Abstract
Many plant-associated microbes synthesize the auxin indole-3-acetic acid (IAA), and several IAA biosynthetic pathways have been identified in microbes and plants. Saccharomyces cerevisiae has previously been shown to respond to IAA by inducing pseudohyphal growth. We observed that IAA also induced hyphal growth in the human pathogen Candida albicans and thus may function as a secondary metabolite signal that regulates virulence traits such as hyphal transition in pathogenic fungi. Aldehyde dehydrogenase (Ald) is required for IAA synthesis from a tryptophan (Trp) precursor in Ustilago maydis. Mutant S. cerevisiae with deletions in two ALD genes are unable to convert radiolabeled Trp to IAA, yet produce IAA in the absence of exogenous Trp and at levels higher than wild type. These data suggest that yeast may have multiple pathways for IAA synthesis, one of which is not dependent on Trp.
THE auxin indole-3-acetic acid (IAA) is best known for its role in plant cell elongation, division, and differentiation (Halliday et al. 2009; Moller and Weijers 2009; Sundberg and Ostergaard 2009; Zazimalova et al. 2009; Abel and Athanosios 2010; McSteen 2010; Scarpella et al. 2010); however, IAA has been identified in numerous plant-associated bacteria (reviewed in Glick et al. 1999a,b) and several fungi, including Rhizopus suinous (Thimann 1935), Rhizoctonia (Furukawa et al. 1996), Colletotrichum (Robinson et al. 1998), and yeast (Nielsen 1931; Gruen 1959). Microbial IAA plays a significant role in plant–microbe interactions (Glick et al. 1999a), both pathogenic and symbiotic (Hirsch et al. 1989; Reineke et al. 2008). Plants infected with pathogenic microbes manifest phenotypes consistent with elevated levels of IAA, such as gall formation (a tumor resulting from cellular proliferation) and lengthening of the stem (Viglierchio 1971; Barash and Manulis-Sasson 2009; Stewart and Nemhauser 2009). The interplay between microbial-derived IAA and plant-derived IAA in plant disease is just beginning to be defined.
Exogenous IAA regulates filamentation in Saccharomyces cerevisiae, a fungus that is primarily associated with plants, by inducing expression of genes that mediate its morphological transition from a vegetative form to a pseudohyphal or filamentous form (Prusty et al. 2004). The fungal transcription factor, Yap1, regulates IAA homeostasis in S. cerevisiae (Prusty et al. 2004) by downregulating auxin permeases (Avt proteins) that import IAA in S. cerevisiae (Prusty et al. 2004). We show here that IAA stimulates filamentation in the human pathogen Candida albicans and that C. albicans Yap1 (Cap1) also mediates IAA phenotypes. Filamentation often underlies the development of virulence of C. albicans. For example, the C. albicans double mutant cph1Δ/Δ efg1Δ/Δ is defective in the MAP kinase pathway through Cph1, as well as in the PKA pathway via Efg1. This mutant fails to switch from vegetative to filamentous form (Lo et al. 1997; Brown et al. 1999; Riggle et al. 1999; Liu 2001; Sohn et al. 2003) and is also avirulent (Dieterich et al. 2002). These studies suggest that the secondary metabolite IAA is a chemical signal that regulates fungal pathogenesis.
Plants have multiple pathways to synthesize, inactivate, and catabolize IAA (Delker et al. 2008; Lau et al. 2008; Normanly 2009). Molecular genetic studies in model systems such as Arabidopsis thaliana (reviewed in Normanly 2009), coupled with precise analytical methods (Barkawi et al. 2008), have helped expose some redundancy within this network. In fungi, IAA has been generally proposed as a metabolite of tryptophan (Trp) (Hazelwood et al. 2008) but this has been conclusively demonstrated only in Ustilago maydis (Reneke et al. 1988) and S. uvarum (Shin et al. 1991). Early studies used activity assays or qualitative colorimetric techniques to indicate the presence of IAA. Thin layer chromatography (TLC) and high performance liquid chromatography (HPLC) were subsequently employed for the detection of IAA, where the bioactive compound was shown to chromatograph with authentic IAA. Definitive isotope dilution quantification of IAA was first carried out with [14C]IAA and extracts from U. zeae tumors (Turian and Hamilton 1960).
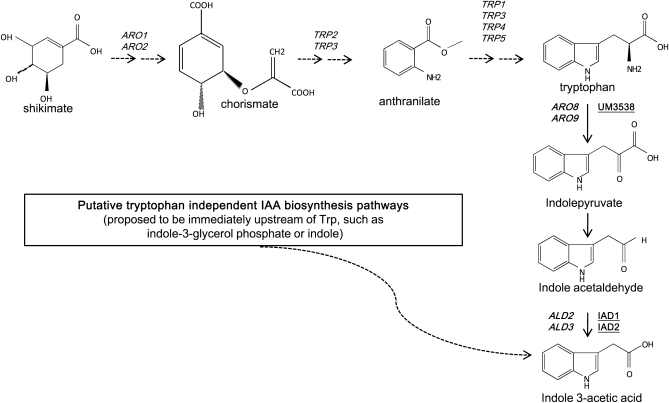
Here, we used gas-chromatography mass spectrometry (GC-MS) coupled with stable isotope dilution to demonstrate that S. cerevisiae synthesizes IAA. We identified genes homologous to the aldehyde dehydrogenase that functions in a Trp-dependent IAA biosynthetic pathway in U. maydis (Figure 1) (Basse et al. 1996; Reineke et al. 2008). Our results are consistent with the presence of a Trp-independent IAA biosynthetic pathway in yeast as well.
Figure 1.—
The IAA biosynthetic pathway identified in this study (in boldface type) and the analogous pathway identified in U. maydis (right, underlined) where the homologs of Ald2 and Ald3 have been shown to catalyze the conversion of indole-3-acetaldehyde to indole-3-acetic acid.
MATERIALS AND METHODS
Strains, media, and growth conditions:
Table 1 lists the strains used in this study. Deletion strains were derived from the yeast-deletion set (Winzeler et al. 1999) and subsequently reconstructed by replacement of the relevant ORF with a dominant drug resistance marker (Wach et al. 1994). Analytical and phenotypic studies were performed in cognate deletion mutants, made in the Σ1278b background. A [14C]Trp incorporation assay was performed to verify that phenotype observed in the library strain could be recapitulated in the newly constructed Σ1278b strain. Typically three independent transformants were isolated, confirmed by PCR, and used for further studies. Standard culture conditions were used (Sherman et al. 1986) and analysis of IAA-associated phenotypes was performed as described earlier (Prusty et al. 2004).
