356 nm, λem = 448 nm) was monitored over 16 min. The change of fluorescence over time was used to assess mant–GDP incorporation into Rho4 in the presence and absence of the GEF. Similar conditions were also used in recent publications by Yeh et al. (2007) and Hlubek et al. (2008). The latter authors also used equal amounts of GEF and GTPase. In the kinetics presented in Figure 4B, the mixing of Bud3 and mant–GDP with Rho4–GDP is defined as time-point zero. After 16 min the measurement was stopped and the resulting emission curves were further analyzed. To allow comparison between independently prepared biological samples of each GEF and GTPase, we used the linear range of the slope from each individual experiment. The kinetics of two independent GEF and of two independent GTPase preparations each performed in duplicate measurements was determined and the background fluorescence of mant–GDP without added proteins was subtracted. The mean value of the slopes calculated for Rho4 in the absence of Bud3 represents the intrinsic activity of Rho4 and was set to 100%. To determine the exchange activity of Rho4 in the presence of the GEF relative to its intrinsic activity, the mean slope value calculated for emission curves of Rho4 in the presence of the GEF was divided by the mean slope value calculated for the intrinsic activity of Rho4 and the resulting value was multiplied by 100 to obtain the relative value displayed in Figure 4A (relative exchange activity of Rho4 in the presence of Bud3 = (mean value of slopeRho4 + slopeBud3/mean value of slopeRho4 × 100).
Figure 4.—
AnBuD3 is a Rho4 exchange factor. In vitro guanine nucleotide exchange activity was determined by measuring binding of mant–GDP to purified Rho4 in the presence or absence of the putative GEF AnBud3 construct containing the GEF domain. The diagram indicates the mean values ± SD of at least two independent Rho protein and two GEF purifications with each experiment performed in duplicate. The intrinsic Rho activity is set to 100% (A). An example of in vitro kinetics for mant–GDP binding to purified AnRho4, AnBud3, and both is shown. The exchange activity of AnRho4 is stimulated by AnBud3 (B).
Conidiation experiments:
Conidiophore development was monitored using the sandwich cover slip method described by Lin and Momany (2003). Briefly, 1 ml of melted MAGUU media was placed on a cover slip that was then transferred to the surface of a 4% water agar plate. The cover slip was inoculated with spores once the media had solidified, whereupon a second cover slip was placed on top. After 3–4 days, conidiophores had formed and become attached to the top cover slip, which was then dipped into 100% ethanol and mounted for differential interference contrast (DIC) microscopy. For Calcofluor staining, the cover slips were fixed and stained after ethanol treatment.
sepA1 and sepH 1 experiments:
The sepA1 GFP–AnBud3 strain AHS51 was generated by crossing the GFP–AnBud3 strain AHS41 with the sepA1 strain ASH630 and screening at restrictive temperature (42°) on selective media. The sepH 1 GFP–AnBud3 strain AHS62 was generated by transforming the sepH 1 strain AHS61 with the same GFP fusion construct used to generate AHS41. The DNA replication inhibitor hydroxyurea (HU) was used to arrest the nuclear division cycle. A total of 50 mm HU was added to liquid cultures 1 hr prior to shiftdown, and cultures were maintained in the presence of HU for an additional 2 hr once returned to 28°. The strain AHS53 (sepA1 tpmA∷gfp) was used to assess the effect of the sepA1 mutation on the formation of contractile actin rings at the semipermissive temperature of 37°.
Microscopy:
Digital images of plates were collected with an Olympus C-3020ZOOM digital camera. DIC and fluorescent images were collected with either an Olympus BX51 microscope with a reflected fluorescence system fitted with a Photometrics CoolSnap HQ camera or an Olympus Fluoview confocal laser-scanning microscope. Images were processed with IPLab Scientific Image Processing 3.5.5 (Scanalytics, Fairfax, VA) and Adobe Photoshop 6.0 (Adobe Systems, San Jose, CA).
Δbud3 suppressor screen:
A suspension of 106 conidia from the strain AHS3 was plated on MNUU plates and irradiated with UV to a survival rate of ∼10%. Plates were incubated for 6 days at 28°. The faint green colonies that emerged were patched in grids on master MNUU plates and for retesting. In addition, retention of the Δbud3 mutation was verified by PCR. Candidates for further study were picked on the basis of restoration of septum formation (as observed by Calcofluor staining). Standard genetic analysis was used to determine that suppressor mutations were not linked to bud3 and defined a single gene.
RESULTS
The A. nidulans homolog of Bud3 is required for septum formation:
Our original annotation of the A. nidulans genome revealed the existence of potential homologs of the axial budding markers Bud4 and Axl2 (Harris and Momany 2004; Galagan et al. 2005). Subsequent annotation using the cognate proteins from Ashbya gossypii (AgBud3; Wendland 2003) and C. albicans (CaO19.7079) as additional queries for BLASTp and PSI–BLAST searches also uncovered a potential homolog of Bud3. AnBud3 (ANID_00113.1) is a predicted 1538-amino-acid (aa) protein with a RhoGEF domain located between aa 250 and 450 (Figure S1). Homologs of AnBud3 (>40% identity over their entire lengths) exist in all sequenced euascomycete genomes (e.g., Figure S1). AnBud3 only possesses limited homology to S. cerevisiae and A. gossypii Bud3 (21% identity over the first ∼550 aa, which corresponds to the predicted RhoGEF domain). Our description and functional characterization of the Bud4 and Axl2 homologs will be presented elsewhere.