TABLE 1.
Strains used in this study
| Strain | Description | Source |
|---|---|---|
| BY4741 | MATa; his3Δ1; leu2Δ0; met15Δ0; ura3Δ0 | YDLa |
| ald2Δ | ald2Δ in BY4741 | YDL |
| ald3Δ | ald3Δ in BY4741 | YDL |
| ald4Δ | ald4Δ in BY4741 | YDL |
| ald5Δ | ald5Δ in BY4741 | YDL |
| ald6Δ | ald6Δ in BY4741 | YDL |
| ald2Δ ald3Δ | ald2Δ ald3Δ in BY4741 | This study |
| Σ1278b | MATa; ura3Δ0 | J. Heitman, Duke University |
| ald2Δ/Δ ald3Δ/Δ | ald2Δ ald3Δ in Σ1278b, MATa/α | This study |
| Caf2-1 | Candida albicans wild type | G. Fink, MIT |
| cph1Δ/Δ | Homozygous cph1Δ in Caf2-1 | G. Fink, MIT |
| efg1Δ/Δ | Homozygous efg1Δ in Caf2-1 | G. Fink, MIT |
| cph1Δ/Δ efg1Δ/Δ | Homozygous cph1Δ efg1Δ in Caf2-1 | G. Fink, MIT |
| cap1Δ/+ | Heterozygous cap1Δ in Caf2-1 | M. Raymond, University of Montreal |
| cap1Δ/Δ | Homozygous cap1Δ in Caf2-1 | M. Raymond, University of Montreal |
Yeast Deletion Library.
[14C]Trp incorporation assay:
Yeast strains were grown in 5-ml overnight cultures with aeration at 30° in synthetic complete medium (Sigma, St. Louis) (Guthrie and Fink 1991). To estimate cell density, the absorbance at 600 nm was measured and the culture was adjusted to an OD600 of 1 (∼2 × 107 cfu/ml). Cells (1 ml) were harvested by centrifugation at 3000 rpm for 5 min on an Eppendorf table-top microfuge at room temperature. Cells were washed twice by resuspending pellets in water and then harvested by centrifugation. Cell pellets were resuspended in 200 μl of SD medium supplemented with auxotrophic amino acids (Guthrie and Fink 1991). Samples were incubated with rocking (Thermolyne, speci mix) at 30° for ∼18 hr in media containing 400 μm Trp and 0.5 μCi of [14C]Trp (Trp L-[side chain-3-14C], specific activity 50 mCi/mmol; American Radiochemicals). Cells were removed by centrifugation (3000 rpm in an Eppendorf table-top microfuge) at room temperature and the conditioned medium (CM) was transferred to new tubes for TLC. Control samples were prepared identically but without the addition of cells to the SD medium. Ten microliters of the CM was spotted on TLC plates. The [14C]Trp metabolites in the CM were resolved on a silica gel 60 F254 (20 × 20 cm, 250 μm thick, precoated) TLC plate (EMD Chemicals). A mixture of 85% chloroform, 14% methanol, and 1% water was used as the eluting solvent. IAA that had incorporated label from [14C]Trp was visualized by autoradiography. Commercially available [14C]IAA (American Radiochemicals) was used as a standard. To screen the yeast deletion set, this assay was adapted for use in 96-well microtiter dishes by scaling down the reaction volume to 50 μl containing 0.1 μCi [14C]Trp.
Quantification of IAA from yeast:
To confirm that IAA was present in the CM, 5-ml cultures were harvested and stored at −80°. The supernatants were thawed on ice and 38.4 ng of [13C6]IAA (99 atom%, Cambridge Isotope Laboratories) in 10 μl of 2-propanol was added as an internal standard. Additionally, 500 μl of 0.2 m imidazole (pH 7.0) was added. The sample was mixed and left to equilibrate on ice for 1 hr. The sample was loaded onto a 200-mg NH2 solid phase extraction (SPE) column (aka amino columns, Alltech) that was preconditioned with sequential applications of 2 ml each hexane, acetonitrile, and water and 0.2 m imidazole (pH 7.0) followed by 6 ml of water on a vacuum manifold (Fisher Scientific, Pittsburgh, PA). After loading the sample, the column was allowed to aspirate under vacuum for an additional 30 sec at 3–5 psi. Next, the column was washed with sequential additions of 1 ml each of hexane, ethyl acetate, acetonitrile, and methanol. IAA was eluted in ∼6 ml of methanol that was 5% acetic acid. Dried samples were resuspended in 1.3 ml of a mixture (∼6:1 to reach a pH between 3 and 3.5) of 0.25% phosphoric acid and 0.1 m succinic acid, pH 6.0. The sample was placed in a 2-ml capacity 96-well plate and subjected to an additional SPE step with polymethymethacrylate epoxide resin, using a Gilson 215 SPE automated liquid handler (ALH) as described in Barkawi et al. (2008). The epoxide SPE column eluate was transferred to 2-ml amber vials, and ∼1 ml of ethereal diazomethane (prepared as described in Cohen 1984) was added. After 5 min incubation at room temperature, the sample was dried to a residue under a stream of N2 gas in a 45° sand bath. The methylated IAA was resuspended in 45 μl of ethyl acetate and subjected to GC-MS analysis as described in Barkawi et al. (2008), except that a full scan spectrum was obtained. For mutant analysis this protocol was scaled down to 1-ml cultures containing the same amount of [13C6]IAA internal standard but only 0.2 ml of 0.2 m imidazole, pH 7.0.