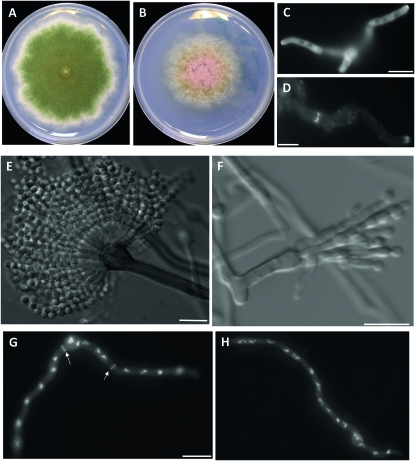
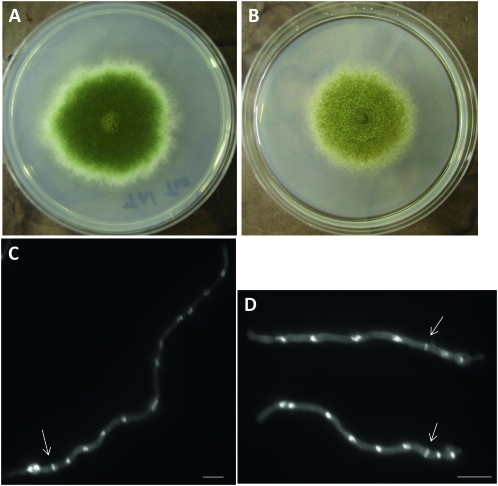
To determine the possible function of AnBud3 during hyphal morphogenesis, a mutant possessing a complete gene deletion was generated using recently described protocols (Yang et al. 2004; Nayak et al. 2006). bud3∷pyroAA.f. deletion mutants (hereafter referred to as Δbud3) formed colonies that were slightly smaller than wild type and were notably devoid of conidia (Figure 1, A and B). On minimal media, Δbud3 mutants produced ∼520-fold fewer conidia/ml compared to its parental strain TN02A3. A similar effect (i.e., ∼75-fold reduction compared to TN02A3) was observed on rich media. To determine the possible basis of the conidiation defect, conidiophores from the mutant as well as wild-type controls were imaged using a previously described “sandwich slide” protocol (Lin and Momany 2003). A range of defects was noted, included elongated metulae and phialides, as well as conidiospores that apparently failed to undergo cytokinesis (Figure 1, E and F). Because a stage-specific arrest was not observed, it seems likely that AnBud3 is required at multiple steps during conidiophore development.
Figure 1.—
Effects of the Δbud3 deletion on colony morphology, septum formation, and conidiation. (A and B) Colony morphologies of strains TNO2A3 (wild type; A) and AHS3 (Δbud3; B) grown on minimal medium (MNUU) for 9 days. SepA–GFP localizes to hyphal tips, but not septa, in ΔAnbud3 mutants. (D) SepA–GFP localization at septa in wild-type hyphae. For C and D, ΔAnbud3 and wild-type strains possessing sepA∷gfp (AHS30 and AKS70, respectively) were grown in YGV media for 12 hr prior to imaging. (E) Wild-type conidiophore. (F) Δbud3 conidiophore. Fused metulae and phialides bearing a few spores were observed. (G and H) Wild-type (G) and Δbud3 (H) hyphae grown in YGVUU for 12 hr. Note the absence of septa in the Δbud3 mutant. Septa and nuclei were visualized in fixed hyphae using Calcofulor and Hoechst 33258, respectively. Arrows indicate septa. Bar, 10 μm.
Cover slip cultures were used to examine Δbud3 mutants for defects in hyphal morphogenesis. The timing and pattern of spore polarization was indistinguishable from wild type, and the resulting hyphae displayed no obvious defects in polarized growth (Figure 1, G and H). On the other hand, septum formation was completely abolished, as no septa were observed in Δbud3 mutants (Figure 1, G and H; n > 1000 hyphae grown on YGVUU). To gain further insight into the nature of the septation defect in Δbud3 mutants, strains possessing a SepA–GFP fusion were analyzed. As noted previously (Sharpless and Harris 2002), SepA is a component of the contractile actin ring that forms at septation sites (Figure 1D). In Δbud3 sepA∷gfp strains (AHS30), SepA–GFP exhibited normal localization at hyphal tips, but no rings were observed (n > 1000 hyphae grown on YGV; Figure 1C). Accordingly, AnBud3 appears to be required for an early step in septum formation that precedes the formation of the contractile actin ring.
These observations demonstrate that AnBud3 is not required for the establishment or maintenance of hyphal polarity, but is needed for normal septation. Notably, the defects in septum formation may account for the abnormal development observed in Δbud3 mutants, as reduced conidiation has previously been associated with defects in septum formation (Harris et al. 1994).
AnBud3 functions as a GEF for Rho4:
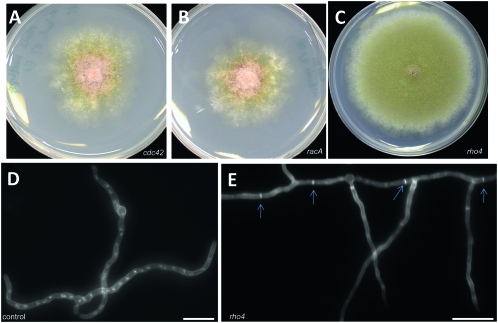
The presence of a predicted Rho–GEF domain in AnBud3 suggested that its role in septation might be to locally activate a Rho GTPase by promoting GDP–GTP exchange. A genetic approach was used to identify candidate Rho GTPase targets for AnBud3. This approach is based on the premise that increased levels of a target GTPase can compensate for defects in its associated GEF. For example, in S. cerevisiae, the GTPases Cdc42 and Rho1 function as dosage suppressors of mutations affecting their cognate GEFs, Cdc24, and Rom1, respectively (Bender and Pringle 1989; Ozaki et al. 1996). Accordingly, we predicted that one of the six annotated Rho GTPases from A. nidulans (Harris et al. 2009) might function as a dosage suppressor of Δbud3. We were particularly interested in ANID_02687.1, a predicted homolog of Neurospora crassa rho-4, which is required for septum formation and assembly of the contractile actin ring (Rasmussen and Glass 2005). Candidate GTPases were amplified and cotransformed into a Δbud3 pyrG89 strain along with the autonomously replicating plasmid pRG3–AMA1. For each GTPase, multiple Pyr+ transformants were picked and tested for suppression of the conidiation defects caused by deletion of bud3. Neither cdc42 nor racA could suppress Δbud3, however multicopy rho4 was capable of restoring conidiation (Figure 2, A–C). In addition, these transformants were also able to form septa (Figure 2, D and E). Two observations demonstrate that suppression was due to the presence of rho4. First, the entire rho4 coding region could be amplified from pRG3–AMA1-based plasmids rescued from the original transformants. Second, retransformation experiments showed that rescued plasmids containing rho4 were able to suppress Δbud3. On the basis of this genetic evidence, we conclude that AnBud3 likely functions as a GEF for the Rho GTPase Rho4.