RESULTS
S. cerevisiae secretes IAA:
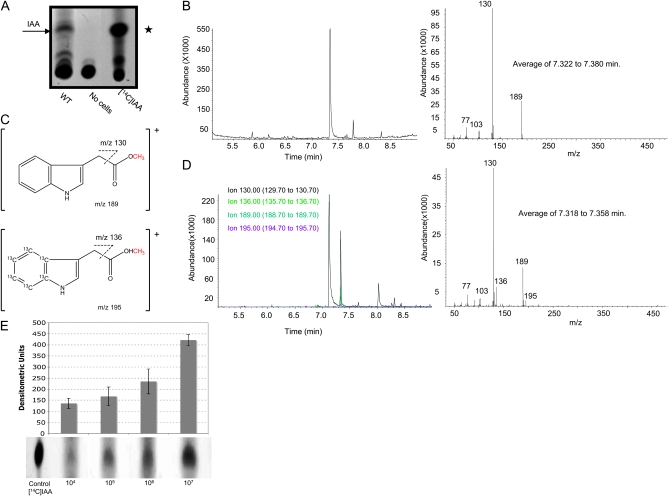
To confirm that S. cerevisiae synthesizes IAA, we analyzed CM from yeast cultures. Thin layer chromatography of CM from S. cerevisiae grown in the presence of [14C]Trp revealed a radiolabeled product that comigrated with commercially available [14C]IAA (Figure 2A). UV shadow of the fluor-impregnated TLC plate showed a UV absorbing compound with the same retention profile as the pure unlabeled IAA that was used as a standard (data not shown but position marked with asterisk in Figure 2A). GC-MS analysis of IAA that was extracted from the CM along with [13C6]IAA internal standard and methylated for GC analysis (Figure 2, B–D) confirmed the presence of IAA in the CM. Figure 2B (left) shows the total ion chromatogram (TIC) of pure methyl (Me)-IAA, which shows a GC retention time for authentic Me-IAA to be between 7.322 and 7.380 min. Figure 2B (right) shows the full scan spectrum corresponding to this retention time. The predominant ions for pure Me-IAA (also shown in Figure 2C, top) are m/z 189 (intact Me-IAA, aka molecular ion) and mass to charge ration (m/z) 130 (fragment ion). The observed fragment ions of m/z 103 and 77 were consistent with pure Me-IAA as well, but were lower in abundance and not typically used for quantification. Figure 2C (bottom) shows the molecular and fragment ions for Me-[13C6 ]IAA. Figure 2D (left) shows the TIC of IAA that was extracted from 5 ml of CM (to which [13C6]IAA had been added), methylated, and run on GC-MS. The four predominant ions for Me-IAA and Me-[13C6]IAA are shown. Figure 2D (bottom) shows the full scan spectrum for the retention time that corresponds to authentic Me-IAA, demonstrating that yeast secretes IAA.
Figure 2.—
(A) S. cerevisiae produces a molecule that comigrates with commercially available IAA. Wild-type yeast cells were incubated with [14C]Trp, and products of the conditioned media were resolved by thin layer chromatography (TLC). Commercially available IAA and [14C]IAA were used as controls. The position (marked with the asterisk) of the nonradiolabeled IAA control was determined by UV shadowing. (B) Total ion chromatogram (TIC, left) and full scan spectrum (right) of authentic methyl-IAA. (C) Top, methyl-IAA molecular ion m/z 189 and fragment ion m/z 130 (the site of fragmentation to form the fragment ion is indicated by dashed lines). Bottom, methyl-[13C6]IAA molecular ion m/z 195 and fragment ion m/z 136. For each compound, the derivatization moiety (the methyl group) is shown in red. (D) TIC (left) and corresponding full-scan spectrum (right) of IAA (methylated prior to GC-MS analysis) that was purified from the culture medium of wild-type yeast that had been grown in the presence of Trp. The TIC shows four selected ions; m/z 130 and m/z 189 are the fragment ion and the molecular ion, respectively, of endogenous IAA (methylated prior to GC-MS analysis). Ions with m/z 136 and 195 are the fragment ion and the molecular ion, respectively, of [13C6]IAA (methylated prior to GC-MS analysis) that was added to the yeast culture medium supernatant prior to extraction of IAA. The large peak in the TIC (left) with a retention time of ∼7.15 min was determined to be tryptophol by full-scan spectra analysis (not shown). (E) The CM taken from a high-density culture contained a much greater concentration of IAA than CM from a low-density culture as determined by TLC (bottom) and densitometry of the autoradiograph (top).
The accumulation of IAA in the CM reached its highest level after cultures entered stationary phase. To correlate the production of IAA with cell density, cells from a high-density culture (108 cells/ml) were diluted to either low (5 × 105 cells/ml) or high density (5 × 107 cells/ml) in fresh medium. IAA secreted into the medium was assessed by TLC (Figure 2E). After normalizing for the difference in cell number, we found that CM taken from a high-density culture contained more IAA than CM from a low-density culture (Figure 2E), indicating that IAA accumulation is directly proportional to cell density. In S. cerevisiae, IAA is perceived, is imported, and stimulates diploid pseudohyphal growth and haploid invasive growth by regulating the cell surface glycoprotein Flo11. Together these studies suggest that IAA accumulates in the growth environment of yeast where it may act as a chemical signal that regulates virulence traits.
A genomic scale screen for IAA homeostasis mutants:
To identify genes involved in IAA synthesis, specifically the conversion of Trp to IAA, we initiated an unbiased, systematic genomic screen of the yeast deletion library (Brachmann et al. 1998; Winzeler et al. 1999). The haploid deletion library in S. cerevisiae consists of ∼4940 clones representing every single viable gene disruption. A [14C]Trp incorporation assay was developed and optimized to facilitate a large-scale screen using microtiter plates. An aliquot of the CM from each reaction was loaded onto a TLC plate and components of the CM were resolved and compared with a 14C-IAA standard. A total of 1425 deletion strains (29% of the library) have been screened to date. A secondary screen was performed in triplicate on putative mutants and related gene families, using the [14C]Trp incorporation assay.