Figure 2.—
Dosage suppression of Δbud3 growth and septation defects by rho4. (A–C) Colony morphology of Δbud3 strain AHS3 transformed with multiple copies of cdc42 (A), rac1 (B), or rho4 (C) and grown on selective MN media. Only rho4 functions as a dosage suppressor and restores conidiation. Hyphal morphology of Δbud3 strain AHS3 transformed with vector control (D) or rho4 (E). Hyphae were grown on YGV medium for 14 hr and stained with Calcofluor and Hoechst 33258 to visualize septa and nuclei, respectively. The presence of rho4 restored septum formation (arrows) to the ΔAnbud3 mutant. Bar, 10 μm.
Because GEFs activate their target GTPase, mutational inactivation of the GTPase would typically be expected to cause the same phenotypes as loss of its GEF. Accordingly, we generated a rho4 deletion and tested for defects in septum formation and conidiation that resemble those observed in Δbud3 mutants. As shown in Figure 3, A and B, Δrho4 mutants displayed similar colony morphology as Δbud3 mutants. Furthermore, Δrho4 mutants were completely defective in septation and formed aberrant conidiophores (Figure 3, C–G). These observations implicate Rho4 in the same morphological processes as Bud3. To test this, we generated Δbud3∷pyrGA.f. Δrho4∷pyroAA.f. double mutants (AHS25) by a standard cross, and found that they exhibited the same phenotype as the two parent single mutants (Figure 3, H and I). This epistatic interaction provides additional support for the notion that AnBud3 and Rho4 function in the same pathway that regulates septum formation.
Figure 3.—
Effects of the Δrho4 mutation on growth, hyphal morphology, and development. (A and B) Colony morphology of wild-type (TNO2A3; A) and Δrho4 (AHS5; B) strains following growth on MNUU medium for 9 days. (C and D) Hyphal morphology of wild-type (TNO2A3; C) and Δrho4 (AHS5; D) strains following growth for 14 hr on YGVUU at 28°. Arrows indicate septa. Bar, 10 μm. (E–G) Conidiophore morphology of wild-type (TNO2A3; E) and Δrho4 (AHS5; F–G) strains following growth for 3 days on MAGUU. Arrow in F indicates abnormal formation of a secondary conidiophore. Arrows in G indicate fused metulae and phialides. (H) Hyphal morphology of Δbud3 Δrho4 double mutant strain (AHS25) after 14 hr of growth in YGVUU at 28°. (I) Conidiophore morphology of Δbud3 Δrho4 double mutant strain (ASH25) following growth for 3 days on MAGUU. The arrow indicates abnormal formation of a secondary conidiophore generated from a phialide fused to its subtending metulae. Bars, 10 μm except for E–G, where bars = 3 μm.
To provide further evidence for the relationship between AnBud3 and Rho4, we used in vitro assays to determine whether AnBud3 exhibited GEF activity toward Rho4 (see materials and methods). As shown in Figure 4, a fragment that encompasses the predicted GEF domain of AnBud3 specifically stimulated the GDP–GTP exchange activity of Rho4. Furthermore, the same fragment was also capable of promoting the exchange activity of the heterologous Rho4 from N. crassa (Figure S2). Accordingly, when coupled with the genetic interactions and phenotypic similarities displayed by the respective mutants, these results strongly implicate AnBud3 as a Rho4 GEF in A. nidulans.
AnBud3 and Rho4 localization patterns at septa:
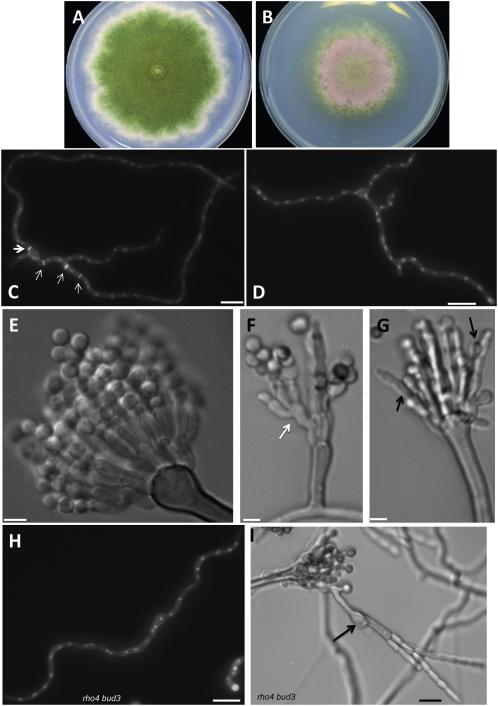
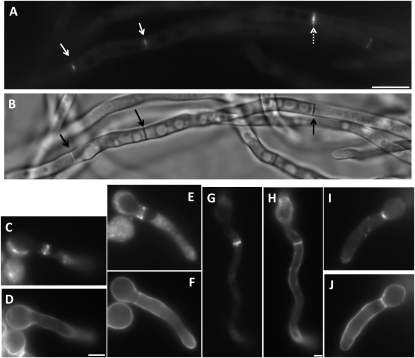
As a further test for the function of AnBud3, we used a GFP fusion protein to characterize its localization pattern. We constructed strains in which the sole functional source of AnBud3 was supplied by a GFP∷bud3 fusion expressed under control of native promoter sequences. As expected, GFP–AnBud3 localized to septation sites, where it formed a constricting ring (Figure 5, A and B). Notably, GFP–AnBud3 first localized to incipient septation sites prior to the appearance of any detectable Calcofluor-stained septum (i.e., 21/109 GFP–AnBud3 rings were not associated with a septum; Figure 5, C and D). GFP–AnBud3 localization at septation sites remained unchanged as septa first appeared (i.e., 22/109 rings colocalized with a thin septum; Figure 5, E and F) and then as they began to thicken (i.e., 54/109 rings colocalized with a thick septum; Figure 5, G and H). However, GFP–AnBud3 rings ultimately constricted (i.e., 12/109 rings were constricted and colocalized with a thick septum; Figure 5, I and J), suggesting that AnBud3 is a likely component of the contractile actin ring.