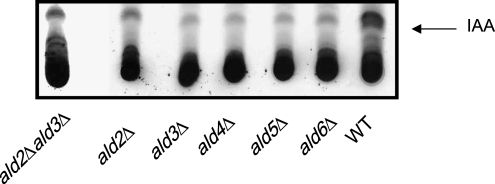
This screen identified three genes, ALD2, ARO9, and ADH2, representing families of particular interest with respect to IAA biosynthesis in yeast: the ALDehyde dehydrogenases, the AROmatic transaminases, and the Alcohol DeHydrogenases (Figure 1). In S. cerevisiae, the aromatic transaminases Aro8 and Aro9 have been implicated in the conversion of Trp to indole pyruvate (IPA) (Chen and Fink 2006). As expected, aro8Δ and aro9Δ mutants show decreased conversion of labeled Trp to labeled IAA compared to the cognate wild type but are not the focus of this study (data not shown). Alcohol dehydrogenases are proposed to convert indole acetaldehyde (IAAld) to indole-3-ethanol (aka tryptophol) (Chen and Fink 2006). Interestingly, adh2Δ, identified in the screen, was the only member of the ADH family to show decreased [14C]IAA accumulation (data not shown). One explanation for this result is that Adh2 preferentially catalyzes the conversion of ethanol to acetylaldehyde. Therefore adh2Δ mutants are unable to convert indole-3-ethanol to IAAld, ultimately leading to decreased IAA accumulation. Deletion mutants of members of the ALD family accumulated lower levels of radioactive IAA from radioactive Trp than did wild type. We focused our study on the aldehyde dehydrogenase (ALD) genes hypothesized to catalyze the ultimate step in the production of IAA and set out to test whether altering IAA production affects filamentation. Multiple sequence alignment and phylogenetic analysis (data not shown) indicate that S. cerevisiae Ald2 and Ald3 share identity with U. maydis Iad1. Ald2 and Ald3 are nearly identical to each other and have 50% (Ald3) and 49% (Ald2) protein sequence identity with U. maydis Iad1, a NAD-dependent aldehyde dehydrogenase. Ald2 and Ald3 have less sequence identity with the NADH-dependent aldehyde dehydrogenases such as lez O (data not shown). Single and double deletions of the ALD genes showed decreased IAA production from [14C]Trp when compared with wild-type cells on TLC (Figure 3). These results together with previous enzymatic studies in U. maydis (Reineke et al. 2008) suggest that these genes are involved in IAA synthesis. ALD2 and ALD3 are also required for synthesis of a nonproteinogenic amino acid, β-alanine in S. cerevisiae (White et al. 2003).
Figure 3.—
Products of the CM of ald single deletion mutants and ald2Δald3Δ double deletion incubated with [14C]Trp were resolved by TLC and compared to the isogenic wild-type strain. Each experiment was performed a minimum of three times. Three and two independent transformants were tested for the single and double mutants, respectively. One representative transformant for each mutant is shown.
The ald2Δald3Δ deletion mutant exhibits virulence traits:
IAA regulates dimorphic transition in S. cerevisiae by inducing adhesion and filamentation (Prusty et al. 2004). The ability of a fungus to perceive a small molecule signal that causes it to differentiate into an invasive form has important implications for host–pathogen interactions. To test the hypothesis that mutants with aberrant IAA accumulation also affect dimorphism, we examined diploid filamentation and haploid invasive growth in all ald single mutants and selected combinations of double mutants. The ald2Δald3Δ double mutant demonstrated increased filamentation (Figure 4A) and invasive growth (Figure 4B) as compared wild type. We also tested a previously reported growth inhibition phenotype associated with exposure to IAA (Figure 4C). This IAA-associated growth inhibition phenotype exhibits a direct proportionality between IAA concentration and growth inhibition. Deletion of both ald2 and ald3 caused an increase in sensitivity to IAA (Figure 4C, right) whereas single deletion of an ALD gene did not affect IAA sensitivity when compared with wild-type cells (data not shown). Together, these data suggest that a perturbation in the IAA secretion profile alters substrate adhesion and filamentation of S. cerevisiae. However, these phenotypes are consistent with ald2Δald3Δ mutants producing more IAA than isogenic wild-type strains.
Figure 4.—
(A) A representative diploid ald2Δ/Δ ald3Δ/Δ colony was grown on filamentation-inducing media and photographed after 3 days of growth (bar, 1 μm). (B) Haploid ald2Δald3Δ strains were spotted onto SC media and washed. Before wash, unwashed plates; after wash, the plates after washing. (C) A filter disk saturated with IAA (right) was placed on a lawn of ald2Δald3Δ mutant cells (bottom) and compared to the wild-type cells (top). Control disks (left) do not contain IAA. Plates were incubated for 3 days in the dark. The clear area around the IAA-containing filter disks indicates a zone of growth inhibition.
The ald2Δald3Δ mutant uncovers an IAA biosynthetic pathway that is independent of exogenous Trp:
The ALD genes were identified on the basis of a radiolabeled [14C]Trp incorporation assay. IAA accumulation in the CM of the double mutant was quantified using GC-MS and [13C6]IAA as an internal standard. These measurements revealed that the CM of the ald2Δald3Δ deletion mutant contained fourfold more IAA (240.3 ng/ml ± 71.9 ng/ml) than the wild type (59.8 ng/ml ± 3.8 ng/ml). The amount of IAA present in the conditioned media is adequate to induce filamentation in an in vitro plate assay. Together these analytical data correlate well with the phenotypic data, suggesting that the ald2Δald3Δ double mutant makes more IAA and thus exhibits enhanced virulence traits as compared to its wild-type counterpart.
While the radiolabeled [14C]Trp incorporation assay detects the pool of IAA synthesized from labeled Trp, the GC-MS analysis allowed us to detect any unlabeled (endogenous) IAA that was present. We grew the ald2Δald3Δ double mutant in the absence of exogenous Trp and quantified IAA from the CM using GC-MS. These measurements revealed that the ald2Δald3Δ mutant was able to synthesize a modest amount of IAA (9.48 ng/ml ± 0.22 ng/ml) in the absence of exogenous Trp. Wild-type yeast also produced similar amount of IAA in the absence of Trp (9.81 ng/ml ± 0.77 ng/ml).
IAA induces filamentation in C. albicans:
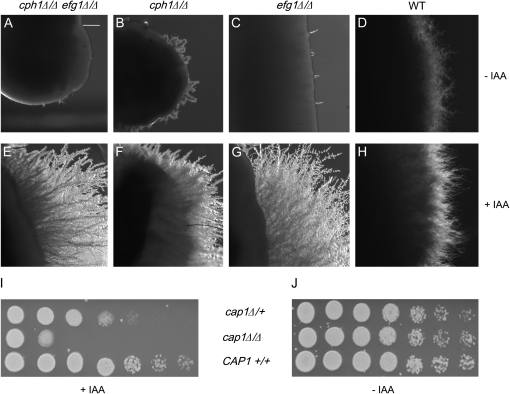
The effects of the secondary metabolites identified in fungi appear to be largely species specific (Chen and Fink 2006). Previous work suggests that IAA induces invasive growth in S. cerevisiae (Prusty et al. 2004). To test whether the IAA effects could cross species barriers, we exposed wild-type Candida albicans, a human pathogen, as well as attenuated mutants in the mitogen-activated protein (MAP) kinase and the cAMP-dependent protein kinase pathways (Figure 5) to IAA. The cph1Δ/Δ efg1Δ/Δ double mutant, which fails to switch from the vegetative to the filamentous form, was filamentous in the presence of IAA (compare Figure 5A with 5E). The single mutants efg1Δ/Δ or cph1Δ/Δ that normally show reduced filamentation also showed a robust filamentation when exposed to IAA (compare Figure 5B with 5F and 5C with 5G). Wild-type strains also filamented more when treated with IAA as compared to untreated cells (compare Figure 5D and 5H). These results indicate that IAA enhances filamentation of the human pathogen C. albicans. Furthermore, the IAA-mediated filamentation signal does not require components of the MAPK or PKA pathways. The cph1Δ/Δ efg1Δ/Δ double mutant, which is nonfilamentous under standard laboratory conditions and avirulent in mice, filaments in the oral cavity of immunosuppressed piglets and when embedded in agar (Riggle et al. 1999). Together these results suggest that IAA-mediated filamentation in C. albicans occurs via an Efg1p- and Cph1p-independent mechanism and confirm prior findings of the existence of an alternate filamentation pathway in C. albicans.
Figure 5.—
The human pathogen Candida albicans was exposed to IAA [experimental plates (E–H) contain 50 μm IAA and control plates (A–D) contain no IAA; bar in A, 10 μm]. A–H show the edge of a patch of C. albicans (A and E, cph1efg1; B and F, cph1; C and G, efg1; and D and H, isogenic wild-type control strains) growing on synthetic low ammonium media with xylose as a carbon source. Plates were incubated in the dark to prevent photodegradation of IAA. I and J show the IAA sensitivity profile of a cap1 homozygous deletion mutant as compared to an isogenic wild-type and a heterozygous mutant [experimental plates (I) contain 120 μm IAA, and control plates (J) contain no IAA].
To test if other aspects of IAA regulation in S. cerevisiae were also conserved in C. albicans, we tested Cap1, the C. albicans homolog of Yap, for its sensitivity to IAA. The amino acid auxin permeases genes are upregulated in the yap1 mutant, which is sensitive to growth on IAA because it retains more IAA (Prusty et al. 2004). Heterozygous and homozygous deletion mutants of CAP1 (Alarco and Raymond 1999) (obtained from M. Raymond, University of Montreal) to grew less well on media containing IAA as compared to the isogenic wild type (Figure 5I), suggesting that the cap1Δ/Δ mutant was more sensitive to IAA. The heterozygous mutant, cap1Δ/+ exhibited an intermediate sensitivity to IAA as compared to the wild-type CAP1+/+ strain or the homozygous cap1Δ/Δ deletion strain. These results suggest that cap1 mutants are hypersensitive to IAA, further supporting our hypothesis that the molecular mechanism of IAA response is likely to be conserved between S. cerevisiae and C. albicans.
DISCUSSION
The quantitative GC-MS analysis in this study confirmed that S. cerevisiae synthesizes and secretes IAA into the culture environment where it is available to function as a signal that regulates filamentation. Filamentation is a pathogenic trait because it contributes directly to virulence of pathogenic fungi like C. albicans. Pathogenic bacteria and fungi are known to produce IAA, but a direct link to pathogenicity has not been demonstrated in these pathogens.
IAA is a small molecule capable of stimulating the developmental transition from the vegetative yeast form to the filamentous form in S. cerevisiae (Prusty et al. 2004). The current study provides strong support for a connection between fungal dimorphism and IAA synthesis, because the ald2Δald3Δ strain that accumulates more IAA is also more filamentous. IAA was also able to stimulate dimorphic transition in the human pathogen C. albicans. Deletion of a key regulator of the IAA responses had the same effect in both organisms. Homologs of enzymes that transport and synthesize IAA in S. cerevisiae are present in C. albicans. We suggest that IAA is an important signal that triggers dimorphic transition—a virulence trait.
A genomic scale screen for IAA homeostasis mutants implicated the aldehyde dehydrogenases, Ald2 and Ald3 in the final step of IAA synthesis from Trp. Ald2 and Ald3 share significant sequence similarity with Iad1, the U. maydis aldehyde dehydrogenase that has been shown to catalyze the conversion of IAAld to IAA (Basse et al. 1996; Akamatsu et al. 2000; Mizuno et al. 2006; Pigeau and Inglis 2007; Reineke et al. 2008). The ALD genes are responsible for acetate formation during anaerobic fermentation (Saint-Prix et al. 2004; Pigeau and Inglis 2007) and are hence of interest to the brewing industry. They have previously been implicated in mediating a variety of stress responses and are regulated by general-stress transcription factors Msn2 and -4 (Miralles and Serrano 1995; Navarro-Avino et al. 1999; Aranda and del Olmo 2003). Ald activity is required in the synthesis of two amino acid derivatives, IAA and β-alanine in U. maydis and S. cerevisiae, respectively (White et al. 2003; Reineke et al. 2008). This screen identified other members of the pathway (Aro9), which has previously been implicated in the first step of IAA synthesis (Chen and Fink 2006). In the process of characterizing mutants in a Trp-dependent IAA synthesis pathway, we uncovered another pathway that did not rely on exogenous Trp for IAA biosynthesis. Trp-independent synthesis of IAA has been demonstrated in several plant species, but the intermediates, intermediate steps, and genes involved in this pathway remain undefined (Woodward and Bartel 2005; Normanly 2009). The observation that S. cerevisiae has an analogous pathway provides a much simpler system to employ in the characterization of Trp-independent IAA synthesis.
There is precedence for multiple IAA biosynthetic pathways in microbes, particularly plant-associated bacteria (Clark et al. 1993; Costacurta and Vanderleyden 1995; Glick et al. 1999b; Lambrecht et al. 2000). An interesting example of differential utilization of multiple IAA biosynthetic pathways in microbes is found in Erwinia herbicola, which requires a functional indole acetamide (IAM) pathway (Trp is converted to IAM and then to IAA) to be pathogenic to plants and requires a functional IPA pathway (Figure 1) to exist as a plant epiphyte (Manulis et al. 1998). We note that while aldehyde dehydrogenase has been implicated in IAA synthesis in U. maydis, this pathway is not involved in tumorigenesis (Reineke et al. 2008). This result is consistent with our observation that ALD2 and ALD3 are not necessary for IAA-induced filamentation and that an alternate IAA synthesis pathway likely exists in yeast.