Figure 5.—
Localization of GFP–AnBud3. (A and B) GFP–AnBud3 rings (A, open arrows) and corresponding septa (B, solid arrows) following growth of strain AHS41 on YGVUU for 15 hr at 28°. The dashed arrow indicates a thick ring in the process of constricting. (C–J) Coordination of GFP–AnBud3 ring dynamics with septum deposition. (C, E, G, and I) GFP–AnBud3 localization. (D, F, H, and J) Calcofluor staining to visualize septa and cell walls. (C and D) GFP –AnBud3 localization at septation site prior to appearance of visible septum. (E and F) GFP–AnBud3 rings associated with thin septum. (G and H) Thicker AnBud3 ring associated with more prominent septum. (I and J) Constricting AnBud3 ring and associated septum. Bar, 3 μm, except A and B, where bar = 10 μm.
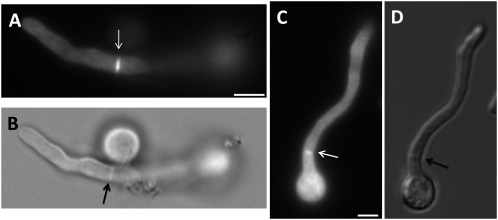
Because our genetic evidence implicates AnBud3 as a putative GEF for Rho4, we surmised that Rho4 would also localize to septa. Accordingly, we constructed a strain in which the sole functional copy of rho4 was fused at its 5′ end to GFP and was expressed under control of the inducible alcA(p) promoter. As expected, the alcA(p)∷gfp∷rho4 strain displayed a growth defect on repressing glucose media, though not as severe as that caused by deletion of rho4 (data not shown). On the other hand, the strain grew no worse than wild type on inducing threonine media. Under these conditions, GFP–Rho4 localized to septa and appeared to undergo constriction (Figure 6). This observation suggests that like AnBud3, Rho4 is also a component of the contractile actin ring.
Figure 6.—
Localization of GFP–Rho4. (A and B) GFP–Rho4 localization (A) and corresponding septum (B; observed using DIC optics) following 13 hr growth of strain AHS43 at 28° on alcA(p) inducible threonine–MNV. Arrow indicates a GFP–Rho4 ring at the septation site. (C and D) A constricting GFP–Rho4 ring (C; white arrow) and corresponding septum (D; black arrow). Bar, 3 μm.
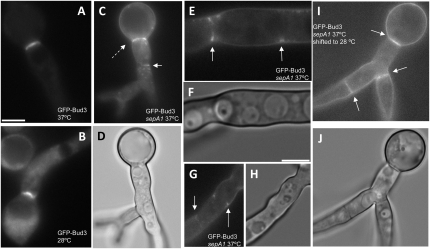
Our genetic analysis supports a model whereby AnBud3 acts as a GEF that locally activates Rho4, which in turn leads to localized recruitment of SepA and assembly of the CAR. According to this model, localization of AnBud3 to septation sites would precede formation of the CAR. To test this prediction, we took advantage of the temperature-sensitive sepA1 mutation. At both restrictive (42°) and semipermissive temperatures (37°–39°), this mutation abolishes assembly of the CAR (Sharpless and Harris 2002; Figure S3). We thus determined whether AnBud3 localizes to septation sites in sepA1 mutants incubated under these conditions. As expected, GFP–AnBud3 localized to rings in wild-type hyphae at 37° (Figure 7, A and B). Notably, in sepA1 mutants, GFP–AnBud3 localization was also observed at septation sites (Figure 7, C–H). In most cases, GFP–AnBud3 accumulated at cortical sites or appeared to form incipient rings (Figure 7, E–H), though rare examples of a complete ring were occasionally observed (Figure 7, C and D). These results suggest that assembly of the CAR is not a prerequisite for the recruitment of AnBud3 to septation sites.
Figure 7.—
Recruitment of AnBud3 to septation sites does not require presence of the contractile actin ring. (A and B) GFP–AnBud3 localization in wild-type hyphae (AHS41) grown at 37° (A) or 28° (B). (C–H) GFP–AnBud3 localization in the sepA1 mutant (AHS51) at 37° (C, E, and G) and corresponding DIC images (D, F, and H). Dashed arrow (C) indicates a rare example of an intact GFP–AnBud3 ring, whereas solid arrows mark the more prevalent examples of incomplete rings or cortical patches. (I and J) GFP–AnBud3 localization (I) and corresponding DIC image (J) in sepA1 mutant hyphae 2 hr following a shift from 37° to 28°. Solid arrows indicate GFP–AnBud3 rings. Bars, 3 μm.
Roles of nuclear division and the SIN in AnBud3 localization:
We have previously demonstrated that the sepA1 septation defect is reversible. In particular, a shift back to permissive temperature (i.e., 28°) triggers rapid and synchronous formation of septa with appropriate spacing in a manner that is dependent upon nuclear division (Harris et al. 1997; also see Trinci and Morris 1979). We exploited the reversibility of the sepA1 mutation to determine whether nuclear division is required for the appearance of AnBud3 rings. In particular, sepA1 hyphae that express GFP–AnBud3 were shifted to 28° after incubation at 37°. As expected, numerous AnBud3 rings appeared within 2 hr, and in many cases, multiple rings formed in a single hypha (Figure 7, I and J). However, when shifted down in the presence of 50 mm hydroxyurea, which blocks DNA replication and subsequent nuclear division, the localized recruitment of AnBud3 and the formation of rings were abolished (Figure S4). A similar treatment is known to prevent formation of septa upon shiftdown of sepA1 mutants (Harris et al. 1997). Thus, nuclear division appears to be generally required for the localization of AnBud3 to septation sites and the subsequent formation of rings.
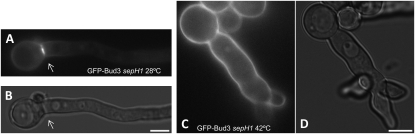
A potential pathway that might link nuclear division to AnBud3 localization is the SIN, which is required for assembly of the CAR in A. nidulans (Bruno et al. 2001; Kim et al. 2006) and functions upstream of SepA (Sharpless and Harris 2002). To test this notion, we transformed our GFP∷bud3 construct into a strain possessing the temperature-sensitive sepH 1 mutation. This mutation, which affects the A. nidulans homolog of the S. pombe Cdc7/S. cerevisiae Cdc15 protein kinase, abolishes CAR assembly and septation at restrictive and semipermissive temperatures (Bruno et al. 2001; Sharpless and Harris 2002). At permissive temperature (28°), AnBud3 localization and septum formation were indistinguishable from wild type in the sepH 1 mutant (Figure 8, A and B). However, under semipermissive conditions (39°), no evidence for AnBud3 recruitment to septation sites was observed (Figure 8, C and D). Note that although the presence of AnBud3–GFP clearly altered the morphology of sepH 1 mutants, hyphae were large enough to form septa. These data suggest that the SIN could coordinate CAR assembly via recruitment of AnBud3 to septation sites. Nevertheless, if indeed this is the case, it is not the sole mechanism by the SIN acts, because rho4 could not function as a dosage suppressor of the sepH 1 mutation in the same manner as it suppresses ΔAnbud3 (H. Si and S. D. Harris, unpublished observation).
Figure 8.—
Localization of GFP–AnBud3 in the sepH 1 mutant. GFP–AnBud3 localization at 28° (A) and 42° (C) following 14-hr growth of strain AHS62 on YGV. GFP–AnBud3 localization to septation sites was not observed at 42°. B and D are corresponding DIC images. Arrows indicate septation sites. Bar, 5 μm.
The presumptive GAP Msb1 does not regulate septum formation:
Annotation of the A. nidulans genome revealed almost all predicted Rho GTPase activating proteins (Rho GAPs) could be matched to a Rho GTPase by analogy to known modules in S. cerevisiae and S. pombe (Harris et al. 2009; S. D. Harris, unpublished data). The sole exception is ANID_02983.1, which is a presumptive homolog of S. cerevisiae Msb1. Both proteins are predicted to possess full-length Rho–GAP domains at their N terminus. Furthermore, results from genetic analyses in budding yeast implicate Msb1 in both Cdc42 and Rho1 signaling pathways (Bender and Pringle 1989; Sekiya-Kawasaki et al. 2002), though it is not known if it possesses GAP activity. Accordingly, we reasoned that AnMsb1 might function as a GAP for Rho4, and tested this idea by deleting it in both wild-type and ΔAnbud3 strains. Our expectation was that loss of a Rho4 GAP would lead to a hyperactive Rho GTPase, which would result in increased septum formation in a wild-type background and would also be potentially capable of suppressing the loss of septation in Δbud3 mutants. However, deletion of msb1 only had minor effects on colony growth (i.e., reduced conidiation) and did not affect septum formation (Figure 9). Furthermore, Δmsb1 did not restore septation to any extent to Δbud3 mutants (data not shown). These observations suggest that AnMsb1 alone is not likely to function as a GAP for Rho4.
Figure 9.—
Effects of the Δmsb1 mutation on growth and hyphal morphology. (A and B) Colony morphology of wild-type (TNO2A3; A) and Δmsb1 (AHS7; B) strains following growth on MNVUU medium for 6 days and 7 days, respectively. (C and D) Hyphal morphology of wild-type (TNO2A3; C) and Δmsb1 (AHS7; D) strains following growth for 13 hr on MNVUU at 28°. Arrows indicate septa. Bar, 10 μm.
As an alternative approach to the identification of candidate GAPs for Rho4, we isolated a set of extragenic suppressors of Δbud3 (Figure S5), which were then tested to determine whether they harbored mutations in any of the annotated Rho GAPs (Harris et al. 2009). However, no predicted GAP was capable of complementing the suppressor mutation (i.e., restoring the original Δbud3 phenotype) when amplified and cotransformed with the pRG3–AMA1 plasmid. Thus, the nature, or even the existence, of the Rho4 GAP remains unresolved.
DISCUSSION
The formation of septa in A. nidulans hyphae requires the formin-dependent assembly of a CAR (Sharpless and Harris 2002). Although Rho GTPases are known to activate formins (e.g., Dong et al. 2003; Martin et al. 2007), the identity of the relevant GTPase(s) that direct CAR assembly has remained unknown. Whereas our previous results show that Cdc42 has no detectable role in septation (Virag et al. 2007), the results presented here demonstrate that the Bud3–Rho4 GTPase module is required for CAR assembly and formin recruitment to septation sites.
The Bud3–Rho4 GTPase module:
Our observations demonstrate that AnBud3 and Rho4 are required for septum formation in A. nidulans hyphae. The loss of either protein abolishes septation; in the case of bud3 mutants, this appears to be caused by the failure to recruit the formin SepA, which is required for CAR assembly, to septation sites. Furthermore, both proteins localize to septation sites, where they form constricting rings. Characterization of the AnBud3 localization pattern in particular reveals that it first appears prior to the formation of a detectable septum, then constricts in a manner consistent with the notion that it is a component of the CAR. Finally, our genetic and biochemical analyses clearly establish that AnBud3 serves as a GEF that promotes activation of Rho4. A similar relationship between Bud3 and Rho4 has recently been described for another filamentous ascomycete fungus, N. crassa (Justa-Schuch et al. 2010). In this case, Bud3 also acts as a GEF for Rho4, which in turn directs assembly of the CAR at septation sites (Rasmussen and Glass 2005). Collectively, these results define Bud3 and Rho4 as essential components of the GTPase modules that direct CAR assembly during septation in those filamentous fungi that belong to the euascomycetes. Among the questions that still need to be addressed is the identity of the relevant Rho4 GAP. Although AnMsb1 appeared to be a reasonable candidate, our results suggest that even if it does have GAP activity, it is not the sole GAP for Rho4. Instead, it seems likely that multiple GAPs might act in a redundant manner to regulate Rho4.