The coexistence of both Trp-dependent and Trp-independent IAA-biosynthetic pathways has been documented in plants (Normanly et al. 2004; Woodward and Bartel 2005) and microbes (Prinsen et al. 1993). In plants, Trp-independent IAA synthesis is proposed to branch from either indole or indole glycerol phosphate, both precursors of Trp (Normanly et al. 2004). One of the proposed Trp-dependent IAA biosynthetic pathways for plants converts Trp to IPA (reviewed in Woodward and Bartel 2005 and Normanly 2009). The Arabidopsis TAA-1 protein can convert Trp to IPA in vitro, and mutations in the TAA-1 gene produce less IAA when the plant is subjected to simulated shade (Tao et al. 2008), high temperature (Yamada et al. 2009), or ethylene (Stepanova et al. 2008). IAAld has been proposed as an intermediate of the Trp-dependent IAA synthetic pathway in plants, but this has yet to be confirmed, and plant orthologs of ALD genes have not been identified. One putative aldehyde oxidase from Arabidopsis shows a substrate preference for IAAld in vitro, but the relevance of this gene to IAA biosynthesis in vivo has yet to be confirmed (Seo et al. 1998). Future studies will involve using differential stable isotope labeling coupled with genetic mutants to identify components of alternate IAA biosynthetic pathways in S. cerevisiae.
Secondary metabolites are recognized as important signals. Aspergillus fumigatus hyphae release a small molecule, gliotoxin, which can exacerbate the pathogenesis of invasive aspergillosis (Sutton et al. 1996). Pseudomonas aeruginosa produces a signaling molecule, homoserine lactone, which inhibits C. albicans filamentation (Hogan et al. 2004). Two predominant types of small molecules, acyl homoserine lactones (AHLs) (Fuqua et al. 2001; Danhorn et al. 2004; Akimkina et al. 2006) and modified oligopeptides (Kleerebezem et al. 1997), are used by gram-negative and gram-positive bacteria, respectively, to regulate phenotypes that lead to virulence such as antibiotic production and biofilm formation. C. albicans has been shown to produce secondary metabolites such as tyrosol and farnesol that regulate dimorphic transition (Shchepin et al. 2003; Chen et al. 2004). Aromatic alcohols such as tryptophol and phenylalanol, a catabolic product of Phe, are produced by both S. cerevisiae and C. albicans but exert different effects on their morphogenesis, suggesting that they have distinct species-specific effects. IAA differs from these previously described signaling molecules because its effects appear to cross species barriers. Diverse fungal species respond to IAA; therefore, defining the pathways by which IAA regulates filamentation in C. albicans will yield a better understanding of its pathogenesis and potentially the development of broad-spectrum antifungal therapies. Furthermore, auxin permeases that import IAA in S. cerevisiae are homologous to the Arabidopsis IAA importer, Aux1 (Prusty et al. 2004). Therefore, defining IAA synthesis and regulation in yeast, a simple eukaryote, will yield a better understanding of IAA regulation in plants.
Acknowledgments
The authors thank G. Fink, J. Celenza, M. Lorenz, and J. Cohen for critical reading of the manuscript and helpful discussions and acknowledge C. Jain and M. Lewandowski for assistance with sample preparation and GC-MS analysis, respectively. This work was supported in part by Worcester Polytechnic Institute and National Science Foundation funds MCB 0517420 to J.N.
References
- Abel, S., and T. Athanosios, 2010. Odyssey of auxin, in Cold Spring Harbor Perspectives in Biology. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [DOI] [PMC free article] [PubMed]
- Akamatsu, S., H. Kamiya, N. Yamashita, T. Motoyoshi, N. Goto-Yamamoto et al., 2000. Effects of aldehyde dehydrogenase and acetyl-CoA synthetase on acetate formation in sake mash. J. Biosci. Bioeng. 90 555–560. [PubMed] [Google Scholar]
- Akimkina, T., K. Yook, S. Curnock and J. Hodgkin, 2006. Genome characterization, analysis of virulence and transformation of Microbacterium nematophilum, a coryneform pathogen of the nematode Caenorhabditis elegans. FEMS Microbiol. Lett. 264 145–151. [DOI] [PubMed] [Google Scholar]
- Alarco, A. M., and M. Raymond, 1999. The bZip transcription factor Cap1p is involved in multidrug resistance and oxidative stress response in Candida albicans. J. Bacteriol. 181 700–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aranda, A., and M. L. del Olmo, 2003. Response to acetaldehyde stress in the yeast Saccharomyces cerevisiae involves a strain-dependent regulation of several ALD genes and is mediated by the general stress response pathway. Yeast 20 747–759. [DOI] [PubMed] [Google Scholar]
- Barash, I., and S. Manulis-Sasson, 2009. Recent evolution of bacterial pathogens: the gall-forming Pantoea agglomerans case. Annu. Rev. Phytopathol. 47 133–152. [DOI] [PubMed] [Google Scholar]
- Barkawi, L. S., Y. Y. Tam, J. A. Tillman, B. Pederson, J. Calio et al., 2008. A high-throughput method for the quantitative analysis of indole-3-acetic acid and other auxins from plant tissue. Anal. Biochem. 372 177–188. [DOI] [PubMed] [Google Scholar]
- Basse, C. W., F. Lottspeich, W. Steglich and R. Kahmann, 1996. Two potential indole-3-acetaldehyde dehydrogenases in the phytopathogenic fungus Ustilago maydis. Eur. J. Biochem. 242 648–656. [DOI] [PubMed] [Google Scholar]
- Brachmann, C. B., A. Davies, G. J. Cost, E. Caputo, J. Li et al., 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14 115–132. [DOI] [PubMed] [Google Scholar]
- Brown, Jr., D. H., A. D. Giusani, X. Chen and C. A. Kumamoto, 1999. Filamentous growth of Candida albicans in response to physical environmental cues and its regulation by the unique CZF1 gene. Mol. Microbiol. 34 651–662. [DOI] [PubMed] [Google Scholar]
- Chen, H., and G. R. Fink, 2006. Feedback control of morphogenesis in fungi by aromatic alcohols. Genes Dev. 20 1150–1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H., M. Fujita, Q. Feng, J. Clardy and G. R. Fink, 2004. Tyrosol is a quorum-sensing molecule in Candida albicans. Proc. Natl. Acad. Sci. USA 101 5048–5052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark, E., S. Manulis, Y. Ophir, I. Barash and Y. Gafni, 1993. Cloning and Characterization of iaaM and iaaH from Erwinia herbicola pathovar gypsophilae. Mol. Plant Pathol. 83 234–240. [Google Scholar]
- Cohen, J. D., 1984. Convenient apparatus for the generation of small amounts of diazomethane. J. Chromatogr. 303 193–196. [Google Scholar]
- Costacurta, A., and J. Vanderleyden, 1995. Synthesis of phytohormones by plant-associated bacteria. Crit. Rev. Microbiol. 21 1–18. [DOI] [PubMed] [Google Scholar]
- Danhorn, T., M. Hentzer, M. Givskov, M. R. Parsek and C. Fuqua, 2004. Phosphorus limitation enhances biofilm formation of the plant pathogen Agrobacterium tumefaciens through the PhoR-PhoB regulatory system. J. Bacteriol. 186 4492–4501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delker, C., A. Raschke and M. Quint, 2008. Auxin dynamics: the dazzling complexity of a small molecule's message. Planta 227 929–941. [DOI] [PubMed] [Google Scholar]
- Dieterich, C., M. Schandar, M. Noll, F. J. Johannes, H. Brunner et al., 2002. In vitro reconstructed human epithelia reveal contributions of Candida albicans EFG1 and CPH1 to adhesion and invasion. Microbiology 148 497–506. [DOI] [PubMed] [Google Scholar]
- Fuqua, C., M. R. Parsek and E. P. Greenberg, 2001. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu. Rev. Genet. 35 439–468. [DOI] [PubMed] [Google Scholar]
- Furukawa, T., J. Koga, T. Adachi, K. Kishi and K. Syono, 1996. Efficient conversion of L-tryptophan to indole-3-acetic acid and/or tryptophol by some species of Rhizoctonia. Plant Cell Physiol. 37 899–905. [Google Scholar]
- Glick, B. R., C. L. Patten, G. Holguin and D. M. Penrose, 1999. a Auxin production, pp. 86–133 in Biochemical and Genetic Mechanisms Used by Plant Growth Promoting Bacteria, edited by B. R. Glick, C. L. Patten, G. Holguin and D. M. Penrose. Imperial College Press, London.