In addition to A. nidulans and N. crassa, Bud3 and Rho4 homologs have been implicated in septum formation in filamentous fungi that belong to the hemiascomycetes. In A. gossypii, Bud3 serves as a landmark for future septation events and also functions to properly position the CAR (Wendland 2003). It remains unknown whether this Bud3 homolog, or for that matter the founding S. cerevisiae homolog, possess GEF activity. In C. albicans, Rho4 appears to regulate deposition of the septum during both yeast and hyphal phases of growth (Dunkler and Wendland 2007). At this time, no relationship between Bud3 and Rho4 has been described for any hemiascomycete. We speculate that Bud3 activation of Rho4 may represent an ancestral interaction that was lost in the hemiascomycete lineage. This could presumably account for lack of pronounced sequence similarity between euascomycete Bud3 homologs and S. cerevisiae Bud3, and for the observation that the euascomycete Rho4 homologs form a distinct clade of fungal Rho GTPases that apparently lack hemiascomycete members (Rasmussen and Glass 2005), although the relationship of C. albicans Rho4 to this clade is uncertain (Dunkler and Wendland 2007). An investigation of the possible interaction between Bud3 and Rho4 homologs in archiascomycetes such as the yeast S. pombe might help to further clarify how the Bud3–Rho4 GTPase module has evolved in fungi.
The Bud3–Rho4 pathway:
Our results show that the Bud3–Rho4 GTPase module controls assembly of the CAR during septation in A. nidulans. A likely effector of Rho4 that mediates this function is the formin SepA, which is no longer recruited to septation sites in ΔAnbud3 mutants even though its localization at hyphal tips is unaffected. Furthermore, our results show that AnBud3 still accumulates at septation sites in the absence of SepA, thereby implying that its function lies upstream of SepA. We envision the following scenario on the basis of our observations. In response to signals emanating from the nucleus (see below), AnBud3 localizes to presumptive septation sites, where it activates Rho4 to initiate the process of assembling the CAR. Activated Rho4 accomplishes this task by locally recruiting SepA and other regulators of actin filament dynamics. Moreover, AnBud3 and Rho4 remain associated with the assembled CAR during the process of constriction. By doing so, AnBud3, and by inference activated Rho4, may control additional steps beyond recruitment of SepA, such as maintenance of the CAR or coordination of septum deposition with ring constriction. (e.g., Nakano et al. 2003; Santos et al. 2003).
One of the distinct features of septum formation in filamentous fungi such as A. nidulans is the uncoupling of cell division from nuclear division (Clutterbuck 1970; Harris 2001; Walther and Wendland 2003), which implies the existence of unique regulatory mechanisms that coordinate CAR assembly with mitosis (i.e., not every dividing nucleus is capable of triggering formation of a CAR). Because Rho4 appears to serve as a pivotal regulator of CAR assembly during septation, it seems likely that it would be responsive to signals emanating from dividing nuclei. Moreover, by analogy to the well-characterized Rho/Cdc42 GTPase modules in S. cerevisiae (Park and Bi 2007), the GEFs and/or GAPs that regulate Rho4 are potential targets for these signals. Our observations that nuclear division and a functional SIN pathway are required for AnBud3 localization at septation sites are consistent with this idea. Future efforts will focus on determining whether the protein kinase constituents of the SIN (SepH, SepL, and SidB) (Bruno et al. 2001; Kim et al. 2006, 2009) interact directly with AnBud3 to control its localization and/or activity. Notably, in S. pombe, Orb6, which like SidB is a member of the NDR kinase family, spatially regulates polarized growth by restricting the localization of the Cdc42 GEF Gef1 (Das et al. 2009). Finally, it should also be noted that our data imply that the Bud3–Rho4 module is not the sole target of the SIN. Instead, we envision the SIN acting via multiple targets to coordinate CAR assembly and function with nuclear division.
Acknowledgments
This work was supported by the Nebraska Research Foundation (S.H.), National Science Foundation grant IOS-0920504 (S.H.), and the Deutsche Forschungsgemeinschaft Priority Program SPP1111 (S.S.).
Supporting information is available online at http://www.genetics.org/cgi/content/full/genetics.110.114165/DC1.
References
- Abe, K., K. L. Rossman, B. Liu, K. D. Ritola, D. Chiang et al., 2000. Vav2 is an activator of Cdc42, Rac1, and RhoA. J. Biol. Chem. 275 10141–10149. [DOI] [PubMed] [Google Scholar]
- Balasubramanian, M. K., E. Bi and M. Glotzer, 2004. Comparative analysis of cytokinesis in budding yeast, fission yeast and animal cells. Curr. Biol. 14 R806–R818. [DOI] [PubMed] [Google Scholar]
- Bauer, Y., P. Knechtle, J. Wendland, H. Helfer and P. Philippsen, 2004. A Ras-like GTPase is involved in hyphal growth guidance in the filamentous fungus Ashbya gossyppii. Mol. Biol. Cell 15 4622–4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender, A., and J. R. Pringle, 1989. Multicopy suppression of the cdc24 budding defect in yeast by CDC42 and three newly identified genes including the ras-related gene RSR1. Proc. Natl. Acad. Sci. USA 86 9976–9980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno, K. S., J. L. Morrell, J. E. Hamer and C. J. Staiger, 2001. SEPH, a Cdc7p orthologue from Aspergillus nidulans, functions upstream of actin ring formation during cytokinesis. Mol. Microbiol. 42 3–12. [DOI] [PubMed] [Google Scholar]
- Chant, J., 1999. Cell polarity in yeast. Annu. Rev. Cell Dev. Biol. 15 365–391. [DOI] [PubMed] [Google Scholar]
- Chant, J., and I. Herskowitz, 1991. Genetic control of bud site selection in yeast by a set of gene products that constitute a morphogenetic pathway. Cell. 65 1203–1212. [DOI] [PubMed] [Google Scholar]
- Chant, J., and J. R. Pringle, 1995. Patterns of bud-site selection in the yeast Saccharomyces cerevisiae. J. Cell Biol. 129 751–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chant, J., M. Mischke, E. Mitchell, I. Herskowitz and J. Pringle, 1995. Role of Bud3p in producing the axial budding pattern of yeast. J. Cell Biol. 129 767–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clutterbuck, A. J., 1970. Synchronous nuclear division and septation in Aspergillus nidulans. J. Gen. Microbiol. 60 133–135. [DOI] [PubMed] [Google Scholar]
- Das, M., D. J. Wiley, X. Chen, K. Shah and F. Verde, 2009. The conserved NDR kinase Orb6 controls polarized cell growth by spatial regulation of the small GTPase Cdc42. Curr. Biol. 19 1314–1319. [DOI] [PubMed] [Google Scholar]
- Dong, Y., D. Pruyne and A. Bretscher, 2003. Formin-dependent actin assembly is regulated by distinct modes of Rho signaling in yeast. J. Cell. Biol. 161 1081–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunkler, A., and J. Wendland, 2007. Candida albicans Rho-type GTPase-encoding genes required for polarized cell growth and cell separation. Eukaryot. Cell. 6 844–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efimov, V. P., 2003. Roles of NUDE and NUDF proteins of Aspergillus nidulans: insights from intracellular localization and overexpression effects. Mol. Biol. Cell. 14 871–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freifelder, D., 1960. Bud position in Saccharomyces cerevisiae. J. Bacteriol. 80 567–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galagan, J. E., S. E. Calvo, C. Cuomo, L. J. Ma, J. R. Wortman et al., 2005. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438 1105–1115. [DOI] [PubMed] [Google Scholar]
- Gao, X. D., L. M. Sperber, S. A. Kane, Z. Tong, A. H. Tong et al., 2007. Sequential and distinct roles of the cadherin domain-containing protein Axl2p in cell polarization in yeast cell cycle. Mol. Biol. Cell. 18 2542–2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gull, K., 1978. Form and function of septa in filamentous fungi, pp. 78–93 in The Filamentous Fungi, Developmental Mycology, edited by J. E. Smith and D. R. Berry. John Wiley & Sons, New York.
- Harkins, H. A., N. Page, L. R. Schenkman, C. De Virgilio, S. Shaw et al., 2001. Bud8p and Bud9p, proteins that may mark sites for bipolar budding in yeast. Mol. Biol. Cell 12 2497–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, S. D., 2001. Septum formation in Aspergillis nidulans. Curr. Opin. Microbiol. 4 736–739. [DOI] [PubMed] [Google Scholar]
- Harris, S. D., and M. Momany, 2004. Polarity in filamentous fungi: moving beyond the yeast paradigm. Fungal Genet. Biol. 41 391–400. [DOI] [PubMed] [Google Scholar]
- Harris, S. D., J. L. Morrell and J. E. Hamer, 1994. Identification and characterization of Aspergillus nidulans mutants defective in cytokinesis. Genetics 136 517–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, S. D., L. Hamer, K. E. Sharpless and J. E. Hamer, 1997. The Aspergillus nidulans sepA gene encodes an FH1/2 protein involved in cytokinesis and the maintenance of cellular polarity. EMBO J. 16 3474–3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, S. D., G. Turner, V. Meyer, E. A. Espeso, T. Specht et al., 2009. Morphology and development in Aspergillus nidulans: a complex puzzle. Fungal Genet. Biol. 46 S82–S92. [DOI] [PubMed] [Google Scholar]
- Hausauer, D. L., M. Gerami-Nejad, C. Kistler-Anderson and C. A. Gale, 2005. Hyphal guidance and invasive growth in Candida albicans require the Ras-like GTPase Rsr1p and its GTPase-activating protein Bud2p. Eukaryot. Cell 4 1273–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hlubek, A., K. O. Schink, M. Mahlert, B. Sandrock and M. Bolker, 2008. Selective activation by the guanine nucleotide exchange factor Don1 is a main determinant of Cdc42 signaling specificity in Ustilago maydis. Mol. Microbiol. 68 615–623. [DOI] [PubMed] [Google Scholar]
- Justa-Schuch, D., Y. Heilig, C. Richthammer and S. Seiler, 2010. Septum formation is regulated by the RHO4-specific exchange factors BUD3 and RGF3 and by the landmark protein BUD4 in Neurospora. Mol. Microbiol. (in press). [DOI] [PubMed]
- Kafer, E., 1977. Meiotic and mitotic recombination in Aspergillus and its chromosomal aberration. Adv. Genet. 19 33–131. [DOI] [PubMed] [Google Scholar]