- Glick, B. R., C. L. Patten, G. Holguin and D. M. Penrose, 1999. b Biochemical and Genetic Mechanisms Used by Plant Growth Promoting Bacteria. Imperial College Press, London.
- Gruen, H. E., 1959. Auxins and fungi. Annu. Rev. Plant Physiol. 10 405–441. [Google Scholar]
- Guthrie, C., and G. R. Fink, 1991. Guide to yeast genetics and molecular biology. Methods Enzymol. 194 1–863. [PubMed] [Google Scholar]
- Halliday, K. J., J. F. Martinez-Garcia and E.-M. Josse, 2009. Integration of light and auxin signaling. Cold Spring Harb. Perspect. Biol. (in press). [DOI] [PMC free article] [PubMed]
- Hazelwood, L. A., J. M. Daran, A. J. van Maris, J. T. Pronk and J. R. Dickinson, 2008. The Ehrlich pathway for fusel alcohol production: a century of research on Saccharomyces cerevisiae metabolism. Appl. Environ. Microbiol. 74 2259–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch, A. M., T. V. Bhuvaneswari, J. G. Torrey and T. Bisseling, 1989. Early nodulin genes are induced in alfalfa root outgrowths elicited by auxin transport inhibitors. Proc. Natl. Acad. Sci. USA 86 1244–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan, D. A., A. Vik and R. Kolter, 2004. A Pseudomonas aeruginosa quorum-sensing molecule influences Candida albicans morphology. Mol. Microbiol. 54 1212–1223. [DOI] [PubMed] [Google Scholar]
- Kleerebezem, M., L. E. Quadri, O. P. Kuipers and W. M. de Vos, 1997. Quorum sensing by peptide pheromones and two-component signal-transduction systems in Gram-positive bacteria. Mol. Microbiol. 24 895–904. [DOI] [PubMed] [Google Scholar]
- Lambrecht, M., Y. Okon, A. Vande Broek and J. Vanderleyden, 2000. Indole-3-acetic acid: a reciprocal signalling molecule in bacteria-plant interactions. Trends Microbiol. 8 298–300. [DOI] [PubMed] [Google Scholar]
- Lau, S., G. Jurgens and I. De Smet, 2008. The evolving complexity of the auxin pathway. Plant Cell 20 1738–1746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, H., 2001. Transcriptional control of dimorphism in Candida albicans. Curr. Opin. Microbiol. 4 728–735. [DOI] [PubMed] [Google Scholar]
- Lo, H. J., J. R. Kohler, B. DiDomenico, D. Loebenberg, A. Cacciapuoti et al., 1997. Nonfilamentous C. albicans mutants are avirulent. Cell 90 939–949. [DOI] [PubMed] [Google Scholar]
- Manulis, S., A. Haviv-Chesner, M. T. Brandl, S. E. Lindow and I. Barash, 1998. Differential involvement of indole-3-acetic acid biosynthetic pathways in pathogenicity and epiphytic fitness of Erwinia herbicola pv. gypsophilae. Mol. Plant-Microbe Interact. 11 634–642. [DOI] [PubMed] [Google Scholar]
- McSteen, P., 2010. Auxin and monocot development. Cold Spring Harb. Perspect. Biol. (in press). [DOI] [PMC free article] [PubMed]
- Miralles, V. J., and R. Serrano, 1995. A genomic locus in Saccharomyces cerevisiae with four genes up-regulated by osmotic stress. Mol. Microbiol. 17 653–662. [DOI] [PubMed] [Google Scholar]
- Mizuno, A., H. Tabei and M. Iwahuti, 2006. Characterization of low-acetic-acid-producing yeast isolated from 2-deoxyglucose-resistant mutants and its application to high-gravity brewing. J. Biosci. Bioeng. 101 31–37. [DOI] [PubMed] [Google Scholar]
- Moller, B., and D. Weijers, 2009. Auxin control of embryo patterning. Cold Spring Harb. Perspect. Biol. (in press). [DOI] [PMC free article] [PubMed]
- Navarro-Avino, J. P., R. Prasad, V. J. Miralles, R. M. Benito and R. Serrano, 1999. A proposal for nomenclature of aldehyde dehydrogenases in Saccharomyces cerevisiae and characterization of the stress-inducible ALD2 and ALD3 genes. Yeast 15 829–842. [DOI] [PubMed] [Google Scholar]
- Nielsen, N., 1931. Über Wuchsstoffe der Hefe. Biochem. Z. 237 244–246. [Google Scholar]
- Normanly, J., 2009. Approaching Cellular and Molecular Resolution of Auxin Biosynthesis and Metabolism. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [DOI] [PMC free article] [PubMed]
- Normanly, J., J. Sovin and J. Cohen, 2004. Auxin Metabolism in Plant Hormones: Biosynthesis, Signal Transduction, Action. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- Pigeau, G. M., and D. L. Inglis, 2007. Response of wine yeast (Saccharomyces cerevisiae) aldehyde dehydrogenases to acetaldehyde stress during icewine fermentation. J. Appl. Microbiol. 103 1576–1586. [DOI] [PubMed] [Google Scholar]
- Prinsen, E., A. Costacura, K. Michiels, J. Vanderleyden and H. Van Onckelen, 1993. Azospirillum brasilense indole-3-acetic acid biosynthesis: evidence for a non-tryptophan-dependent pathway. Mol. Plant-Microbe Interact. 6 609–615. [Google Scholar]
- Prusty, R., P. Grisafi and G. R. Fink, 2004. The plant hormone indoleacetic acid induces invasive growth in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 101 4153–4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke, G., B. Heinze, J. Schirawski, H. Buettner, R. Kahmann et al., 2008. Indole-3-acetic acid (IAA) biosynthesis in the smut fungus Ustilago maydis and its relevance for increased IAA levels in infected tissue and host tumour formation. Mol. Plant Pathol. 9 339–355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reneke, J. E., K. J. Blumer, W. E. Courchesne and J. Thorner, 1988. The carboxy-terminal segment of the yeast alpha-factor receptor is a regulatory domain. Cell 55 221–234. [DOI] [PubMed] [Google Scholar]
- Riggle, P. J., K. A. Andrutis, X. Chen, S. R. Tzipori and C. A. Kumamoto, 1999. Invasive lesions containing filamentous forms produced by a Candida albicans mutant that is defective in filamentous growth in culture. Infect. Immun. 67 3649–3652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, M., J. Riov and A. Sharon, 1998. Indole-3-acetic acid biosynthesis in Colletotrichum gloeosporioides f. sp. aeschynomene. Appl. Environ. Microbiol. 64 5030–5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saint-Prix, F., L. Bonquist and S. Dequin, 2004. Functional analysis of the ALD gene family of Saccharomyces cerevisiae during anaerobic growth on glucose: the NADP+-dependent Ald6p and Ald5p isoforms play a major role in acetate formation. Microbiology 150 2209–2220. [DOI] [PubMed] [Google Scholar]
- Scarpella, E., M. Barkoulas and M. Tsiantis, 2010. Control of leaf and vein development by auxin. Cold Spring Harb. Perspect. Biol. (in press). [DOI] [PMC free article] [PubMed]
- Seo, M., S. Akaba, T. Oritani, M. Delarue, C. Bellini et al., 1998. Higher activity of an aldehyde oxidase in the auxin-overproducing superroot1 mutant of Arabidopsis thaliana. Plant Physiol. 116 687–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shchepin, R., J. M. Hornby, E. Burger, T. Niessen, P. Dussault et al., 2003. Quorum sensing in Candida albicans: probing farnesol's mode of action with 40 natural and synthetic farnesol analogs. Chem. Biol. 10 743–750. [DOI] [PubMed] [Google Scholar]
- Sherman, F., G. R. Fink and J. B. Hicks, 1986. Laboratory Course Manual for Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Shin, M., T. Shinguu, K. Sano and C. Umezawa, 1991. Metabolic fates of L-tryptophan in Saccharomyces uvarum (Saccharomyces carlsbergensis). Chem. Pharm. Bull. (Tokyo) 39 1792–1795. [DOI] [PubMed] [Google Scholar]
- Sohn, K., C. Urban, H. Brunner and S. Rupp, 2003. EFG1 is a major regulator of cell wall dynamics in Candida albicans as revealed by DNA microarrays. Mol. Microbiol. 47 89–102. [DOI] [PubMed] [Google Scholar]
- Stepanova, A. N., J. Robertson-Hoyt, J. Yun, L. M. Benavente, D. Y. Xie et al., 2008. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133 177–191. [DOI] [PubMed] [Google Scholar]
- Stewart, J. L., and J. L. Nemhauser, 2009. Do trees grow on money? Auxin as the currency of the cellular economy. Cold Spring Harb. Perspect. Biol. (in press). [DOI] [PMC free article] [PubMed]
- Sundberg, E., and L. Ostergaard, 2009. Distinct and dynamic auxin activities during reproductive development. Cold Spring Harb. Perspect. Biol. (in press). [DOI] [PMC free article] [PubMed]
- Sutton, P., P. Waring and A. Mullbacher, 1996. Exacerbation of invasive aspergillosis by the immunosuppressive fungal metabolite, gliotoxin. Immunol. Cell. Biol. 74 318–322. [DOI] [PubMed] [Google Scholar]
- Tao, Y., J. L. Ferrer, K. Ljung, F. Pojer, F. Hong et al., 2008. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133 164–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimann, K. V., 1935. On the plant growth hormone produced by Rhizopus suinus. J. Biol. Chem. 109 279–291. [Google Scholar]
- Turian, G., and R. H. Hamilton, 1960. Chemical detection of 3-indolylacetic acid in Ustilago zeae tumors. Biochim. Biophys. Acta 41 148–150. [DOI] [PubMed] [Google Scholar]
- Viglierchio, D. R., 1971. Nematodes and other pathogens in auxin-related plant-growth disorders. Bot. Rev. 37 1–21. [Google Scholar]
- Wach, A., A. Brachat, R. Pohlmann and P. Philippsen, 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast 10 1793–1808. [DOI] [PubMed] [Google Scholar]
- White, W. H., P. L. Skatrud, Z. Xue and J. H. Toyn, 2003. Specialization of function among aldehyde dehydrogenases: the ALD2 and ALD3 genes are required for beta-alanine biosynthesis in Saccharomyces cerevisiae. Genetics 163 69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler, E. A., D. D. Shoemaker, A. Astromoff, H. Liang, K. Anderson et al., 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285 901–906. [DOI] [PubMed] [Google Scholar]
- Woodward, A. W., and B. Bartel, 2005. Auxin: regulation, action, and interaction. Ann. Bot. (Lond) 95 707–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada, M., K. Greenham, M. J. Prigge, P. J. Jensen and M. Estelle, 2009. The transport inhibitor response2 (TIR2) gene is required for auxin synthesis and diverse aspects of plant development. Plant Physiol. (in press). [DOI] [PMC free article] [PubMed]
- Zazimalova, E., A. S. Murphy, K. Hoyerova and P. Hosek, 2009. Auxin transporters—Why so many? Cold Spring Harb. Perspect. Biol. (in press). [DOI] [PMC free article] [PubMed]