- Kang, P. J., A. Sanson, B. Lee and H.-O. Park, 2001. A GDP/GTP exchange factor involved in linking a spatial landmark to cell polarity. Science 292 1376–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, P. J., E. Angerman, K. Nakashima, J. R. Pringle and H.-O. Park, 2004. Interactions among Rax1p, Rax2p, Bud8p, and Bud9p in marking cortical sites for bipolar bud-site selection in yeast. Mol. Biol. Cell 15 5145–5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J. M., L. Lu, R. Shao, J. Chin and B. Liu, 2006. Isolation of mutations that bypass the requirement of the septation initiation network for septum formation and conidiation in Aspergillus nidulans. Genetics 173 685–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, J. M., C. J. Zeng, T. Nayak, R. Shao, A. C. Huang et al., 2009. Timely septation requires SNAD-dependent spindle pole body localization of the septation initiation network components in the filamentous fungus Aspergillus nidulans. Mol. Biol. Cell 20 2874–2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapp, A., and V. Simanis, 2008. An overview of the fission yeast septation initiation network (SIN). Biochem. Soc. Trans. 36 411–415. [DOI] [PubMed] [Google Scholar]
- Krappmann, A. B., N. Taheri, M. Heinrich and H. U. Mosch, 2007. Distinct domains of yeast cortical tag proteins Bud8p and Bud9p confer polar localization and functionality. Mol. Biol. Cell 18 3323–3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, X., and M. Momany, 2003. The Aspergillus nidulans swoC1 mutants shows defects in growth and development. Genetics 165 543–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord, M., F. Inose, T. Hiroko, T. Hata, A. Fujita et al., 2002. Subcellular localization of Axl1, the cell type-specific regulator of polarity. Curr. Biol. 12 1347–1352. [DOI] [PubMed] [Google Scholar]
- Martin, S. G., S. A. Rincon, R. Basu, P. Perez and F. Chang, 2007. Regulation of the formin for3p by cdc42p and bud6p. Mol. Biol. Cell 18 4155–4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momany, M., and J. E. Hamer, 1997. Relationship of actin, microtubules, and crosswall synthesis during septation in Aspergillus nidulans. Cell Motil. Cytoskeleton 38 373–384. [DOI] [PubMed] [Google Scholar]
- Nakano, K., T. Mutoh, R. Arai and I. Mabuchi, 2003. The small GTPase Rho4 is involved in controlling cell morphology and septation in fission yeast. Genes Cells 8 357–370. [DOI] [PubMed] [Google Scholar]
- Nayak, T., E. Szewczyk, C. E. Oakley, A. Osmani, L. Ukil et al., 2006. A versatile and efficient gene targeting system for Aspergillus nidulans. Genetics 172 1557–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmani, A. H., B. R. Oakley and S. A. Osmani, 2006. Identification and analysis of essential Aspergillus nidulans genes using the heterokaryon rescue technique. Nat. Protocols 1 2517–2526. [DOI] [PubMed] [Google Scholar]
- Ozaki, K., K. Tanaka, H. Imamura, T. Hihara, T. Kameyama et al., 1996. Rom1p and Rom2p are GDP/GTP exchange proteins (GEPs) for the Rho1p small GTP binding protein in Saccharomyces cerevisiae. EMBO J. 15 2196–2207. [PMC free article] [PubMed] [Google Scholar]
- Park, H.-O., and E. Bi, 2007. Central roles of small GTPases in the development of cell polarity and beyond. Microbiol. Mol. Biol. Rev. 71 48–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson, C. L., K. Xu, K. E. Sharpless and S. D. Harris, 2004. MesA, a novel fungal protein required for the stabilization of polarity axes in Aspergillus nidulans. Mol. Biol. Cell 15 3658–3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippsen, P., A. Kaufmann and H. P. Schmitz, 2005. Homologues of yeast polarity genes control the development of multinucleated hyphae in Ashbya gossypii. Curr. Opin. Microbiol. 8 370–377. [DOI] [PubMed] [Google Scholar]
- Rasmussen, C. G., and N. L. Glass, 2005. A Rho-type GTPase, rho-4, is required for septation in Neurospora crassa. Eukaryot. Cell 4 1913–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos, B., J. Gutierrez, T. M. Calonge and P. Perez, 2003. Novel Rho GTPase involved in cytokinesis and cell wall integrity in the fission yeast Schizosaccharomyces pombe. Eukaryot. Cell 2 521–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiya-Kawasaki, M., M. Abe, A. Saka, D. Watanabe, K. Kono et al., 2002. Dissection of upstream regulatory components of the Rho1p effector, 1,3-beta-glucan synthase, in Saccharomyces cerevisiae. Genetics 162 663–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharpless, K. E., and S. D. Harris, 2002. Functional characterization and localization of the Aspergillus nidulans formin SEPA. Mol. Biol. Cell 13 469–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taheri-Talesh, N., T. Horio, L. Araujo-Bazan, X. Dou, E. A. Espeso et al., 2008. The tip growth apparatus of Aspergillus nidulans. Mol. Biol. Cell 19 1439–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinci, A. P.J., and N. R. Morris, 1979. Morphology and growth of a temperature-sensitive mutant of Aspergillus nidulans which forms aseptate mycelia at non-permissive temperatures. J. Gen. Microbiol. 114 53–59. [Google Scholar]
- Virag, A., M. P. Lee, H. Si and S. D. Harris, 2007. Regulation of hyphal morphogenesis by cdc42 and rac1 homologues in Aspergillus nidulans. Mol. Microbiol. 66 1579–1596. [DOI] [PubMed] [Google Scholar]
- Vogt, N., and S. Seiler, 2008. The RHO1-specific GTPase-activating protein LRG1 regulates polar tip growth in parallel to Ndr kinase signaling in Neurospora. Mol. Biol. Cell 19 4554–4569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther, A., and J. Wendland, 2003. Septation and cytokinesis in fungi. Fungal Genet. Biol. 40 187–196. [DOI] [PubMed] [Google Scholar]
- Wendland, J., 2003. Analysis of the landmark protein Bud3 of Ashbya gossypii reveals a novel role in septum construction. EMBO Rep. 4 200–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfall, P. J., and M. Momany, 2002. Aspergillus nidulans septin AspB plays pre- and postmitotic roles in septum, branch, and conidiophore development. Mol. Biol. Cell 13 110–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolkow, T. D., S. D. Harris and J. E. Hamer, 1996. Cytokinesis in Aspergillus nidulans is controlled by cell size, nuclear positioning and mitosis. J. Cell Sci. 109 2179–2188. [DOI] [PubMed] [Google Scholar]
- Yang, L., L. Ukil, A. Osmani, F. Nahm, J. Davies et al., 2004. Rapid production of gene replacement constructs and generation of a green fluorescent protein-tagged centromeric marker in Aspergillus nidulans. Eukaryot. Cell 3 1359–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh, B. J., R. J. Rutigliano, A. Deb, D. Bar-Sagi and W. A. Lim, 2007. Rewiring cellular morphology pathways with synthetic guanine nucleotide exchange factors. Nature 447 596–600. [DOI] [PubMed] [Google Scholar]